Abstract
The DNA flanking the 5′ sequence of the mouse 1α-hydroxylase gene has been cloned and sequenced. A TATA box has been located at −30 bp and aCCAAT box has been located at −79 bp. The gene’s promoter activity has been demonstrated by using a luciferase reporter gene construct transfected into a modified pig kidney cell line, AOK-B50. Parathyroid hormone stimulates this promoter-directed synthesis of luciferase by 17-fold, whereas forskolin stimulates it by 3-fold. The action of parathyroid hormone is concentration-dependent. 1,25-Dihydroxyvitamin D3 does not suppress basal promoter activity and marginally suppresses parathyroid hormone-driven luciferase reporter activity. The promoter has three potential cAMP-responsive element sites, and two perfect and one imperfect AP-1 sites, while no DR-3 was detected. These results indicate that parathyroid hormone stimulates 25-hydroxyvitamin D3-1α-hydroxylase by acting on the promoter of the 1α-hydroxylase gene.
Keywords: vitamin D, calcium, bone, kidney, cyclic AMP
Vitamin D is a major actor in calcium homeostasis of higher animals (1). To carry out these functions, vitamin D must be metabolized to its biologically active hormonal form, 1,25-dihydroxyvitamin D3 (1,25-(OH)2D3). This is a two-step process requiring 25-hydroxylation in the liver and 1α-hydroxylation in the kidney. The resulting hormone, 1,25-(OH)2D3, then binds to a nuclear vitamin D receptor (VDR) in target tissues, and the liganded receptor acts as a transcription factor to modulate the expression of specific genes encoding proteins that bring about the actions of vitamin D (2).
The importance of the 1α-hydroxylase enzyme is emphasized by the occurrence of a genetic disorder of vitamin D metabolism, vitamin D dependency rickets type I (VDDRI) (3). This disorder is characterized by very low serum 1,25-(OH)2D3 levels despite normal vitamin D intakes (4) and is thought to be the result of a defect in the 1α-hydroxylase gene (5). This link has recently been reinforced with the identification of the gene for human 1α-hydroxylase (6). The gene was mapped to chromosomal region 12q13.1-q13.3, which contains the VDDRI disease locus.
The 1α-hydroxylation step is the most tightly regulated step in vitamin D metabolism. Several physiological factors interact to regulate 1α-hydroxylase activity that, in turn, determines serum and tissue levels of 1,25-(OH)2D3. These regulators include parathyroid hormone (PTH) and hypophosphatemia, which stimulate 1α-hydroxylase activity, and 1,25-(OH)2D3, which suppresses it (7–9).
PTH is a major regulator of 1α-hydroxylase activity. Garabedian et al. demonstrated that parathyroidectomized animals are unable to synthesize 1,25-(OH)2D3, whereas PTH administration restores this activity (7). Horiuchi et al. showed that cyclic AMP (cAMP) mediates this action of PTH (10). This conclusion is further supported by the observation that forskolin, a cAMP inducer (11), can stimulate 1α-hydroxylation in vitro in primary cultures of chicken proximal tubule cells (12). In vivo and in vitro studies have indicated that actinomycin D blocks the increase in 1α-hydroxylase activity observed in response to PTH (13, 14). This implies that transcription is needed in this process.
One mode of action by which cAMP modulates transcription is through cAMP-responsive elements (CRE) in the promoters of target genes (15). cAMP-dependent protein kinase A (PKA) phosphorylates CRE-binding protein (CREB), increasing its transactivational activity. Although a consensus CRE has been described, there is controversy over whether it is sufficient for mediating a response to cAMP. It is becoming clear that the promoter context is also important (16). Other pathways of cAMP modulation of transcription may include transactivation through AP-1 and AP-2 sites in target genes, but these mechanisms are not yet clearly defined (17).
Recently, a major advance in technology of the 1α-hydroxylase has been achieved by the cloning of the mouse (18), rat (6, 19), and human (6, 29) cDNAs encoding the 1α-hydroxylase, an accomplishment first described by St. Arnaud et al. (20). This provided an opportunity for our group to clone the mouse gene including its promoter. Herein we describe the sequence, structure, and responsiveness of this promoter to PTH and forskolin. Our results suggest that PTH, through cAMP, activates the 1α-hydroxylase promoter.
