Abstract
We report the x-ray crystal structure of the methylesterase CheB, a phosphorylation-activated response regulator involved in reversible modification of bacterial chemotaxis receptors. Methylesterase CheB and methyltransferase CheR modulate signaling output of the chemotaxis receptors by controlling the level of receptor methylation. The structure of CheB, which consists of an N-terminal regulatory domain and a C-terminal catalytic domain joined by a linker, was solved by molecular replacement methods using independent search models for the two domains. In unphosphorylated CheB, the N-terminal domain packs against the active site of the C-terminal domain and thus inhibits methylesterase activity by directly restricting access to the active site. We propose that phosphorylation of CheB induces a conformational change in the regulatory domain that disrupts the domain interface, resulting in a repositioning of the domains and allowing access to the active site. Structural similarity between the two companion receptor modification enzymes, CheB and CheR, suggests an evolutionary and/or functional relationship. Specifically, the phosphorylated N-terminal domain of CheB may facilitate interaction with the receptors, similar to the postulated role of the N-terminal domain of CheR. Examination of surfaces in the N-terminal regulatory domain of CheB suggests that despite a common fold throughout the response regulator family, surfaces used for protein–protein interactions differ significantly. Comparison between CheB and other response regulators indicates that analogous surfaces are used for different functions and conversely, similar functions are mediated by different molecular surfaces.
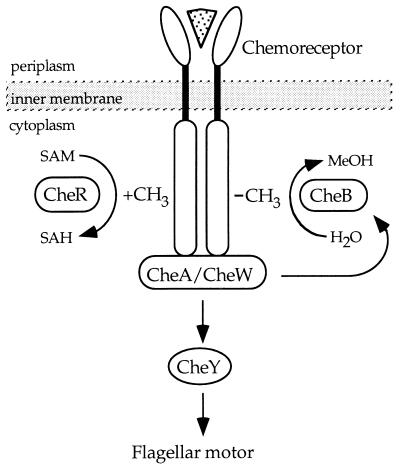
The methylesterase CheB is a member of a large and functionally diverse family of proteins known as response regulators. These proteins are involved in a wide variety of phosphotransfer-dependent signal transduction pathways found in prokaryotes and eukaryotes (1, 2). In the bacterial chemotaxis system, reversible methylation of transmembrane chemoreceptors plays an important role in ligand-dependent signaling and subsequent cellular adaptation (3, 4). In the enteric bacteria Escherichia coli and Salmonella typhimurium, CheB functions together with the methyltransferase CheR to control the level of receptor methylation. Phosphorylated CheB catalyzes deamidation of specific glutamine residues in the cytoplasmic region of the chemoreceptors and demethylation of specific methyl glutamate residues introduced into the chemoreceptors by CheR (Fig. 1). Like many members of the response regulator family, CheB has a multidomain architecture. It consists of an N-terminal regulatory domain and a C-terminal effector domain joined by a linker region.
Figure 1.
Receptor mediated signaling in bacterial chemotaxis. A family of dimeric transmembrane chemoreceptors sense changes in external levels of chemical attractants or repellents (▿) in the surrounding medium. The opposing activities of the S-adenosylmethionine-dependent methyltransferase CheR and the phosphorylation-regulated methylesterase CheB control receptor methylation levels, which in turn influence the signaling output of the chemoreceptor–histidine kinase CheA–coupling protein CheW complex. CheA autophosphorylation and subsequent transfer of the phosphoryl group to the response regulators CheY or CheB result in stimulation of effector responses: phospho-CheY controls the direction of flagellar motor rotation, whereas phospho-CheB serves to attenuate the response by hydrolysis of specific methyl glutamate residues on the receptor. SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; MeOH, methanol.
Regulation of CheB involves numerous protein–protein interactions. The autophosphorylating histidine kinase CheA serves as a phosphoryl donor to two response regulators, CheY and CheB (5, 6), suggesting the possibility that similar surfaces of response regulator domains are involved in recognition of their cognate histidine kinase. The receptor modification enzymes, CheR and CheB, catalyze methylation/demethylation at the same sites on the chemoreceptors, raising the interesting question of evolution of these two companion enzymes. Furthermore, activation by means of phosphorylation in the response regulator family is thought to involve communication between phosphorylated regulatory domains and associated or downstream effector domains. Previous studies of CheB have implied that interdomain interactions are important for regulation of methylesterase activity. Removal of the N-terminal regulatory domain by either genetic manipulation or proteolysis results in unregulated activation of C-terminal methylesterase activity (7, 8). The N-terminal domain thus appears to control C-terminal methylesterase activity in an autoinhibitory manner. Presumably, a phosphorylation-induced conformational change relieves this inhibition by altering the juxtaposition of the regulatory and catalytic domains. The structures of the domains of CheB are known from previous x-ray crystallographic studies of the C-terminal catalytic domain (9) and from homology modeling of the N-terminal regulatory domain with other response regulators (10, 11). The central question regarding CheB, and in fact all multidomain response regulators, is how the regulatory and effector domains are oriented with respect to each other and how this positioning relates to regulation of effector function.
