Abstract
Proper control of the mammalian cell cycle requires the function of cyclin-dependent kinase (CDK) inhibitors. The p21 family currently includes three distinct genes, p21, p27Kip1, and p57Kip2, that share a common N-terminal domain for binding to and inhibiting the kinase activity of CDK-cyclin complexes. The p21 protein also binds to proliferating cell nuclear antigen (PCNA) through a separate C-terminal domain affecting DNA replication and repair. The p27 and p57 proteins also each contain unique C-terminal domains whose functions are unknown. Here we show that the human p57 protein, like p21, contains a PCNA-binding domain within its C terminus that, when separated from its N-terminal CDK-cyclin binding domain, can prevent DNA replication in vitro and S phase entry in vivo. Disruption of either CDK/cyclin or PCNA binding partially reduced p57’s ability to suppress myc/RAS-mediated transformation in primary cells, while loss of both inhibitory functions completely eliminated p57’s suppressive activity. Thus, control of cell cycle and suppression of cell transformation by p57 require both CDK and PCNA inhibitory activity, and disruption of either or both functions may lead to uncontrolled cell growth.
The primary control of the eukaryotic cell cycle is provided by the activity of a family of serine/threonine protein kinases, cyclin-dependent kinases (CDKs) (1). The enzymatic activity of a CDK can be positively regulated by the binding of a cyclin and negatively regulated by the binding of a CDK inhibitor. In mammalian cells, there exists at least two distinct families of CDK inhibitors, represented by the two prototypic CDK inhibitors p16 and p21 (2). The p16/INK4 family of CDK inhibitors currently includes three additional genes: p15INK4b, p18INK4c, and p19INK4d (2). Members of the INK4 gene family are related in sequence and evolution and specifically bind in a binary fashion to two closely related CDK proteins, CDK4 and CDK6, thus suppressing cell growth in a pRb-dependent manner (3). In contrast, the p21 family that currently contains two other genes, p27Kip1 and p57Kip2, broadly inhibits CDK activity by forming a ternary complex with the CDK and the cyclin. A unique feature of CDK inhibitors is that their expression can be induced by or is correlated with a wide range of cell growth inhibitory signals, that include mitogen starvation, cell-cell contact inhibition, DNA damage, antiproliferative cytokine treatment, terminal cell differentiation, and cellular senescence (2, 3).
p21, initially identified in normal human fibroblasts as a component of quaternary cyclin D-CDK complexes that also contain proliferating cell nuclear antigen (PCNA), was subsequently found to contain two separate binding activities: a CDK-cyclin binding domain at the N terminus and a C-terminally located PCNA binding site, respectively (reviewed in ref. 2). Two additional members of the p21 family, p27 and p57 share with p21 a common N-terminal domain for binding to a wide range of CDK-cyclin complexes. The p27 and p57 proteins each contain a unique C-terminal domain known as the QT domain, and p57 additionally contains a central PAPA repeat (4, 5). The functions of the PAPA repeat and the QT domain are unknown.
The p57 gene is located on human chromosome 11p15.5 (5), a region predicted to contain a tumor suppressor gene(s) involved in the development of several human cancers, including those of the breast, bladder, lung, ovary, kidney, and testicle (6). In particular, the gene for Beckwith–Wiedemann syndrome (BWS) is localized in the 11p15.5 as determined by the linkage analysis of autosomal dominant pedigrees. BWS is characterized by numerous growth abnormalities and an increased risk of childhood tumors. Specific loss of the maternal 11p15 allele and uniparental disomy (paternal) suggest that the BWS gene is genomically imprinted, and the p57 gene is maternally expressed and imprinted in both human and mouse (7, 8). Consistent with its suggested role in tumor growth suppression, mice lacking p57 function displayed altered cell proliferation and differentiation, increased apoptosis and phenotypes seen in patients with BWS (9, 10). More direct evidence for the involvement of the p57 gene in human cancer was obtained by the finding that four of 24 (17%) BWS patients examined contain mutations in the p57 gene that altered the structure of the p57 protein (11, 12). Intriguingly, of the p57 mutations identified, three retained the N-terminal CDK-cyclin binding domain: one lost the central PAPA repeat and C-terminal domain, and two contained deletions in the C-terminal QT domain. These findings strongly implicate a separate activity located in the C terminus of p57 whose disruption may contribute to altered cell growth control. In this report, we demonstrate that human p57 contains a PCNA-binding domain in this region that may result in such an alteration.
MATERIALS AND METHODS
p57 Constructs.