MATERIALS AND METHODS
Genomic Cloning and Sequence Analysis of the 5′ Flanking Domain.
We used the PCR to amplify mouse genomic DNA with primers designed from the rat 1α-hydroxylase cDNA (19). The sequences of the primers were 5′-CTAAAGGCTGTGATCAAAGAAGTG-3′ (nucleotides 1110–1133) and 5′-TCGAGCTGGATTAAAAGAGTT-3′ (nucleotides 1278–1298). The PCR product obtained was subcloned into pCR 2.1 vector by using a TA cloning kit (Invitrogen) and sequenced by automated sequencing (Pharmacia). A mouse P1 genomic library was then screened by Genome Systems (St. Louis) by using a PCR product generated from oligonucleotides designed from the partial mouse genomic sequence obtained. The sense oligonucleotide 5′-TGTCCCAGACAGAGACATCC-3′ and the antisense oligonucleotide 5′-GTGTGGGGTCCCTTGAAG-3′ generate a PCR product of 480 bp. One P1 clone was identified. Recombinant P1 DNA was prepared according to the instructions of Genome Systems. The P1 plasmid containing the 1α-hydroxylase genomic clone was digested with various restriction enzymes (Pharmacia). Digestion products were run in an 0.8% agarose gel and transferred to a nylon membrane (Hybond N+, Amersham) for Southern analysis. The membrane was hybridized with full-length rat cDNA probe labeled with [α-32P]dCTP (Amersham) by random priming (Prime-A-Gene, Promega).
SacI digestion of the P1 clone resulted in a single 14.5-kb fragment that was identified with the cDNA probe. The SacI fragment was subcloned into pBluescript KS(−) (Stratagene). A 2.5-kb EcoRI-SalI fragment of this clone hybridized with a cDNA probe specific to the 5′ region of the rat cDNA (nucleotides 1–196) (19). The EcoRI-SalI DNA fragment was subcloned into pBluescript KS(−). This clone was sequenced by automated sequencing and found to contain the 5′ end of the reported mouse cDNA sequence (18) as well as 1.7 kb of 5′ flanking sequence. The sequence was analyzed for putative cis-acting regulatory elements by using the genetics computer group sequence analysis Software Package (Madison, WI).
Primer Extension Analysis.
Total RNA was extracted from op/op mouse kidney by using the LiCl-urea method (21). Mice were maintained on a −D, normal calcium, normal phosphorus diet (Diet 11) (22, 23). Poly(A)+ mRNA was purified by using the Oligotex mRNA midi kit (Qiagen).
Primer extension was performed by using the avian myeloblastosis virus (AMV)–reverse transcriptase primer extension system (Promega). The following oligonucleotides were used: m1a1, cggtgaaaaactctggaggcgagcttgactgcctg; m1a2: ggagcccagcgaggcatccagctgcagaggcaggt; m1a3, gagggcccagggatgtcagacaagctccggagaac; m1a4, ctgccagaccatattggcccgtaccgcgcagcgcc; m1a5, tgtagggtgggcaacgtaaactgtgcgaagtgtcc.
Oligonucleotides were purified on a 15% acrylamide denaturing gel containing 7 M urea and 1× TBE (90 mM Tris-borate/2 mM EDTA, pH 8.0). Oligonucleotide bands were visualized on a fluorescent thin-layer chromatography plate under a UV lamp. Oligonucleotides were eluted from the gel by a 24-h incubation in elution buffer (0.5 M ammonium acetate, pH 7/1 mM EDTA/0.1% SDS), ethanol precipitated two times, and resuspended in water. Each oligonucleotide was labeled at its 5′ terminus with [γ-32P] ATP (50 μCi; 1 Ci = 37 GBq) (Amersham) by T4 polynucleotide kinase. Primer extension reactions were performed with 10 fmol labeled oligonucleotide and purified mRNA equivalent to 200 μg total RNA. The labeled primers were incubated with mRNA for 15 min at 68°C, then annealed by cooling the mixture for 20 min at room temperature. Reverse transcription was performed for 30 min at 42°C with AMV reverse transcriptase. Reaction products were ethanol precipitated in the presence of 0.5 M ammonium acetate, pH 7.0, and 30 μg of glycogen. The RNA–DNA products were fractionated in an 8% acrylamide sequencing gel in the presence of ΦX174 DNA/HinfI molecular weight markers (Promega). Autoradiography was performed at −80°C for 7 days by using two intensifying screens.