We have determined the x-ray crystal structure of intact S. typhimurium CheB at a resolution of 2.4 Å. The model suggests a structural basis for phosphorylation-dependent activation of effector function and provides insights into recognition of the phosphoryl donor CheA and interaction with chemoreceptor substrates.
EXPERIMENTAL METHODS
Crystallization and Data Collection.
CheB was purified by a modified version of the protocol described by Simms et al. (7). Purified CheB was stored as a precipitate in 50% saturated (NH4)2SO4. Immediately prior to crystallization, the protein was resuspended in and dialyzed against 0.1 M morpholinoethanesulfonic acid (Mes), pH 6.0. CheB was concentrated to 12 mg/ml by use of a Centricon-10 (Amicon). Crystals were grown by the vapor diffusion hanging-drop method with microseeding by using 1 μl of protein solution, 1 μl of finely crushed CheB crystals in 1.5 M Li2SO4/0.1 M imidazole at pH 6.5 and 2 μl of reservoir solution consisting of 1.45 M Li2SO4, 0.1 M imidazole at pH 6.5, 10 mM MgCl2, and 5 mM NaN3. Diffraction images indicated crystal twinning, with a majority of reflections belonging to the C2 space group (unit cell dimensions of a = 163.8 Å, b = 100.5 Å, c = 53.1 Å, β = 98.6°). A small number of the total reflections (about 10%) were contributed by a separate minor lattice. Only those reflections belonging to the major lattice (C2) were integrated and merged. The intensity distribution of the C2 lattice is consistent with distributions observed in single crystals. There are two molecules of intact CheB in the asymmetric unit.
Structure Solution and Refinement.
The structure was solved by the molecular replacement method, using the two independent search models for each of the domains in the CheB molecule (9, 12) (Table 1). A crystal of intact S. typhimurium CheB was used to collect a native data set to a resolution of 2.4 Å. The crystal was equilibrated in cryoprotectant solution [crystallization solution with the addition of 30% (wt/vol) ethylene glycol] for 2 hr and flash-frozen in a nitrogen gas stream (Oxford Cryosystems, Oxford, U.K.) at 100 K. Data were collected on an RAXIS-IV detector (Rigaku International, Tokyo) using mirror-focused and Ni-filtered Cu Kα radiation. Space group determination and data integration were done with denzo (13). All data were merged, scaled, and truncated with the rotavata/agrovata and truncate programs in the CCP4 suite (14). Initial phase estimates were obtained by the molecular replacement method with x-plor (15). Searches were made by using the crystal structure of the CheB C-terminal domain (residues 154–347) (9) and a polyalanine model of CheY based on the crystal structure (12). The self-rotation function indicated the presence of local twofold symmetry. The top two peaks obtained from the self-rotation function were at ψ, φ values of 0.0, 0.0 and 90.0, 81.5 and are indicative of local symmetry axes being parallel to the crystallographic a and c axes. The two peaks were 5 and 2 σ values, respectively, above the background. A cross-rotation search combined with Patterson correlation refinement yielded two unambiguous solutions for the orientation of the C-terminal domain. These two solutions for the orientations of the C-terminal domains were related to each other by ψ, φ, κ values of 90.1, 81.0, 182.3, consistent with the self-rotation function results. A translation search was than performed by using one C-terminal domain. The top solution from that search was then fixed in a combined translation search for the second C-terminal domain. A unique solution for the combined position of the two C-terminal domains in the asymmetric unit was 5 σ values over the background. In the electron density maps calculated with phases derived from the C-terminal domain alone the orientation of the central β-sheet for the N-terminal domain region was also evident. The top two peaks from the cross-rotation/Patterson correlation refinement search with the polyalanine model of CheY were related to each other by local symmetry and were consistent with the electron density obtained by using phases calculated from the C-terminal domain. Although a translation search performed with the polyalanine CheY model of the N-terminal domain did not yield a distinct solution, the position of the N-terminal domain was apparent in the electron density map calculated with phases from the C-terminal domain. After rigid body refinement of the two C-terminal and two N-terminal domains the R-factor was 48%. After positional refinement of that model (R-factor 38%) the calculated electron density map revealed the position of the interdomain linker that was not included in either of the search models (Fig. 2). Further refinement employed the simulated annealing protocol in x-plor (15) while model rebuilding was done by using turbo (16). The 18-residue linker was built in dm averaged electron density maps (17). The model was confirmed by use of simulated annealing omit maps (18). In the initial stages of refinement, noncrystallographic symmetry restraints were applied to all residues not involved in crystal packing contacts. The final model was refined by refmac (14) to a crystallographic R value of 22.2% with Rfree value of 28.8%.