For the yeast two-hybrid screen, a C-terminal cDNA fragment containing the C-terminal 55 amino acid residues of human p27 (codons 142–198) and the C-terminal 79 residues of human p57 (codons 236–316) were generated by PCR and inserted into pGBT8 as a fusion protein with the DNA-binding domain of Gal4p (amino acids 1–147). Yeast two-hybrid screening was conducted in a human WI-38 fibroblasts cDNA library following the manufacturer’s instruction (CLONTECH). We estimate that ≈5 × 106 transformants were screened. Various human p57 mutants were generated by oligonucleotide-mediated mutagenesis by using either Quick-Change (Stratagene) or direct ligation and were verified by DNA sequencing. Individual PCNA binding mutants are listed (see Fig. 2). The CDK/cyclin binding mutant used in cell transformation assays (p57CKmut) contains two substitutions (R31A and F34D) in the cyclin binding domain and two (W61A and F65D) in the CDK binding region. Wild-type and mutant p57 genes were placed under the control of a cytomegalovirus promoter for the analysis of cell cycle arrest (pcDNA3, Invitrogen) or inserted in the pVNic retroviral vector for the cell transformation assay (13). Expression of each protein was first verified by in vitro translation in a TNT rabbit reticulocyte lysate system (Promega). Expression of each mutant protein in vivo and loss of CDK and cyclin binding activity in p57CKmut was confirmed by coupled immunoprecipitation and Western blot analysis following cotransfection of pVNic-p57CKmut and pCMV-cyclin D1 or pCMV-CDK2 into Saos-2 cells (confirmatory results not shown).
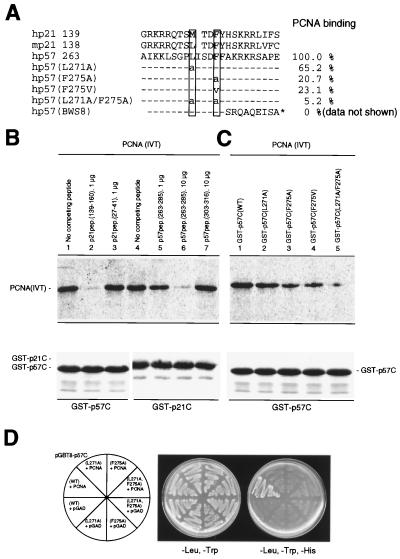
Figure 2.
p57 and p21 binds to PCNA via similar sequences. (A) A small region of human p21 (residues 139–160) sufficient for binding to PCNA (17–19) contain two critical residues, Met-147 and Phe-150 (boxed), are conserved in both mouse p21 and human p57. Mouse p57 lacks a complete corresponding region and predictably the ability to bind PCNA. A series of wild-type (residues 263–285) and mutant p57 peptides used in this study are listed. The hp57(BWS8) corresponds to a mutant p57 identified in a BWS patient (#8) (11). Comparison of binding between the p57 proteins and PCNA determined from experiments shown in Fig. 2C are shown on the right side and binding efficiency between wild-type p57 protein and PCNA was set at 100%. (B) p21 and p57 bind to PCNA via similar sequences. 35S-labeled PCNA protein (20 μl) was incubated with the various p21 or p57 peptides and then added to an equal amount of GST-p57C or GST-p21C fusion proteins attached to glutathione-agarose beads. GST-fusion proteins and bound PCNA were recovered from the different mixtures and resolved by SDS/PAGE. The gel was stained with Coomassie blue (Lower) to verify equal recovery of the GST fusion protein prior to autoradiography (Upper). (C) Mapping of PCNA binding site in vitro. 35S-labeled PCNA protein (20 μl) was incubated with 2 μg wild-type or various mutant GST-p57C proteins. GST-p57C protein and bound PCNA were recovered from the different mixtures and resolved by SDS/PAGE. (D) Mapping of PCNA binding site in vivo. Yeast HF7c cells were simultaneously transformed with the parental pGAD or plasmid expressing a GAL4bd fusion protein with a wild-type or a mutant p57 C-terminal domain and with a plasmid expressing GAL4ad fusion with PCNA protein. Cells were streaked on nonselective medium with histidine (-Leu, -Trp) or selective medium without histidine (-Leu, -Trp, -His).
Peptides, Antibodies, and Protein Expression.
All peptides were purified through HPLC and characterized on a mass spectrometer. A rabbit polyclonal antibody specific to p57 was generated by using a synthetic peptide derived from the human p57 C terminus (GVGSVEQTPRKRLR). This antibody cross reacts efficiently with mouse p57. The monoclonal anti-PCNA antibody (PC10) was purchased from PharMingen. Histidine-tagged proteins, the C terminus of human p57 (the last 79 residues), mouse p57 (the last 53 residues), human p27 (the last 55 residues), and full-length human p21 and p57, were purified following the manufacturer’s instruction (Qiagen, Chatsworth, CA). For the production of glutathione S-transferase (GST) fusion proteins, the C termini of human p21 (the last 63 residues) and human p57 (the last 79 residues) were fused with GST and purified by using glutathione Sepharose 4B (Pharmacia). Recombinant baculoviruses expressing hexahistidine tagged C-terminal p57 were constructed by using the pVL1393 baculovirus transfer vector (PharMingen). Other recombinant baculoviruses have been described (5, 14). Procedures for metabolic labeling and immunoprecipitations were as described (14, 15).
In Vitro Binding Assays.