Construction of Reporter Plasmids.
A 1.7-kb EcoRI-XhoI fragment corresponding to nucleotides −1652 to +15 relative to the determined transcriptional start site of the mouse 1α-hydroxylase gene was subcloned into a pGL2 basic vector (Promega). The natural 3′ XhoI site and a 5′ XhoI linker were used to insert the fragment at a XhoI site upstream of a luciferase reporter gene. The construct is designated pGL2–1αPr.
Cell Culture, Transfection, and Luciferase Assays.
AOK-B50 cells were maintained in DMEM supplemented with 7% fetal calf serum (FCS). Eighteen to 24 h before transfection, cells were plated at 50–60% confluency on six-well plates. Cells were transfected with 2.5 μg of pGL2–1αPr (or empty pGL2b vector as a negative control) by using lipofectin (Life Technologies). The DNA–lipofectin mixture was removed after 24 h, and cells were fed with DMEM containing 7% FCS and treated with hormones or vehicle as indicated for an additional 24 h. Cells were harvested in 150 μl lysis buffer (Promega), and luciferase assays were performed by using the Monolight 2010 (Analytical Luminescence Laboratory, San Diego). Luciferase measurements were normalized for protein by using the Bradford procedure (24) (Bio-Rad).
Chemicals.
Parathyroid hormone was obtained from Peninsula Laboratories, forskolin was purchased from Sigma, and 1,25(OH)2D3 was acquired from Tetrionics (Madison, WI).
RESULTS
Identification and Sequence Analysis of Mouse 1α-Hydroxylase Flanking Sequence.
The mouse 1α-hydroxylase gene was cloned by PCR screening of a P1 genomic library. From the P1 plasmid we isolated a 14.5-kb SacI fragment that hybridized with the full-length rat 1α-hydroxylase cDNA (19). Further analysis of this DNA revealed that a 2.6-kb EcoRI-SalI fragment hybridized with a probe specific to the most 5′ region of the rat 1α-hydroxylase cDNA. This EcoRI-SalI fragment was subcloned, and sequence analysis confirmed that this clone contained sequence encoding the 5′ terminal end of the mouse 1α-hydroxylase cDNA (18) as well as 1.7 kb of upstream sequence. The 5′ flanking sequence of the mouse 1α-hydroxylase gene is presented in Fig. 1. A TATA box matching the consensus sequence (TATA(A/T)A(A/T)) (25) and CCAAT box are indicated. A computer search was performed to locate additional putative regulatory elements. Three CRE-like sequences and three consensus AP-1 sites were identified. In addition, a Pu Box and two imperfect AP-2 sites were found.
Figure 1.
Nucleotide sequence of the 5′ flanking region of the mouse 1α-hydroxylase gene. The transcriptional start site is designated as +1. Putative cis regulatory elements are underlined. The translational start codon is indicated in bold. An asterisk (∗) indicates the end of the published mouse 1α-hydroxylase cDNA.
Determination of the Transcriptional Start Site.