Table 1.
Summary of crystallographic analysis
| Measurement | Value |
|---|---|
| Data collection | |
| Resolution limit, Å | 2.4 |
| Total reflections | 97,667 |
| Unique reflections | 32,281 |
| Completeness, % | 96.9 |
| Rsym,* % | 5.4 |
| Refinement | |
| Resolution, Å | 10-2.4 |
| Number of protein atoms | 5,200 |
| Number of water molecules | 411 |
| Rcryst,† % | 22.2 |
| Rfree,‡ % | 28.8 |
| Average B, Å2 | 23.5 |
| rmsd bond length, Å | 0.008 |
| rmsd bond angles, degrees | 2.0 |
rmsd, Root mean square deviation.
Rsym = Σ |Ihi − 〈Ih〉|/Σ 〈Ih〉, where I is the scaled intensity of the given reflection h.
Rcryst = Σh |Foh − Fch|/Σh Foh, where Foh and Fch are the observed and calculated structure factor amplitudes for reflection h.
Value of Rcryst for 5% of randomly selected reflections excluded from the refinement.
Figure 2.
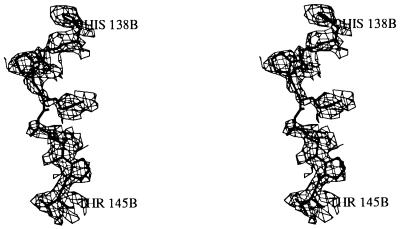
Stereo view of a representative portion of the initial electron density map calculated after positional refinement of the molecular replacement solution overlaid with the final refined model of CheB. The electron density map was calculated to a resolution of 2.7 Å by using phases calculated from the model containing the C-terminal domain and the polyalanine model of the CheY structure in place of the CheB N-terminal domain. This model does not contain CheB residues 134–153. For the sake of clarity, only the density associated with linker residues His-138 to Thr-145 are displayed. Even at this stage the initial electron density map exhibited distinct features of side-chain density and allowed for sequence assignment for the interdomain linker. The figure was generated by using turbo (16).
RESULTS AND DISCUSSION
Overall Topology.
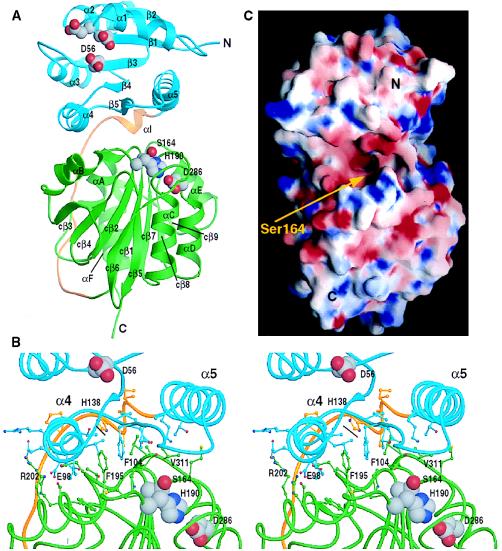
The overall structure of CheB, depicted by a ribbon diagram in Fig. 3A, shows the N-terminal regulatory domain (residues 1–134) to be positioned over the C-terminal edge of the central seven-stranded parallel β-sheet of the C-terminal methylesterase domain (residues 153–347). The two domains are connected by an 18-residue linker that, except for a single helical turn (residues 135–138), does not exhibit repetitive secondary structure. Electron density for the linker was clearly identified in the initial electron density maps (Fig. 2). The structure for the C-terminal catalytic domain is essentially identical to the structure determined from crystals of CheBc alone (9) (rms deviation for backbone atoms is 0.69 Å) and can be described as a modified doubly wound α/β protein containing a long antiparallel β-hairpin insertion (cβ5–cβ6). With respect to the mechanism of methyl ester hydrolysis, CheB belongs to the class of serine hydrolases that contain active site catalytic triads consisting of serine, histidine, and aspartate residues. In the catalytic domain of CheB, the active site residues, Ser-164, His-190, and Asp-286, are located in a cleft formed by loops at the carboxy ends of β-strands cβ1, cβ2, and cβ7 (Fig. 3A).