The histidine-tagged proteins (1 μg) attached to a Ni-nitrilotriacetate (NTA) resin were mixed with purified PCNA in 200 μl of Nonidet P-40 buffer (15) for 1 h at 4°C. The Ni-NTA resins were washed three times with 1 ml of Nonidet P-40 buffer containing 50 mM imidazole. For in vitro competition assays (Fig. 2B), 20 μl of 35S-labeled PCNA produced by using the reticulocyte in vitro transcription-translation system was incubated with the p21 or p57 derived peptides for 30 min at room temperature, then added to 200 μl Nonidet P-40 buffer containing 2 μg GST-p57C or GST-p21C fusion proteins attached to glutathione-agarose beads. The beads were incubated for 1 h at 4°C, then washed three times with 1 ml of Nonidet P-40 buffer. Bound proteins were released by boiling in SDS sample buffer, and electrophoresed on 15% denaturing polyacrylamide gels followed by autoradiography.
DNA Replication Assays.
Reaction mixtures (10 μl), assembled at 0°C, contained 40 mM Tris⋅HCl, pH 7.5; 0.5 mM DTT; 1 μg of BSA; 7 mM magnesium acetate; 2 mM ATP; 100 μM each of dATP, dGTP, and dTTP; 20 μM [α-32P]dCTP (17, 100 cpm/pmol); 8.7 fmol of singly-primed M13 DNA; 240 ng of human single-stranded DNA binding protein; 100 fmol of human RFC; 0.1 unit of human pol δ and either 5 or 50 ng of human PCNA, as indicated. Where specified, inhibitors were added to reaction mixtures lacking RFC and pol δ, and reactions were incubated for 10 min at 0°C. RFC and pol δ were then added and reactions were incubated at 37°C for 30 min. The amount of acid-insoluble material formed was determined with an aliquot (1 μl) after termination with 20 mM EDTA. Loading dye was added, and the mixtures were subjected to agarose gel electrophoresis in buffer containing 30 mM NaOH and 1 mM EDTA for 16 h at 35 V. Gels were dried and autoradiographed for 1 h at −80°C.
Cell Culture, Transfection, Fluorescence-Activated Cell Sorter Analysis, and Transformation Assay.
Mammalian cells were cultured in a 37°C incubator with 5% CO2 in DMEM supplemented with 10% fetal bovine serum. Sf9 cells were cultured in Grace’s medium supplemented with 10% heat-inactivated fetal bovine serum at 27°C. Cell transfections were carried out by using the Lipofectamine reagent according to the manufacturer’s instructions (GIBCO/BRL). For cell cycle inhibition analysis, Saos-2 cells at 20–30% confluence were cotransfected with 10 μg of individual plasmids and 1 μg of pcDNA3-GFP. Forty hours posttransfection, cells were harvested with trypsin, washed with PBS, then fixed in 5 ml of PBS containing 3.6% paraformaldehyde for 30 min at 4°C. After washing cells once with PBS containing 1% BSA, the DNA was stained with propidium iodide (50 μg/ml) containing 250 μg/ml ribonuclease A and 0.1% Triton X-100. Flow cytometry analysis was conducted by using a Becton Dickinson FACScan. DNA content in 10,000 green fluorescent protein (GFP)- positive cells is presented in the DNA histograms. Preparation of early passage rat embryo fibroblast and transfection by the calcium phosphate precipitation were described in (16). 0.8 × 106 rat embryo fibroblasts were seeded and transfected the following day by the standard calcium-phosphate method with DNA mixtures containing 30 μg of genomic carrier DNA and 2 μg each of the myc, ras, and indicated p57 expression constructs or empty vector. The foci were verified as transformed by microscopic inspection and counted 8 and 12 days after transfection. The cells were then fixed and stained with Giemsa for photographic documentation.
RESULTS
Human p57 Interacts with PCNA.
