Abstract
Peptide nucleic acids (PNAs) can bind to single-stranded DNA by Watson–Crick base pairing and can form triple helices via Hoogsteen bonding to DNA/PNA duplexes. A single dimeric PNA molecule can form a clamp via both double- and triple-helix formation. We designed PNAs to bind as clamps to a site in the supFG1 mutation reporter gene carried within a chromosomally integrated, recoverable λ phage shuttle vector in mouse fibroblasts. The PNAs were introduced into the cells via permeabilization with streptolysin-O, and cellular uptake was confirmed by fluorescein labeling and fluorescent microscopy. PNAs specific for either an 8- or a 10-bp site in the supFG1 gene were found to induce mutations at frequencies in the range of 0.1%, 10-fold above the background. PNAs with three or four mismatches showed poor in vitro target site binding and were ineffective in the mutagenesis assay. No increased mutagenesis was detected with any of the PNAs in the nontargeted cII gene, also carried within the λ vector, further indicating the specificity of the PNA-induced mutagenesis. DNA sequence analysis revealed that the majority of the mutations were located within the PNA-binding site and consisted mostly of single base pair insertions and deletions within the poly G:C run there, suggesting that a high affinity PNA clamp constitutes a mutagenic lesion that may provoke replication slippage errors. The ability to direct mutations to a target site in chromosomal DNA by using PNAs may provide a useful tool for research and therapeutic applications.
Keywords: triplex, gene targeting, supF, shuttle vector
Reagents that bind DNA in a sequence-specific manner are attractive candidates as tools for genetic manipulation and have the potential to be developed into gene-based therapeutics. Several classes of molecules with DNA binding properties are under study as “anti-gene” agents. Triplex-forming oligonucleotides , comprised of DNA, RNA, or related analogs, bind to DNA in the major groove via Hoogsteen or reverse Hoogsteen bonds and have been shown to inhibit transcription and induce site-specific mutagenesis and recombination in mammalian cells (1–6). Small molecules such as pyrrole-imidazole polyamides have been designed to bind with sequence specificity in the minor groove of DNA and can inhibit transcription factor binding (7, 8).
Another class of DNA binding reagents are peptide nucleic acids (PNAs) (9–11). PNAs are oligonucleotide analogs in which the deoxyribose phosphate backbone is replaced by an uncharged polyamide backbone (9, 10). PNAs can bind to DNA or RNA via Watson–Crick complementarity, with binding affinities significantly higher than those of the corresponding DNA oligomers (11). It has been shown that homopyrimidine PNAs bind to complementary sequences in duplex DNA by strand invasion resulting in the displacement of one DNA strand and formation of a D-loop (12, 13). Dimeric PNAs in which the two PNA strands are connected by a flexible linker can form highly stable PNA/DNA/PNA triplexes at polypurine DNA sequences with extremely high melting temperatures (14). In these molecules, one strand forms Watson–Crick base pairs to the displaced DNA strand in an anti-parallel orientation, and the other strand forms Hoogsteen base pairs to the PNA–DNA duplex in parallel orientation relative to the DNA strand in the duplex (14). As hybrid molecules, PNAs appear to be resistant to both cellular nucleases and proteases (15).
These properties of PNAs are advantageous for use as anti-gene reagents. PNAs have been shown, in vitro, to inhibit transcription initiation and elongation and to block DNA polymerization (16–18). In addition, PNAs can prevent the binding of sequence-specific proteins, such as restriction enzymes, to their target site (19). PNA strand displacement also can create an artificial transcription promoter, at least in vitro (20). So far, however, few studies have reported intracellular effects of PNAs, in part because of the difficulty of cellular PNA delivery. In one approach, translation of the mRNA for the simian virus 40 T antigen was inhibited via microinjection of the PNAs into cells (16). In another study, specific inhibition of transcription of selected target genes was achieved by PNA treatment of human cancer cells mediated by lysolecithin-permeabilization (21).
Here we report experiments showing that PNAs can be introduced effectively into living cells and can cause mutagenesis within a chromosomal target site. Transgenic mouse fibroblasts were generated to contain multiple chromosomally integrated copies of a λ phage shuttle vector carrying the supFG1 mutation reporter gene (2). Previously we had shown that a polypurine tract in this gene is amenable to DNA oligonucleotide-mediated triplex formation in vivo (2, 6). High affinity triplex formation was shown to provoke site-specific mutagenesis via the induction of a repair-dependent pathway (3). To test whether PNA binding might also induce mutagenesis, we synthesized PNAs that can recognize a portion of this sequence and bind tightly to it by forming clamps consisting of PNA/DNA/PNA triplexes. The PNAs were introduced by permeabilization of the cells with streptolysin-O (SLO), and efficient entry was detected by fluorescent and confocal microscopy by using fluorescein-conjugated PNAs. Phage vector rescue and reporter gene analysis revealed the induction of mutations in supFG1 at a frequency 10-fold over background, with the mutations located at or adjacent to the PNA recognition site. These data suggest the potential application of PNAs as anti-gene reagents.