Figure 3.
Structure of the methylesterase CheB. (A) Ribbon representation showing the overall fold of residues 1–347 and the relative positioning of the two domains. The N-terminal regulatory domain is colored blue, the C-terminal catalytic domain is shown in green, and the linker region is colored gold. The acidic cluster of active site residues (Asp-11, Asp-12 ,and Asp-56, the site of phosphorylation) in the N-terminal domain and the catalytic triad residues (Ser-164, His-190, Asp-286) in the C-terminal domain are shown as Corey–Pauling–Koltun (CPK) models with carbon atoms colored gray, oxygen red, and nitrogen, dark blue. For consistency, all of the α-helices and β-strands are labeled analogously to the numbering schemes used previously in descriptions of CheBc (9) and CheY (19). The β-strands within the C-terminal domain are additionally labeled by the prefix “c” (cβ1, cβ2, … ). (B) Stereo diagram of the interdomain region. The molecule is slightly rotated horizontally compared with the orientation in A. Helices and strands are shown as lines with the side chains of residues involved in the interdomain interaction represented by balls and sticks. The coloring scheme and CPK models are as indicated in A. The labels identify residues that show the greatest decrease in solvent accessibility in intact CheB as compared with that calculated for residues in the isolated domains. (C) The electrostatic potential of the molecular surface of CheB shown rotated approximately 90° about the vertical axis relative to the view in A. The view is looking toward the active site. Ser-164 sits within a funnel-like opening formed at the domain interface. In the vicinity of the active site the electrostatic potential is significantly more negative in the intact protein than in the isolated C-terminal domain, and this may have an effect on catalysis. The surface in C was calculated and displayed with the negative surfaces in red, the positive surfaces in blue, and the neutral surfaces in white by using the program grasp (20); A and B and all subsequent figures were prepared by using the program ribbons (21).
The N-Terminal Regulatory Domain.
The N-terminal regulatory domain of CheB shares structural homology with the large family of response regulator proteins, the best-characterized member of this family being the chemotaxis protein CheY (10, 11). In different proteins involved in “two-component” signal transduction pathways, this relatively simple structural module with a parallel five-stranded α/β topology is associated with, and regulates in a phosphorylation-dependent manner, the function of a variety of effector domains. In addition to E. coli and S. typhimurium CheY (19, 22), structures have been determined for Spo0F (23), the regulatory domain of NtrC (24), and intact NarL (25). The most distinctive features of the N-terminal domain of CheB compared with other response regulators are the considerably larger loops connecting β4 to α4 and β5 to α5. The existence of these extended loops has not been predicted in previous sequence alignments of regulatory domains (10, 11). Because of the fact that the electron density associated with the interdomain linker region was readily interpretable (Fig. 2), we were able to assign side chains for the linker and α5 helix and finally the β5-α5 loop. The loops appear to be rather flexible, as they have atomic B values of approximately 50 Å2. Furthermore, the connecting β5–α5 loop (residues 108–118) was strongly influenced by crystal packing and as a result, the loop acquires significantly different conformations in the two CheB molecules present in the asymmetric unit.
Two-component regulatory pathways involve phosphoryl transfer from an autophosphorylated histidine kinase to a specific aspartate residue in the response regulator. In CheB, phosphoryl transfer from the histidine kinase CheA occurs at Asp-56, located atop strand β3 in the N-terminal domain. The phosphoryl transfer reaction is magnesium-dependent and in the structure of Mg2+-CheY, the active site residues, Asp-12, Asp-13, and Asp-57, coordinate the essential metal cation (12). In the regulatory domain of CheB, in addition to the conserved active site aspartate residues (Asp-11, Asp-12 ,and Asp-56), a glutamate residue, Glu-58, is also oriented toward and contributes to the acidic active site cluster. In vitro studies have indicated that the phosphorylated state of CheB is very short lived (t1/2 ≈ 1 sec), and Glu-58 has been reported to be involved both in transferring the phosphoryl group from CheA and in promoting hydrolysis of the aspartyl phosphate, because replacement of Glu-58 with a lysine dramatically reduces phosphotransfer and autophosphatase activity (26).