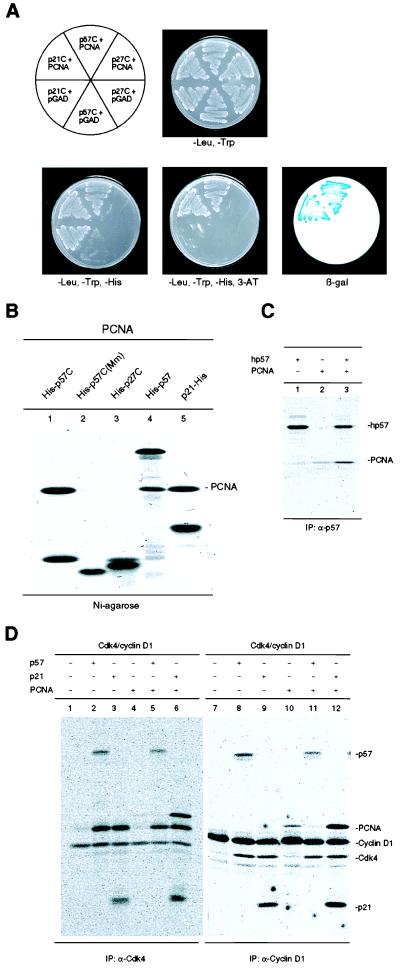
To identify potential biological activities located outside of the CDK-cyclin binding domains of p27 and p57, we searched for cellular proteins that could interact with the C-terminal portion of either protein by using the yeast two-hybrid assay. A 250-bp fragment encoding the 79 C-terminal amino acid residues of human p57 (hereafter referred to as p57C) or a 171 bp fragment encoding the 55 C-terminal amino acid residues of human p27 (p27C) was fused to the yeast Gal4 DNA binding domain. The resulting vectors were cotransformed into yeast HF7c cells with a human WI-38 fibroblasts cDNA library. No positive clones were identified from the screen by using p27C (V. Coffield III and Y.X., unpublished data). Of an estimated 5 × 106 transformants screened by using the C-terminal region of human p57, four clones were obtained that were both His+ and positive for β-galactosidase staining (Fig. 1A and data not shown). Sequencing analysis revealed that two independent clones encoded human PCNA (data not shown). Both were fused in-frame with the GAL4 DNA activation domain, one starting at amino acid residue 2 and the other containing an additional 17 bp of 5′ untranslated region (5′ UTR). To confirm the interaction between p57C and the PCNA protein in a cell-free system, purified recombinant fusion proteins containing six copies of a histidine tag fused either to the C-terminal domain of human p57 (His-p57C, Fig. 1B, lane 1), mouse p57[His-p57C (Mm), lane 2], human p27 (His-p27C, lane 3), full-length human p57 (His-p57, lane 4), or full-length human p21 (p21-His, lane 5) were incubated with purified human PCNA protein and then recovered on nickel-agarose beads. The bound proteins were separated by SDS/PAGE and visualized by Coomassie blue staining (Fig. 1B). Both the full-length and the C-terminal region of human p57, as well as the full-length human p21 protein bound PCNA at approximately a one-to-one molar ratio. The C-terminal domain of mouse p57 containing the last 53 residues did not bind a detectable amount of PCNA (lane 2), and full-length mouse p57 produced by baculoviruses in insect cells also exhibited a much weaker PCNA binding activity than human p57 (data not shown). Whether there exists a significant species difference between p57 proteins in their PCNA binding activity remains to be determined. In two separate assays involving the yeast two-hybrid system and in vitro binding, we did not detect any physical interaction between human p27 (either the C-terminal domain or full-length protein), and PCNA (lane 3, and additional negative data not shown). To further confirm the p57-PCNA interaction, Sf9 insect cells, were coinfected with baculoviruses expressing individual proteins in various combinations, infected cells were metabolically labeled with [35S]-methionine, and protein complexes were either recovered on Ni-agarose or by immunoprecipitation and subsequently analyzed by SDS/PAGE. Both full-length p57 and the p57 C-terminal domain formed stable binary complexes with PCNA in vivo (Fig. 1C).
Figure 1.
Interaction of human p57 with PCNA. (A) Yeast HF7c cells were simultaneously transformed with a plasmid expressing a GAL4bd fusion protein and a plasmid expressing a GAL4ad fusion protein as indicated. Cells were streaked on nonselective medium with histidine (-Leu, -Trp), selective medium without histidine (-Leu, -Trp, -His), and selective medium without histidine but containing 5 mM 3-amino-1,2, 3-triazole (-Leu, -Trp, -His, 3-AT). Staining for β-galactosidase expression, activated from an independent GAL4 responsive promoter, is shown (β-gal, Lower Right). The C-terminal domain of p21 possesses as a trans-activating activity (self-activation) when expressed as a fusion protein with the GAL4 DNA binding domain. (B) Equal amounts of histidine-tagged human p57C (lane 1), mouse p57 (lane 2), human p27 (lane 3), full-length human p57 (lane 4), or human p21 (lane 5) were incubated with 2 μg purified human PCNA protein. Mixtures were recovered on Ni-Sepharose beads, resolved by SDS/PAGE, and stained with Coomassie blue. (C) Exponentially growing Sf9 cells were singly infected with baculoviruses expressing the histidine-tagged C-terminal domain of human p57 (His-p57C), full-length human p57 (hp57), human PCNA, or doubly infected with their combinations as indicated on the top of each lane. Cells were metabolically labeled with [35S]methionine 40 h postinfection and p57 protein complexes were recovered on Ni-Sepharose or immunoprecipitated with an anti-p57 antibody and analyzed by SDS/PAGE and autoradiography. (D) Exponentially growing Sf9 cells were coinfected with baculoviruses expressing various human proteins. CDK4 and cyclin D1 protein complexes were immunoprecipitated with indicated antibodies and analyzed by SDS/PAGE and autoradiography.