MATERIALS AND METHODS
Cells.
Mouse fibroblast cell line 3340 carrying multiple copies of λsupFG1 shuttle vector DNA was established from a skin biopsy from a newborn 3340 supFG1 mouse (22), as described (23). The cells were maintained in DMEM (Life Technologies, Gaithersburg, MD) supplemented with 20% fetal calf serum (Life Technologies) and with 0.2 mg/ml G418 (Life Technologies).
Oligomers.
The DNA oligonucleotides used in this study were synthesized at the W. M. Keck Biotechnology Resource Center at Yale University, desalted by column separation and filtration, and used without further purification. The PNAs were synthesized at PerSeptive Biosystems (Framingham, MA) and purified by HPLC, as described (14). Their composition is detailed in Table 1.
Table 1.
Sequences of PNAs used in this study
| PNA | Sequence |
|---|---|
| PN-8 | H-JJJ TTJ JT-OOO-TCC TTC CC-Lys |
| PN-10 | H-JJJ JJT TJJ T-OOO-TCC TTC CCC C-Lys-Lys-Lys |
| CP-10 | H-JJT TJT JJJ T-OOO-TCC CTC TTC C-Lys-Lys-Lys |
| RP-10 | H-TTC TTT TCC T-OOO-TJJ TTT TJT T-Lys |
| Fl-PN-10 | Flu-O-JJJ JJT TJJ T-OOO-TCC TTC CCC C-Lys-Lys-Lys |
The PNAs are oriented from the N to the C terminus (represented by H and Lys, respectively) such that when the N-terminal end of the PNA faces the 5′ end of the oligonucleotide the complex is termed parallel. J represents pseudoisocytosine that functions as protonated cytosine analog for Hoogsteen base pairing with guanine. The Os represent 8-amino-3,6-dioxaoctanoic acid units which act as a flexible linker. In RP-10, the J-containing PNA segment is at the C-terminal end. In Fl-PN-10, fluorescein is attached at the N-terminal end of the molecule via an O-linker.
PNA Binding Assays.
PNA binding to the target duplex was measured by using a gel mobility shift assay. Two complementary 57-mers containing the sequence corresponding to base pairs 157–213 of supFG1 were synthesized. The non-PNA-binding strand (lower strand in Fig. 1) was end-labeled by using T4 polynucleotide kinase and [γ-32P]ATP. Duplex DNA was prepared by mixing both 57-mers at a ratio of 1:1 in TE buffer (10 mM Tris/1 mM EDTA, pH 8.0) and incubating the solution at 37°C for 3 h. A fixed concentration of duplex DNA (1 × 10−10 M) was incubated with increasing concentrations of the PNA in 10 μl of a solution containing 10 mM Tris (pH 7.5) at 37°C for 3 h. One-third of the reaction was analyzed by electrophoresis in a 10% native polyacrylamide gel containing 89 mM Tris⋅HCl, 89 mM boric acid, and 20 mM MgCl2 at 5 V/cm for 15 h, followed by autoradiography, as described (24).
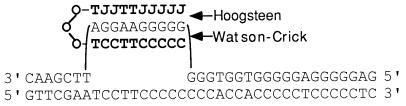
Figure 1.
PNA binding and clamp formation on the polypurine strand within base pairs 167–176 of the supFG1 gene. The binding of the dimeric PNA, PN-10 (Table 1), to the target site is shown, indicating strand displacement, Watson–Crick binding by the C-terminal T- and C-rich segment, and Hoogsteen binding by the N-terminal J- (pseudoisocytosine) and T-containing portion. The linker consisting of 8-amino-3,6-dioxaoctanoic acid units (O) is indicated.
SLO Treatment.
The cells were permeabilized with SLO (Sigma) for delivery of the PNAs into the cells (25). A 500 μl reaction mixture containing permeabilization buffer (137 mM NaCl/100 mM Hepes, pH 7.4/5.6 mM glucose/2.7 mM KCl/2.7 mM EGTA/0.1% BSA), 5 mM DTT, and 400 units/ml of SLO was incubated at room temperature for 15 min. Immediately before treating cells, ATP (to a concentration of 1 mM) and PNA (to a concentration of 1 μM) were added. The mixture was overlaid onto a cell monolayer of ≈2.5 × 105 cells in a 60-mm tissue culture dish, previously washed with PBS. After 5-min incubation, growth medium was added and the cells were allowed to grow for 3–4 days, until confluent. Control cells were treated with SLO in the absence of PNA.
Shuttle Vector Rescue and Analysis.