The Interdomain Interface.
The regulatory and effector domains interact with each other, with each domain contributing a surface area of approximately 1,000 Å2. The extent of this interface and the number of contributing residues (≈35) are very similar to the interacting surfaces seen in protease/protease inhibitor and antigen/antibody complexes (27). The center of the domain interface consists of a small hydrophobic core. The perimeter of the surface involves hydrogen bond interactions and several salt bridges (Fig. 3B). By comparing surface accessibility of residues within the interdomain region in the intact CheB molecule to surface accessibility of the same residues when considering separate individual domains, we have identified those residues that are significantly more buried in intact CheB. These residues contribute the most to the domain–domain interactions, and they therefore may be the sensitive points for destabilization upon phosphorylation. In the central hydrophobic core two phenylalanines, Phe-104 from the N-terminal domain and Phe-195 from the C-terminal domain, lose 75 Å2 and 138 Å2 of solvent accessible surface, respectively, because of the interaction of the two domains. Phe-104 is a highly conserved residue within the family of response regulators and it was shown that the corresponding residue in CheY (Tyr-106) can exist in two distinct conformations, with outward and inward conformations correlating with inactive and active states of CheY (28). If the same type of conformational change involving an inward orientation of Phe-104 occurs on phosphorylation of CheB, this would likely contribute to disruption of the interdomain interface. Even more dramatic changes in solvent accessibility are observed for the residues involved in forming salt bridges. Interestingly, there is specific surface complementarity among the interacting residues from the N-terminal and C-terminal domains. Protruding residues from one domain fit into grooves on the neighboring domain. For example, two glutamate residues, Glu-91 and Glu-98, from the N-terminal domain lose 50 Å2 and 140 Å2, respectively, whereas their partners from the C-terminal domain, Arg-202 and Arg-172, lose 202 Å2 and 16 Å2, respectively, at the domain–domain interface. The active site nucleophile, Ser-164, in the C-terminal domain and the phosphorylation site, Asp-56, in the N-terminal domain are located, respectively, ≈13 Å and ≈11 Å away from the center of the interdomain interface. This arrangement of the two active sites with respect to the interdomain surface has the potential for propagating and magnifying the effects of small localized conformational changes at the site of phosphorylation to the interdomain interface, potentially leading to an opening of the cleft between the two domains. Specifically, phosphorylation of Asp-56 could induce minor reorientation of helices α4 and α5, resulting in disruption of the interdomain interface and allowing a widening of the methylesterase active site cleft.
Interaction with Chemoreceptor Substrates and Structural Relationship with Methyltransferase CheR.
With the molecular structure of CheB available, we can now also address the question of interaction of CheB with its chemoreceptor substrates. The transmembrane chemoreceptors are thought to exist in a closely associated dimeric state with each monomer predicted to form a two-stranded antiparallel α-helical coiled-coil (29–31). The specific glutamate residues that are subject to covalent modification are spaced seven residues apart such that they would lie along one face of the receptor helix (32). The structure of CheB clearly reveals that the N-terminal domain obstructs access of the receptor substrate to the active site of the methylesterase C-terminal domain (Fig. 3A). Even though the residues of the catalytic triad are not directly in contact with residues from the N-terminal domain and their solvent accessibility is the same as that in the C-terminal domain alone, the funnel-shaped opening created by the N-terminal domain, linker, and C-terminal domain interface is not sufficiently wide to allow the effective approach of the substrate chemoreceptors to the active site. In a docking experiment with a simplified substrate model of an α-helix containing a glutamate residue in an extended conformation we found that the carboxylate of the glutamate side chain could not be positioned any closer than 7 Å to the active site serine. The intact chemoreceptor, a dimer of two-stranded antiparallel coiled-coils, would require a much larger entry to the active site than exists in unphosphorylated CheB.