We examined whether p57, like p21, can also form quaternary complexes with CDKs, cyclins and PCNA (14). Quaternary cyclin D1-CDK4-p21-PCNA complexes can be recovered by both anti-cyclin D1 and anti-CDK4 immunoprecipitation (Fig. 1D, lanes 6 and 12). In contrast, repeated attempts failed to detect p57 in either anti-cyclin D1 or anti-CDK4 immunocomplexes, indicating the lack of formation of the quaternary cyclin D1-CDK4-p57-PCNA complex (lanes 5 and 11, and data not shown). In addition, we noticed that the small amount of PCNA in anti-cyclin D1 immunocomplexes (lane 10), presumably due to either direct interaction between cyclin D1 and PCNA and/or an endogenously expressed p21-like protein in Sf9 cells, was diminished by the expression of p57 (lane 11). These observations suggest that, unlike p21, p57 may interact with CDK/cyclin and PCNA in a mutually exclusive manner.
The PCNA binding domain of p21 has been localized to a small region spanning about 20 amino acid residues in the C terminus as analyzed by mutagenesis (17–19). A 20 amino acid region in human p57, but not mouse p57, or mouse and human p27, contains significant similarity (four identical and eight conserved residues) to the PCNA binding domain of p21 (Fig. 2A). This prompted us to determine if human p57 binds to PCNA through this sequence. Equal amounts of fusion proteins consisting of GST and the C-terminal domain of either human p57 (GST-p57C) or p21 (GST-p21C) were incubated with in vitro translated, 35S-labeled PCNA in the presence of various p21 or p57 synthetic peptides. GST fusion proteins were recovered on glutathione-agarose beads, and the bound PCNA protein was analyzed by SDS-PAGE. An excess amount of both GST-p21C and GST-p57C bound nearly all the input PCNA in the absence of a competing peptide (Fig. 2B, lanes 1 and 4). Addition of a molar excess of wild-type p21 peptide containing the PCNA binding region (residues 139–160, lane 2), but not an amino terminal p21 peptide (residues 27–41, lane 3), completely blocked the binding of GST-p57C to PCNA. Conversely, a p57 peptide corresponding to the sequence similar to the PCNA binding domain of p21 (residues 263–285, lanes 5 and 6), but not a p57 peptide outside the conserved region (lane 7), inhibited the binding of GST-p21C to PCNA. Ten-fold more p57 peptide was necessary to completely block the binding of GST-p21C to PCNA as compared with the amount of the p21 peptide necessary to block the binding of GST-p57C to PCNA (compare lanes 5 and 6 with lane 2). This indicates that p57 binds to PCNA with an affinity that is at least 10-fold lower than p21.
An eight-amino acid motif (144QTSMTDFY151) in the 20-amino acid region, in particular Met-147 and Phe-150, is critical for p21 to bind PCNA (18). At these positions, human p57 contains a conserved leucine (L271) and an identical phenylalanine (F275) residue, respectively. Substitution of either residue with an alanine or valine significantly reduced PCNA binding. Mutation of both residues almost completely abolished the binding of p57 to PCNA even when a large excess was incubated with PCNA (Fig. 2C, lane 5). Mutation of either residue also rendered p57 inactive in binding PCNA in vivo as determined by the yeast two-hybrid assay (Fig. 2D). In addition, a truncated p57 protein (BWS8) that has lost 41 C-terminal residues as the result of a T to AG transversion/addition at codon 276 found in BWS exhibited no detectable PCNA binding activity (data not shown). These results indicate that p21 and p57 interact with PCNA via a similar mechanism that involves a conserved region located at the C terminus of both proteins.
C-Terminal Domain of p57 Inhibits PCNA-Dependent DNA Synthesis.
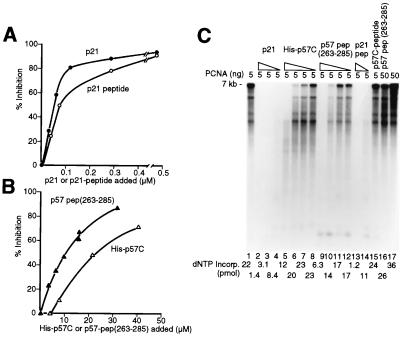
PCNA is essential for rapid and processive synthesis of DNA by DNA polymerases δ and ɛ. The C-terminal domain of p21 possesses a potent inhibitory activity on DNA synthesis that can be reversed by increasing the level of PCNA (20–22). In the presence of limiting levels of PCNA (5 ng, 17 μM monomer), increasing amounts of p57C (His-p57C) or a chemically synthesized p57 peptide (residues 263 to 285) markedly inhibited nucleotide incorporation in the elongation of singly-primed M13 DNA in a pol δ holoenzyme reaction, reaching 50% reduction in the presence of 20 μM and 10 μM quantities of these two derivatives, respectively (Fig. 3B). Consistent with its higher affinity in binding to PCNA, both full-length p21 protein and a p21 peptide (residues 139–160) were more effective inhibitors: 50% inhibition was observed with 0.05 to 0.1 μM (Fig. 3A). The influence of the inhibitors on the lengths of the DNA products formed in the pol δ holoenzyme catalyzed reaction was examined by alkaline agarose gel electrophoresis. Both purified p57C (His-p57C, Fig. 3C, lanes 5–8) and the p57 peptide (residues 263–285, lanes 9–12) reduced the sizes of the DNA synthesis products. Similar but more potent inhibition was observed with purified p21 (lanes 2–4) and the p21 peptide (lanes 13 and 14). When the level of PCNA added to the reactions was increased 10-fold, the lengths of the products produced in the presence of 6.6 μM of the p57 peptide (residues 263–285) were restored (compare lanes 10 and 16). The addition of a 15-mer p57 control peptide corresponding to a region outside the PCNA binding sequence (residues 303–313), at a concentration of 50 μM, had no detectable effect in the elongation reaction (lane 15). These results demonstrate that the C-terminal domain of p57 can inhibit PCNA-dependent DNA elongation reactions catalyzed by the pol δ holoenzyme.