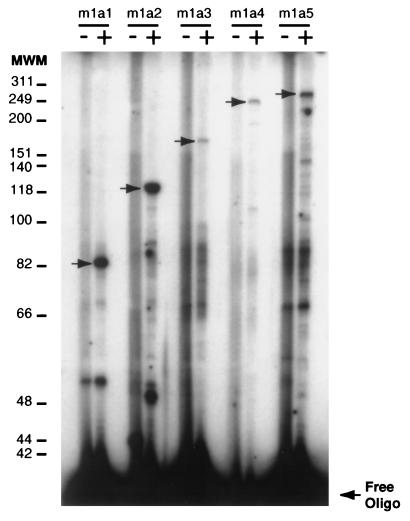
The presence of characteristic promoter regulatory elements in the 5′ flanking sequence led us to perform transcriptional start-site analysis. Primer extension reactions were performed with five independent primers. Each reaction yielded a cDNA that identified the transcription initiation site as the guanine located 42 bp upstream from the translational start codon (Fig. 2), extending the size of the cDNA by 14 bases compared with the published mouse 1α-hydroxylase sequence (18). In our numbering scheme the transcriptional start site is identified as +1. The TATA box is located at −30 bp from this position and the CCAAT box is located at −79 bp (Fig. 1). Both positions correspond well with the sites of these elements in many other eukaryotic gene promoters (25).
Figure 2.
Primer extension analysis of mouse kidney 1α-hydroxylase mRNA population. Primer extension has been performed with op/op mouse kidney mRNA (+ lanes) by using five different labeled primers: m1a1, m1a2, m1a3, m1a4, and m1a5 (see Materials and Methods section for specific sequences). Control lanes (−) represent primer extensions performed with the primer alone. Primer extension products have been fractionated on a sequencing gel. Arrows represent specific 1α-hydroxylase cDNAs. Molecular weight markers (MWM) are indicated in nucleotide bases.
Functional Activity of Mouse 1α-Hydroxylase Gene Promoter.
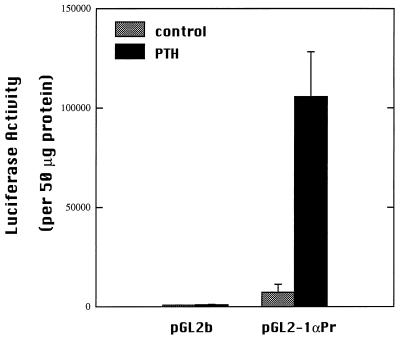
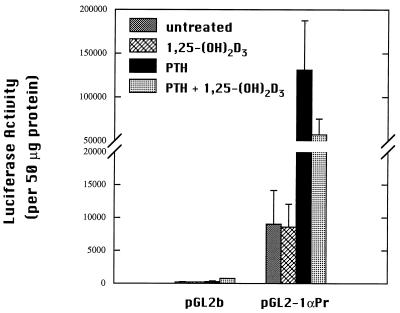
To test the functional activity of the identified promoter, reporter constructs were made. A 1.7-kb fragment of 5′ flanking sequence of the mouse 1α-hydroxylase gene was inserted upstream of a luciferase reporter gene in the pGL2 basic vector. This reporter gene construct, pGL2–1αPr, was transfected into AOK-B50 cells, a porcine kidney cell line. Promoterless pGL2b vector was transfected as a negative control. Cells transfected with pGL2–1αPr displayed basal luciferase activity at levels 60 times the negative control (Fig. 3).
Figure 3.
Transcriptional activity of the mouse 1α-hydroxylase 1.7-kb 5′ flanking sequence. AOK-B50 cells were transiently transfected with the promoter luciferase reporter construct, pGL2–1αPr, or empty pGL2b by using lipofectin. Transfected cells were treated for 24 h with 100 nM PTH or vehicle (water). Luciferase activity was normalized to protein content for each sample. Experiments were repeated three times in duplicate. Values are mean ± SD.
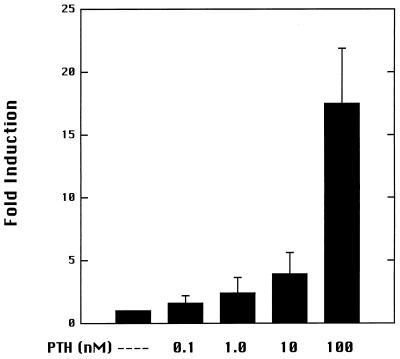
PTH is a potent stimulator of 1α-hydroxylase activity in vivo. The ability of PTH to induce luciferase activity driven by the 1α-hydroxylase promoter was examined by using AOK-B50 cells, which express stably transfected PTH receptors (26). Incubation of transfected cells with 100 nM PTH for 24 h induced luciferase activity 17-fold over activity of untreated transfected cells (Fig. 3). Enhancement of luciferase activity by PTH was concentration-dependent (Fig. 4).