An additional interesting property of the inhibited, unphosphorylated form of CheB was revealed from the calculated electrostatic potential of the molecular surface (Fig. 3C). This view of the molecular surface shows that the area surrounding the funnel-shaped active site is highly negatively charged. In the proposed catalytic mechanism for CheB and other serine hydrolases, the negatively charged environment would be unfavorable for stabilizing the transition state oxyanion. This suggests the possibility that the negative electrostatic potential around the active site in the unphosphorylated form of CheB might also contribute to the negative regulation of methylesterase activity. In contrast, the electrostatic potential of the active site region is substantially less negative when calculated for the isolated C-terminal domain. Additionally, a negative electrostatic environment surrounding the active site might facilitate proper orientation of the enzyme toward the receptor substrate, because the methyl glutamate residues on the chemotaxis receptors are flanked by a negatively charged region consisting of nonmethylated glutamates.
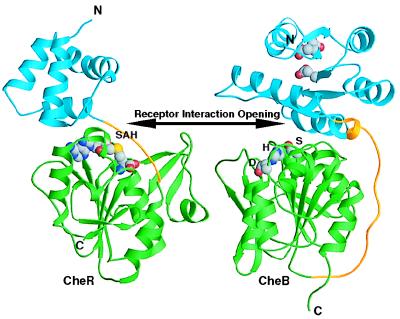
Interaction of CheB with chemotaxis receptors can be further considered on the basis of comparison to the x-ray crystal structure of the other enzyme involved in regulation of chemoreceptor signaling output, the S-adenosylmethionine-dependent methyltransferase CheR (33) (Fig. 4). A structural homology search using dali (34) identified significant similarity between CheR and CheB, raising the possibility of a common evolutionary origin. Fig. 4 shows the two structures that were first overlaid on the basis of dali output and then translated away from each other to more clearly show the specific features of the molecules. In both CheB and CheR, the N-terminal domain is located above the C-terminal edge of the central seven-stranded β-sheet of the C-terminal domain. The active sites of both proteins are positioned in similar locations in the C-terminal domain within the domain–domain interface. However, the proposed receptor interaction clefts occur on different faces of the β-sheet in CheB and CheR. Topological differences in the structures of CheR and CheB may be reflective of their functionally antagonistic interactions with the receptors.
Figure 4.
Structural comparison of the chemoreceptor modification enzymes. Structures of the methyltransferase CheR and the methylesterase CheB were aligned on the basis of similarity of their C-terminal domains by using a structural homology search in dali (34). For both molecules, ribbon diagrams depict the N-terminal domains in blue, linker regions in gold, and C-terminal domains in green. The molecule of S-adenosylhomocysteine (SAH) in CheR and the methylesterase active site residues [Ser-164 (S), His-190 (H), and Asp-286(D)] in CheB are shown as CPK models. The double-headed arrow points toward the active sites and the receptor interaction openings. Functionally antagonistic CheB and CheR contain active sites on opposite faces of the structurally homologous central β-sheets.
Previous biochemical data suggest extensive contacts between the modification enzymes and methylated regions of the receptors. Specific glutamate residues on the chemotaxis receptors that undergo reversible methylation are proposed to align along one face of the receptors (32). Methylation/demethylation rates at each site differ significantly and appear to be influenced by the conformation of the receptor (35, 36). The rates of methylation/demethylation at one site can be influenced by residues located as far as two helical turns away (37, 38). One could rationalize that topological insertions in the doubly wound α/β domains of the receptor modification enzymes, such as the three-stranded β-subdomain in CheR and the β-hairpin motif in CheB, presumably play a more specialized role in substrate recognition and/or catalysis. The two motifs are positioned on the same side edge but on opposite faces of the central β-sheet. In both enzymes the β-motifs and the N-terminal domains form an extended surface surrounding the active site. It seems likely that these surfaces provide the modification enzymes with a mechanism for “read out” of structural information encoded in the extended methylation region of the receptor.
Model for Activation by Phosphorylation.
Structural features of the methylesterase suggest a mechanism for CheB regulation. In our model, the unphosphorylated N-terminal domain inhibits the activity of the methylesterase by obstructing access to the active site and by influencing the electrostatic potential of the active site region. Phosphorylation of the N-terminal domain, at Asp-56, distant from the methylesterase active site, leads to propagation of conformational changes to the interdomain interface. Disruption of the interface results in separation of the domains and a change in electrostatic potential in the esterase active site environment. In addition, we propose that the N-terminal domain plays an active role in interaction with the receptor by providing an additional surface for intermolecular interaction. Enzyme kinetics studies show that phosphorylated intact CheB has significantly higher methylesterase activity than the isolated C-terminal domain (G. Anand and A.M.S., unpublished observations). Thus, the N-terminal domain may have a complex regulatory role. In the unphosphorylated state, it functions as an inhibitor, whereas in the phosphorylated state, it facilitates interaction with the substrate.