Figure 3.
p57 C-terminal domain mediated inhibition of PCNA-dependent DNA synthesis by DNA polymerase δ. (A) Effects of full-length CDK inhibitor p21 and the p21 peptide containing the PCNA binding activity (residues 139–160) on nucleotide incorporation catalyzed by pol δ. (B) Effects of His-p57C and p57 peptide (residues 263–285) on nucleotide incorporation catalyzed by pol δ. (C) Effects of CDK inhibitors p57 and p21 on the length of the DNA products formed in the pol δ holoenzyme catalyzed reaction. All reactions contained 5 ng PCNA except lanes 16 and 17 where 50 ng PCNA protein was added. The levels of inhibitors used were as follows: lanes 2–4 contained 0.3, 0.12 and 0.06 μM p21, respectively; lanes 5–8, 22.5, 9, 4.5 and 2.25 μM p57C, respectively; lanes 9–12, 16.5, 6.6, 3.3, and 16.5 μM p57 peptide (residues 263–285), respectively; lanes 13 and 14, 0.8 and 0.075 μM p21 peptide (residues 139–160). Lane 15 contained 50 μM of a p57 peptide corresponding to the last 15 amino acid residues of human p57 (residues 303–313) outside the conserved PCNA binding sequence. Lane 16 contained 6.6 μM p57 peptide (residues 263–285). Lanes 1 and 17 contained no inhibitors. The reaction described in lane 17 represents the replication activity observed with excess PCNA.
Cell Cycle Arrest by p57 C-Terminal Domain Requires PCNA Binding Activity.
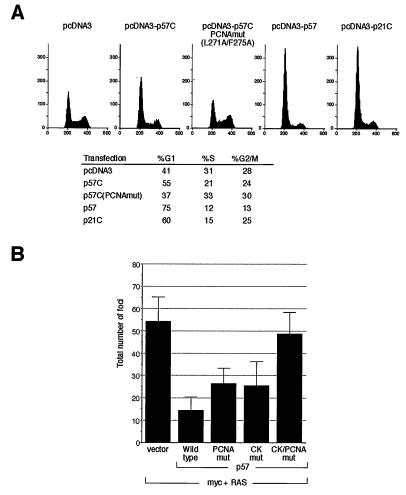
To determine whether the p57 C-terminal domain, when separated from the CDK-cyclin binding domain, could regulate cell cycle progression in vivo in a PCNA binding-dependent manner, we cotransfected p57C or full-length p57 into cultured Saos-2 cells with a plasmid expressing the green fluorescence protein. At 40 hr posttransfection, transfected cells were analyzed for their DNA content by flow cytometry. Ectopic expression of full-length p57 and the C-terminal PCNA binding domains of either p57 or p21, but not the parental pcDNA3 vector, caused cells to accumulate in G1 (Fig. 4A). The mutant p57 (L271A/F275A) that lacks the ability to bind PCNA did not exhibit any inhibitory activity, indicating that G1 cell cycle arrest caused by the p57 C-terminal domain is dependent on its PCNA binding activity. Consistent with its higher affinity for PCNA, the p21 C-terminal domain was more potent than the p57 C-terminal domain in causing cell cycle arrest in vivo (Fig. 4A). The full-length p57 was more potent than the p57 C-terminal domain in causing G1 arrest, suggesting that PCNA binding domain contributes only partially to p57’s cell cycle arrest activity.
Figure 4.
Inhibition of cell cycle and cell transformation by p57. (A) Saos-2 cells were transiently transfected with 1 μg of pcDNA3-green fluorescent protein and 10 μg of indicated plasmids. The percent distribution in different cell cycle stages was analyzed by flow cytometry. (B) Histogram of the average number of foci per plate counted 12 days posttransfection for two individual experiments combined. Twelve plates (six for each experiment) that were split 1:3 on the day following the transfection were counted for each point. Bars = SD.
Suppression of Cell Transformation by p57 Requires Both CDK-Cyclin and PCNA Binding Activities.