Figure 4.
Dose-dependent stimulation of mouse 1α-hydroxylase gene promoter by PTH. AOK-B50 cells were transfected with pGL2–1αPr. Transfected cells were treated for 24 h with the indicated concentrations of PTH. The induction is relative to luciferase activity detected in untreated transfected cells. Experiments were repeated three times in duplicate.
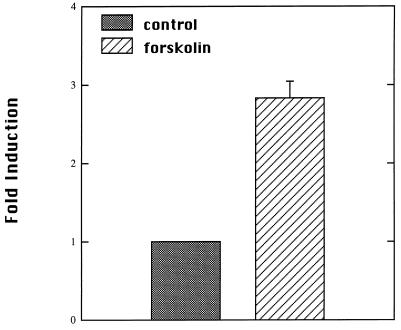
PTH has been demonstrated to stimulate multiple second-messenger pathways in AOK-B50 cells, including activation of adenylate cyclase and release of intracellular calcium (26). Several studies have indicated that stimulation of 1α-hydroxylase activity by PTH can be mimicked by forskolin, a cAMP inducer (10, 12, 27). The ability of forskolin to induce reporter gene expression was examined. Forskolin stimulated luciferase expression 3-fold in AOK-B50 cells (Fig. 5).
Figure 5.
Activation of mouse 1α-hydroxylase promoter by forskolin. AOK-B50 cells were transfected with pGL2–1αPr. Transfected cells were treated for 24 h with 10 μM forskolin or vehicle (ethanol). Data are presented as fold activation relative to vehicle-treated transfected cells. Values were obtained from three independent experiments performed in duplicate.
The hormone 1,25-(OH)2D3 itself is known to suppress 1α-hydroxylase activity (9). Transfected cells incubated with 10 nM 1,25-(OH)2D3 for 24 h showed no decrease in luciferase activity relative to basal expression of untreated transfected control cells (Fig. 6). No change was observed at levels as high as 10−7 M 1,25-(OH)2D3 (data not shown). Interestingly, transfected cells dosed with both PTH and 1,25-(OH)2D3 displayed luciferase activity that was decreased 40% compared with cells treated with PTH alone, but this activity remained substantially above basal luciferase activity (Fig. 6).
Figure 6.
1,25-(OH)2D3 suppression of PTH-stimulated luciferase activity in AOK-B50 cells. Cells were transfected with pGL2–1αPr. Transfected cells were treated with 10 nM 1,25-(OH)2D3, 100 nM PTH, and both 1,25-(OH)2D3 and PTH or vehicle (water and/or ethanol) for 24 h. Luciferase activity is corrected for protein. Experiments were performed at least two times in duplicate. Values are mean ± SD.
DISCUSSION
We have reported the identification of the mouse 1α-hydroxylase gene promoter. 1α-Hydroxylase is the key enzyme in vitamin D metabolism. In a hypocalcemic animal, activity of the 1α-hydroxylase is markedly elevated. This, in turn, increases calcium absorption from the intestine and reabsorption in the kidney. The major, if not only, factor in mediating this response to low calcium is the PTH (7, 9). Studies performed in vivo and in primary cell culture systems showed that induction of 1α-hydroxylase by PTH could be blocked by treatment with actinomycin D, suggesting that a transcriptional mechanism is responsible (27). We have now demonstrated a transcriptional mechanism of 1α-hydroxylase induction by PTH.
There have been multiple reports implicating the cAMP pathway as the mechanism by which PTH modulates 1α-hydroxylase activity. Forskolin, a cAMP inducer, increases luciferase reporter gene activity in our system. This, in combination with the putative CRE sites in the promoter, argues strongly that cAMP is involved. At this time we cannot explain the large difference between the response to PTH and the response to forskolin. PTH has been reported to activate multiple second-messenger pathways in AOK-B50 cells (26), and it is possible that another pathway is contributing to the response. Additionally, AOK-B50 cells may not be highly responsive to forskolin. Additional work will be required to explain this apparent discrepancy.