Comparison to Structures of Other Response Regulators.
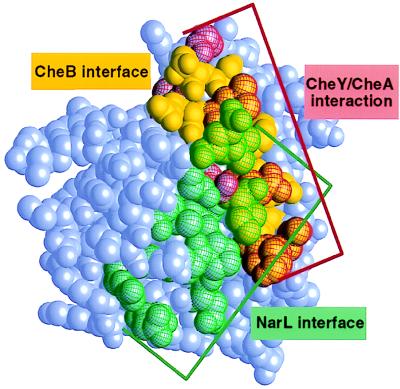
Hundreds of response regulator domains have been identified in prokaryotes and eukaryotes. However, very little is known about the generality of the mechanism of phosphorylation-dependent activation of effector domains. How might the mechanism of regulation by the N-terminal domain proposed for CheB apply to other response regulators? A similar strategy for inhibition of C-terminal domain function by the position of the N-terminal domain has been suggested for the transcriptional regulator NarL, the only other multidomain response regulator with a known three-dimensional structure (25). Despite similarities in regulatory mechanisms, comparison of the N-terminal domain of CheB with other regulatory domains indicates that different molecular surfaces are used for intra- and intermolecular interactions (Fig. 5). In CheB and the transcription factor NarL, the orientations of the regulatory domains with respect to the C-terminal effector domains are very different, being oriented roughly perpendicular to each other with only a small region of overlap. In CheB, residues of the regulatory domain that contribute to the domain interface are located in α4 and at the ends of α5 and β5, whereas in NarL, they are found in the loops connecting α2 and β3, α3 and β4, α4 and β5, and at the end of helix α5. Furthermore, the α4–β5–α5 surface buried in the domain interface of CheB corresponds to a region in CheY that has been proposed to serve as a recognition surface for an interacting region of the kinase CheA, termed P2 (39). Competitive binding of CheB and CheY to CheA suggests that a similar surface of the P2 domain is involved in interaction with both proteins (40). However, it is likely that binding of the regulatory domain of CheB to P2 involves a surface other than the inaccessible α4–β5–α5 region in the domain interface. Thus, CheY and CheB apparently use distinct surfaces for interaction with a single histidine protein kinase. This comparison of CheB, CheY, and NarL indicates that the regulatory domains of response regulators have been adapted to perform similar protein–protein interactions by using different molecular surfaces. Evolution of different protein recognition surfaces among response regulators may contribute to the specificity of phosphotransfer between histidine protein kinase and response regulator pairs.
Figure 5.
CPK model of the N-terminal domain of CheB, showing surfaces that are involved in protein–protein interactions among the response regulators CheB, CheY, and NarL. Residues involved in interaction with the C-terminal domain of CheB are colored yellow. Residues that have been implicated in protein–protein interactions in other response regulators are shown with colored mesh: red for corresponding residues in CheY that are thought to be involved in interaction with the P2 domain of the histidine kinase CheA (39); green for corresponding residues in NarL that interact with its C-terminal DNA-binding domain (25).
Acknowledgments
We thank I. Smalera and Dr. L. Tabernero for assistance with preliminary crystallization of CheB, Dr. G. Parkinson for assistance throughout the structure determination, Dr. E. Martínez-Hackert for valuable discussions, D. Leidich and G. Listner for maintenance of the X-Ray Facility, and Drs. E. Arnold, M. Inouye, and D. van der Helm for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM47958 (A.M.S.), National Science Foundation Grant MCB9258673 (A.M.S.), start-up funds from the University of Oklahoma (A.H.W.), and the W. M. Keck Foundation. A.M.S. is an Assistant Investigator of the Howard Hughes Medical Institute.
ABBREVIATION
- CPK
Corey–Pauling–Koltun
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1a2o).