To determine whether loss of p57’s PCNA inhibitory activity leads to uncontrolled cell growth, we compared the wild-type and PCNA-binding deficient p57 mutants for their ability to suppress myc and RAS mediated cell transformation in rat embryo fibroblasts. Use of full-length p57 bearing different point mutations also allowed an assessment of the relative contributions of the CDK/cyclin- and PCNA-binding activities to p57’s function. Plasmids expressing full-length wild-type p57, p57 mutants lacking the ability to bind to CDKs and cyclins (p57CKmut), to PCNA (p57PCNAmut), or the double mutant (p57CK/PCNAmut) were cotransfected with murine c-myc and an activated [lVal12]Ha-RAS expression construct into early passage rat embryo fibroblasts. The impact of each p57 protein on myc/RAS-mediated cell transformation was assessed by scoring the number of foci generated (Fig. 4B). In two independent experiments, expression of wild-type p57 markedly reduced the number of foci obtained by myc/RAS by ≈75% (Fig. 4B). Disruption of either the PCNA binding or the CDK/cyclin-binding activity led to nearly equivalent reductions in p57’s suppressive potential, suggesting that the CDK/cyclin- and PCNA-binding activities contribute about equally to p57’s in vivo suppressive function. Loss of both binding functions completely eliminated p57’s ability to suppress transformation, as cotransfection of the double mutant with myc/RAS did not significantly affect the number of foci generated in comparison to the empty vector control. This result indicates that cell growth suppression by p57 is primarily dependent on its ability to bind to CDK/cyclin and PCNA and does not involve an as yet unidentified activity in p57.
DISCUSSION
The presence of separate CDK/cyclin and PCNA binding domains in p21 and the ability of either domain alone to inhibit the cell cycle has led to the notion that p21 may uniquely possess two separate activities, each regulating the cell cycle independently via its own mechanism(s). We show here that in addition to inhibiting cyclin-CDK enzymes, p57, like p21, contains a physically separate activity that directly inhibits PCNA-dependent DNA replication in vitro and prevents cell entry into S phase in vivo. Therefore, two common structural and biochemical properties are shared by two members of the p21 CDK inhibitor family: an N-terminal domain for binding to CDK-cyclin complexes and a C-terminal domain for binding to a DNA replication factor, PCNA.
We attribute the partial loss of in vivo cell cycle inhibition by the mutations in the p57 C-terminal domain, L271A and F275A, to the disruption of p57’s binding with PCNA. This interpretation is supported by the observation that mutation of either residue significantly reduced the binding of p57 with PCNA, and mutation of both residues almost completely abolished the binding of p57 to PCNA (Fig. 2). Whether the loss of p57’s ability to arrest the cell cycle when mutated is solely due to the loss of its inhibitory activity on the PCNA-dependent replication must be interpreted cautiously at present. Although the C-terminal domain of p57 can inhibit PCNA-dependent DNA synthesis in vitro (Fig. 3), we have not determined whether these two mutations may potentially affect other biological properties of p57 such as CDK-cyclin binding and cellular localization. In particular, whether loss of PCNA binding activity may affect p57’s stability is an interesting possibility. Both p21 and p27 are short lived proteins with estimated half-lives of 30–60 min and are regulated by ubiquitin-mediated proteolysis (23, 24). Elimination of p27 is promoted by cyclin E-CDK2 phosphorylation of p27 on T187 whose mutation to alanine created a stable p27 protein and caused a G1 arrest resistant to cyclin E overexpression (25). p57 is also a short-lived protein and contains a homologous phosphorylation site, T310, close to the PCNA binding domain that can be phosphorylated by CDK2 (H.W, and Y.X, unpublished data). It is therefore intriguing to speculate that PCNA binding may affect, or be affected by, the T310 phosphorylation that in turn could regulate p57’s degradation by ubiquitin-mediated proteolysis. This postulation is especially relevant in light of the fact that we failed in repeated attempts to detect p57-PCNA complexes by coimmunoprecipitation in mammalian cells despite the presence of readily assembled p57-PCNA complexes. The possibility that PCNA binding may regulate p57’s degradation, together with p57’s lower affinity for PCNA than p21, may provide an explanation for the lack of stable p57-PCNA complexes in mammalian cells.
The localization of the p57 gene to chromosome 11p15.5, its genomic imprinting, and its somatic mutations make it a likely candidate for the tumor suppressor gene postulated to be present at this locus. Supporting this hypothesis is the observation that mice lacking p57 function displayed phenotypes seen in BWS patients (9, 10). The finding that p57 has PCNA-binding activity at its C terminus whose disruption resulted in partial reduction of p57’s ability to suppress cell transformation provides the first evidence that PCNA binding activity has a distinct role in p57’s function. This finding is particularly noteworthy in light of recent reports that three of four p57 mutations identified in BWS patients retained only an intact N-terminal CDK-cyclin binding domain (11, 12): one lost the central PAPA repeat and C-terminal domain, and two contained deletions of the PCNA binding domain (BWS8, and BWS204, Fig. 2A). These findings strongly implicate a C-terminally located function in p57, most likely its PCNA binding ability, whose disruption may directly contribute to BWS development.