High molecular weight DNA was prepared from the tissue culture cells as described (26). λ vector rescue from the cellular DNA was carried out by using λ in vitro packaging extracts (22, 27, 28). Packaging extracts were made as described (28), except that a new Escherichia coli lysogen, NM759 [E. coli K12 recA56 Δ(mcrA) e14° Δ(mrr-hsd-mcr) (λimm434 cIts b2 red3 Dam15 Sam7)/λ] was used instead of BHB2690 for the preparation of the sonicate extract. This lysogen produces extracts that are deficient in methyl-directed restriction activity that would otherwise degrade DNA methylated in the mammalian pattern and reduce the yield of rescued phage. The cellular DNA was incubated in the λ in vitro packaging extracts at a concentration of 0.05 μg/μl for 2 h at 37°C. The packaged phage were diluted in 10 mM Tris (pH 8.0) and 5 mM MgCl2, adsorbed to PG901 [E. coli C1a lacZ125 (am)], and plated in 0.6% top agar on Luria–Bertani plates in the presence of 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) (1.6 mg/ml) and isopropyl β-d-thiogalactoside (1.3 mg/ml), as described. Phage with functional supF genes suppress the nonsense mutation in the host bacteria β-galactosidase gene, allowing synthesis of active enzyme capable of metabolizing X-Gal, thereby yielding blue plaques. Phage with inactivating supF mutations produce colorless plaques.
cII Assay.
Cellular DNA was prepared and packaged as above. A fraction (5%) of the packaged phage was plated on E. coli G1217 hfl+, on which all phage form plaques regardless of cII gene function, allowing quantification of the total number of rescued phage. The remainder were plated on E. coli G1250 hfl−, on which only phage with cII mutations will form plaques, as described (29). The number of plaques formed on G1250 represents the number of mutants, which is divided by the total number calculated from the yield on G1217, to give the mutation frequency.
Mutation Sequencing.
Following plaque purification, PCR amplification of the supFG1 gene sequences was carried out by using the GeneAmp kit (Perkin–Elmer/Cetus). Sequence analysis of the PCR products was performed as described (30).
RESULTS
Experimental Strategy.
To create a chromosomal target site to assay for PNA-induced mutagenesis, the modified supF gene, supFG1, containing an extended polypurine tract (2), was cloned into a λ phage vector. The resulting λsupFG1 vector was used to produce transgenic mice carrying multiple copies of the λ vector DNA (22). A mouse fibroblast cell line, 3340, was established from these mice and immortalized by transfection with a vector expressing the simian virus 40 T antigen (22, 23).
Two dimeric PNA oligomers were designed to bind to the polypurine site in the supFG1 gene by forming clamps on the target DNA sequence (Table 1 and Fig. 1), based on the strategy for PNA clamp formation developed by Egholm et al. (14). PN-8 targets the 8-nt polypurine run from position 167 to 174 in the gene whereas PN-10 is directed at the 10-nt run from 167 to 176 (Fig. 1). We synthesized these dimeric PNA molecules to include two distinct PNA segments connected by a flexible linker composed of three 8-amino-3,6-dioxaoctanoic acid (O-linker) units (Fig. 1, Table 1). Because PNAs can strand invade duplex DNA, we expected the distal (C-terminal or 3′) stretch of PNA to strand invade the target site and form Watson–Crick hydrogen bonds to the displaced complementary DNA strand. The proximal (N-terminal or 5′) PNA stretch, connected by O-linkers, could then fold over to form a triplex by Hoogsteen hydrogen bonds consistent with the parallel pyrimidine motif for triple strand formation (Fig. 1). The cytosines in this triplex-forming portion of the PNAs were replaced by pseudoisocytosine (J), which is a substitute for protonated cytosine and forms stable triplex structures at neutral pH (14). Similar PNA/DNA/PNA clamps have been found to be highly stable structures with melting temperatures of over 70°C (14). As controls, we synthesized two PNAs, CP-10 and RP-10, that do not have the exact target site recognition sequence, with three and four mismatches, respectively (Table 1). In the case of RP-10, the orientation of the potential clamp is reversed. The inclusion of multiple lysine residues has been shown to improve PNA strand invasion (31), and so PNAs PN-10 and CP-10 were also synthesized to contain two additional lysines at the C-terminal end.
PNA Binding to supFG1 Sequences.