References
- 1.Hoch J A, Silhavy T J. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 2.Appleby J L, Parkinson J S, Bourret R B. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 3.Stock J B, Surette M G. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1103–1129. [Google Scholar]
- 4.Morimoto B H, Koshland D E., Jr FASEB J. 1991;5:2061–2067. doi: 10.1096/fasebj.5.7.2010059. [DOI] [PubMed] [Google Scholar]
- 5.Hess J F, Oosawa K, Kaplan N, Simon M I. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 6.Stock A M, Wylie D C, Mottonen J M, Lupas A N, Ninfa E G, Ninfa A J, Schutt C E, Stock J B. Cold Spring Harbor Symp Quant Biol. 1988;53:49–57. doi: 10.1101/sqb.1988.053.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Simms S A, Keane M G, Stock J. J Biol Chem. 1985;260:10161–10168. [PubMed] [Google Scholar]
- 8.Lupas A, Stock J. J Biol Chem. 1989;264:17337–17342. [PubMed] [Google Scholar]
- 9.West A H, Martinez-Hackert E, Stock A M. J Mol Biol. 1995;250:276–290. doi: 10.1006/jmbi.1995.0376. [DOI] [PubMed] [Google Scholar]
- 10.Stock J B, Ninfa A J, Stock A M. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volz K. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 12.Stock A M, Martinez-Hackert E, Rasmussen B F, West A H, Stock J B, Ringe D, Petsko G A. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Sawyer L, Isaacs N, Bayley S, editors. Warrington, U.K.: Science and Engineering Research Council Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 14.Science and Engineering Research Council (U.K.) Collaborative Computational Project, N.4. Acta Cryst D. 1994;50:760–763. [Google Scholar]
- 15.Brünger A T. x-plor, Version 3.1, A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 16.Roussel A, Inisan A G. turbo Manual. Marseille, France: Bio-Graphics; 1992. [Google Scholar]
- 17.Cowtan K. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994. pp. 34–38. [Google Scholar]
- 18.Hodel A, Kim S-H, Brünger A T. Acta Cryst A. 1992;48:851–859. [Google Scholar]
- 19.Stock A M, Mottonen J M, Stock J B, Schutt C E. Nature (London) 1989;337:745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- 20.Nicholls A, Honig B. J Comp Chem. 1991;12:435–445. [Google Scholar]
- 21.Carson M. J Appl Cryst. 1991;24:958–961. [Google Scholar]
- 22.Volz K, Matsumura P. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 23.Madhusudan, Zapf J, Whiteley J M, Hoch J A, Xuong N H, Varughese K I. Structure. 1996;4:679–690. doi: 10.1016/s0969-2126(96)00074-3. [DOI] [PubMed] [Google Scholar]
- 24.Volkman B F, Nohaile M J, Amy N K, Kustu S, Wemmer D E. Biochemistry. 1995;34:1413–1424. doi: 10.1021/bi00004a036. [DOI] [PubMed] [Google Scholar]
- 25.Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R, Dickerson R E. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 26.Stewart R C. J Biol Chem. 1993;268:1921–1930. [PubMed] [Google Scholar]
- 27.Janin J, Chothia C. J Biol Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- 28.Zhu X, Rebello J, Matsumura P, Volz K. J Biol Chem. 1997;272:5000–5006. doi: 10.1074/jbc.272.8.5000. [DOI] [PubMed] [Google Scholar]
- 29.Lupas A, VanDyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 30.Stock J B, Lukat G S, Stock A M. Annu Rev Biophys Biophys Chem. 1991;20:109–136. doi: 10.1146/annurev.bb.20.060191.000545. [DOI] [PubMed] [Google Scholar]
- 31.Le Moual H, Koshland D E., Jr J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 32.Terwilliger T C, Koshland D E., Jr J Biol Chem. 1984;259:7719–7725. [PubMed] [Google Scholar]
- 33.Djordjevic S, Stock A M. Structure. 1997;5:545–558. doi: 10.1016/s0969-2126(97)00210-4. [DOI] [PubMed] [Google Scholar]
- 34.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 35.Stock J B, Koshland D E., Jr J Biol Chem. 1981;256:10826–10833. [PubMed] [Google Scholar]
- 36.Terwilliger T C, Wang J Y, Koshland D E., Jr J Biol Chem. 1986;261:10814–10820. [PubMed] [Google Scholar]
- 37.Shapiro M J, Koshland D E., Jr J Biol Chem. 1994;269:11054–11059. [PubMed] [Google Scholar]
- 38.Shapiro M J, Panomitros D, Koshland D E., Jr J Biol Chem. 1995;270:751–755. doi: 10.1074/jbc.270.2.751. [DOI] [PubMed] [Google Scholar]
- 39.Swanson R V, Lowry D F, Matsumura P, McEvoy M M, Simon M I, Dahlquist F W. Nat Struct Biol. 1995;2:906–910. doi: 10.1038/nsb1095-906. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Swanson R V, Simon M I, Weis R M. Biochemistry. 1995;34:14626–14636. doi: 10.1021/bi00045a003. [DOI] [PubMed] [Google Scholar]