Acknowledgments
We thank Drs. Wade Harper for providing baculoviruses expressing full-length p57; Greg Hannon for providing baculoviruses expressing human PCNA; Lishan Su for providing the green fluorescence protein expression plasmid and helping with the fluorescence-activated cell sorter analysis; Henming Ke for providing PCNA protein; and Bob Duronio, Jennifer Michel, and Chris Jenkins for critical reading of the manuscript. H.W. is supported in part by a Showa University School of Medicine Fellowship. Z.-Q.P. is funded by a grant GM55059 from the National instittes of Health. N.S.A. is a Special Fellow of the Leukemia Society of America. R.A.D. is supported by grants from the National Institutes of Health, as well as the Irma T. Hirschl Award. The Albert Einstein Cancer Center Core support is acknowledged. Y.X. is a recipient of American Cancer Society Junior Faculty Award and a Pew Scholar in Biomedical Science. This study was supported by Public Health Service Grant CA-65572 from The National Institute of Health to Y.X.
ABBREVIATIONS
- CDK
cyclin-dependent kinase
- PCNA
proliferating cell nuclear antigen
- BWS
Beckwith–Wiedemann syndrome
- GST
glutathione S-transferase
References
- 1.Hunter T, Pines J. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 2.Sherr C J, Roberts J M. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 3.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 4.Lee M-H, Reynisdottir I, Massague J. Genes Dev. 1995;9:639–649. doi: 10.1101/gad.9.6.639. [DOI] [PubMed] [Google Scholar]
- 5.Matsuoka S, Edwards M C, Bai C, Parker S, Zhang P, Baldini A, Harper J W, Elledge S J. Genes Dev. 1995;9:650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 6.Hastie N D. Annu Rev Genet. 1994;28:523–558. doi: 10.1146/annurev.ge.28.120194.002515. [DOI] [PubMed] [Google Scholar]
- 7.Hatada I, Mukai T. Nat Genet. 1995;11:204–205. doi: 10.1038/ng1095-204. [DOI] [PubMed] [Google Scholar]
- 8.Matsuoka S, Thompson J S, Edwards M C, Barletta J M, Grundy P, Kalikin L M, Harper J W, Elledge S J, Feinberg A. Proc Natl Acad Sci USA. 1996;93:3026–3030. doi: 10.1073/pnas.93.7.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang P, Liegeois N, Wong C, Finegold M, Hou H, Thompson J C, Silverman A, Harper J W, DePinho R A, Elledge S J. Nature (London) 1997;387:151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 10.Yan Y, Frisen J, Lee M-H, Massague J, Barbacid M. Genes Dev. 1997;11:973–983. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- 11.Hatada I, Ohashi H, Fukushima Y, Kaneko Y, Inoue M, Komoto Y, Okada A, Ohishi S, Nabetani A, Morisaki H, et al. Nat Genet. 1996;14:171–173. doi: 10.1038/ng1096-171. [DOI] [PubMed] [Google Scholar]
- 12.Hatada I, Nabetani A, Morisaki H, Xin Z, Ohishi S, Tonoki H, Niikawa N, Inoue M, Komoto Y, Okada A, et al. Hum Genet. 1997;100:681–683. doi: 10.1007/s004390050573. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber-Agus N, Torres R, Horner J, Lau A, Jamrich M, DePinho R A. Mol Cell Biol. 1993;13:2456–2468. doi: 10.1128/mcb.13.4.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong Y, Hannon G, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins C W, Xiong Y. In: Cell Cycle: Material and Methods. Pagano M, editor. New York: Springer; 1995. pp. 250–263. [Google Scholar]
- 16.Mukerjee B, Morgenbesser S D, DePinho R A. Genes Dev. 1992;6:1480–1492. doi: 10.1101/gad.6.8.1480. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Jackson P K, Kirschner M W, Dutta A. Nature (London) 1995;374:386–388. doi: 10.1038/374386a0. [DOI] [PubMed] [Google Scholar]
- 18.Warbrick E, Lane D P, Glover D M, Cox L S. Curr Biol. 1995;5:275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- 19.Luo Y, Hurwitz J, Massague J. Nature (London) 1995;375:159–161. doi: 10.1038/375159a0. [DOI] [PubMed] [Google Scholar]
- 20.Waga S, Hannon G J, Beach D, Stillman B. Nature (London) 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 21.Flores-Rozas H, Kelman Z, Dean F B, Pan Z-Q, Harper J W, Elledge S J, O’Donell M, Hurwitz J. Proc Natl Acad Sci USA. 1994;91:8655–8659. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbs E, Kelman Z, Gulbis J M, O’Donnell M, Kuriyan J, Burger P M J, Hurwitz J. J Biol Chem. 1997;272:2373–2381. doi: 10.1074/jbc.272.4.2373. [DOI] [PubMed] [Google Scholar]
- 23.Pagano M, Tam S W, Theodoras A M, Beer-Romero P, Sal G D, Chau V, Yew P R, Draetta G F, Rolfe M. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 24.Maki C G, Howley P M. Mol Cell Biol. 1997;17:355–363. doi: 10.1128/mcb.17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]