The hormone 1,25-(OH)2D3 is well established as a suppressor of its own synthesis. In VDR knockout mice, the 1α-hydroxylase transcript is present at 50 times the levels of wild-type littermates (18). In our transient transfection system, 1,25-(OH)2D3 did not suppress basal luciferase activity but did show an ability to reduce PTH-stimulated luciferase activity. A computer search failed to identify a consensus vitamin D-responsive element (DR-3) in the promoter sequence (28). This suggests that 1,25-(OH)2D3 may suppress 1α-hydroxylase expression by an indirect mechanism, or that the vitamin D-responsive site is different from the classical consensus sequence. It is also possible that DNA elements located upstream of the 1.7-kb flanking sequence are required for maximal suppression by 1,25-(OH)2D3. Further investigation will be required to find the mechanism of 1α-hydroxylase suppression by 1,25-(OH)2D3.
The 1α-hydroxylase promoter is unremarkable in terms of sequences. However, it clearly has promoter activity because it increases transcription of luciferase DNA by 60-fold. A TATA box is located as predicted, and there is a CAAT box at −79 bp. Beyond this, two AP1 sites that perfectly match the consensus sequence and one imperfect AP-1 site are located in the distal region of the promoter. Three potential CREB sites are located at positions −167, −941, and −1618 relative to the transcriptional start site. Thus, there are clearly possible sites of PTH activation by known mechanisms. It appears likely that it will now be possible to pinpoint the site of activation of the 1α-hydroxylase promoter by PTH, as well as the mechanism involved, by using the AOK-B50 system and the herein identified promoter.
Acknowledgments
We thank Drs. Henry Kronenberg and John T. Potts of the Massachusetts General Hospital for the AOK-B50 cells and Rebecca Vincenti for her technical assistance. H.L.B. is a graduate student in the Interdepartmental Graduate Degree Program in Nutritional Sciences. This work was supported in part by a program project grant (DK14881) from the National Institutes of Heath (H.F.D.), a fund from the National Foundation for Cancer Research (H.F.D.), and a fund from the Wisconsin Alumni Research Foundation (H.F.D.).
ABBREVIATIONS
- 1
25-(OH)2D3, 1,25-dihydroxyvitamin D3
- VDR
vitamin D receptor
- VDDRI
vitamin D dependency rickets type I
- PTH
parathyroid hormone
- cAMP
cyclic AMP
- CRE
cAMP-responsive element
- CREB
CRE-binding protein
- PKA
protein kinase A
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF041256).
References
- 1.DeLuca H F. The Harvey Lectures. New York: Academic; 1981. , Series 75, pp. 333–379. [Google Scholar]
- 2.Ross T K, Darwish H M, DeLuca H F. Vitam Horm. 1994;49:281–326. doi: 10.1016/s0083-6729(08)61149-8. [DOI] [PubMed] [Google Scholar]
- 3.Prader A, Illig R, Heierli E. Helv Paediatr Acta. 1961;16:452–468. [PubMed] [Google Scholar]
- 4.Scriver C R. N Engl J Med. 1978;299:976–979. doi: 10.1056/NEJM197811022991803. [DOI] [PubMed] [Google Scholar]
- 5.Fraser D, Kooh S W, Kind H P, Holick M F, Tanaka Y, DeLuca H F. N Engl J Med. 1973;289:817–822. doi: 10.1056/NEJM197310182891601. [DOI] [PubMed] [Google Scholar]
- 6.St. Arnaud R, Messerlian S, Moir J M, Omdahl J L, Glorieux F H. J Bone Min Res. 1997;2:1552–1559. doi: 10.1359/jbmr.1997.12.10.1552. [DOI] [PubMed] [Google Scholar]
- 7.Garabedian M, Holick M F, DeLuca H F, Boyle I T. Proc Natl Acad Sci USA. 1972;69:1673–1676. doi: 10.1073/pnas.69.7.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka Y, DeLuca H F. Arch Biochem Biophys. 1973;54:566–574. doi: 10.1016/0003-9861(73)90010-6. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka Y, DeLuca H F. Am J Physiol. 1984;246:E168–E173. doi: 10.1152/ajpendo.1984.246.2.E168. [DOI] [PubMed] [Google Scholar]
- 10.Horiuchi N, Suda T, Takahashi H, Shimazawa E, Ogata E. Endocrinology. 1977;101:969–974. doi: 10.1210/endo-101-3-969. [DOI] [PubMed] [Google Scholar]
- 11.Seamon K B, Daly J W. J Cyclic Nucleic Res. 1982;7:201–224. [PubMed] [Google Scholar]
- 12.Henry H L. Endocrinology. 1985;116:503–510. doi: 10.1210/endo-116-2-503. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka Y, Chen T C, DeLuca H F. Arch Biochem Biophys. 1972;152:291–298. doi: 10.1016/0003-9861(72)90218-4. [DOI] [PubMed] [Google Scholar]
- 14.Korkor A B, Gray R W, Henry H L, Leinman J G, Blumenthanl S S, Garancis J C. J Bone Min Res. 1987;2:517–524. doi: 10.1002/jbmr.5650020608. [DOI] [PubMed] [Google Scholar]
- 15.Lee K A W. Curr Opin Cell Biol. 1991;3:953–959. doi: 10.1016/0955-0674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 16.Deutsch P J, Hoeffler J P, Jameson J L, Lin J C, Habener J F. J Biol Chem. 1988;263:18466–18472. [PubMed] [Google Scholar]
- 17.Karin M. Trends Genet. 1989;5:65–67. doi: 10.1016/0168-9525(89)90027-9. [DOI] [PubMed] [Google Scholar]
- 18.Takeyama K-I, Khanaka S, Sato T, Kobori M, Yanagisawa J, Kato S. Science. 1997;277:1827–1830. doi: 10.1126/science.277.5333.1827. [DOI] [PubMed] [Google Scholar]
- 19.Shinki T, Shimada H, Wakino S, Anazawa H, Hayashi M, Saruta T, DeLuca H F, Suda T. Proc Natl Acad Sci USA. 1997;94:12920–12925. doi: 10.1073/pnas.94.24.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.St. Arnaud R, Moir J M, Messerlian S, Glorieux F H. J Bone Min Res. 1996;11:S124. doi: 10.1359/jbmr.1997.12.10.1552. (abstr.). [DOI] [PubMed] [Google Scholar]
- 21.Auffray C, Rougeon F. Eur J Biochem. 1980;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- 22.Suda T, DeLuca H F, Tanaka Y. J Nutr. 1970;100:1049–1052. doi: 10.1093/jn/100.9.1049. [DOI] [PubMed] [Google Scholar]
- 23.McCary L C, Smith C M, DeLuca H F. J Bone Min Res. 1997;12:1944–1951. doi: 10.1359/jbmr.1997.12.11.1944. [DOI] [PubMed] [Google Scholar]
- 24.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Breathnach R, Chambon P. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- 26.Bringhurst F R, Juppner H, Guo J. Endocrinology. 1993;132:2090–2098. doi: 10.1210/endo.132.5.8386606. [DOI] [PubMed] [Google Scholar]
- 27.Armbrect H J, Forte L R, Wongsurawat N, Zenser T V, Davis B. Endocrinology. 1984;114:644–649. doi: 10.1210/endo-114-2-644. [DOI] [PubMed] [Google Scholar]
- 28.Darwish H M, DeLuca H F. Crit Rev Eukaryotic Gene Expression. 1993;3:89–116. [PubMed] [Google Scholar]
- 29.Monkawa T, Yoshida T, Wakino S, Shinki T, Anazawa H, DeLuca H F, Suda T, Hayashi M, Saruta T. Biochem Biophys Res Commun. 1997;239:527–533. doi: 10.1006/bbrc.1997.7508. [DOI] [PubMed] [Google Scholar]