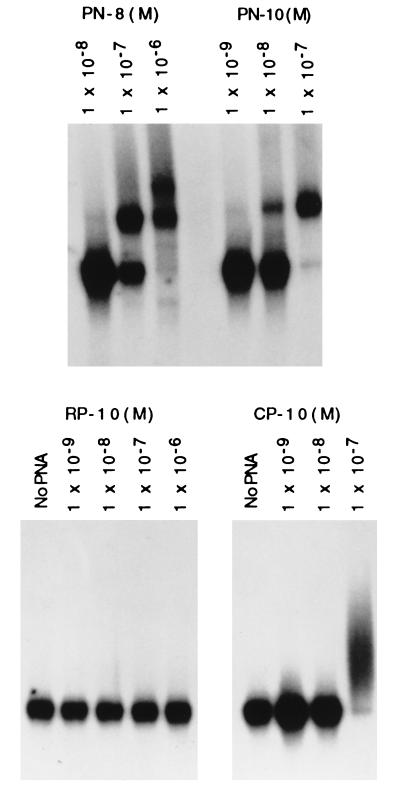
To examine PNA binding to the polypurine site in supFG1, a gel mobility shift assay was used (Fig. 2). Synthetic DNA fragments of 57 bp (matching base pairs 157–213 in the supFG1 gene) were used as duplex targets for PNA binding. Increasing concentrations of each PNA were incubated with fixed concentrations of duplex target DNA in which the nonpurine strand was radioactively labeled. The samples were analyzed for binding by gel mobility shift assay via native PAGE and autoradiography (Fig. 2).
Figure 2.
Binding of PNAs to the supFG1 gene target sequence. A gel mobility shift assay was used to detect binding by the PNAs, PN-8, PN-10, RP-10, and CP-10 (Table 1), to a synthetic fragment containing the polypurine/polypyrimidine site in supFG1. A fixed concentration of the 32P-labeled 57-bp duplex (1 × 10−10 M) was incubated with increasing concentrations of each PNA, as indicated, for 3 h at 37°C. The samples were analyzed by electrophoresis in a 10% polyacrylamide gel, followed by autoradiography. PNA binding generates a complex of reduced mobility relative to the duplex.
As the concentration of each of the specific PNAs, PN-8 and PN-10, was increased, the proportion of the target duplex bound increased, as manifest by the reduced mobility of the PNA–DNA complex (Fig. 2). PN-10 showed somewhat better binding than PN-8, and both were superior to the control PNAs. In the case of PN-8, a second lower mobility species was detected at a PNA concentration of 1 × 10−6 M. This result may represent a quaternary complex in which a second PNA contributes the triplex-forming strand to the DNA:PNA duplex, but this remains to be determined. CP-10 showed little binding except for the smear seen at the 1 × 10−7 M concentration, indicative of a weak binding interaction and in keeping with the 3 mismatches out of 10 to the supFG1 sequence. No binding was detected at all with RP-10. These results should be interpreted only in terms of relative binding of the PNAs to supFG1. Because PNA binding depends on the kinetics of strand invasion, which in turn depends on duplex melting to allow access, these data are not sufficient to generate equilibrium binding affinities.
Cellular Uptake of PNAs.
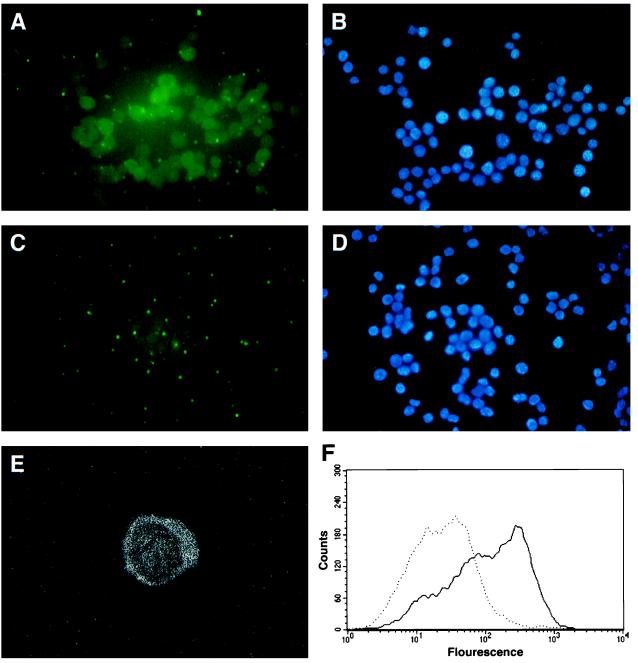
By using the mouse fibroblast cell line 3340, containing the integrated λsupFG1 vector, we carried out experiments to study intracellular chromosome targeting by using the PNA oligomers. The PNAs were introduced into the cells by permeabilization of the cellular membrane following treatment with SLO. To detect PNA internalization into the cells, we used a fluorescein-labeled PNA oligomer Fl-PN-10, and followed its uptake into the cells by both standard fluorescent microscopy and by confocal microscopy (Fig. 3). Within 15 min, ≈50% of the cells were seen to internalize the PNA, showing diffuse cytoplasmic and nuclear fluorescence. Cells not treated with SLO showed no detectable PNA uptake, indicating membrane permeabilization is required for PNA delivery. The percentage of cells taking up the fluorescein-tagged PNA was also measured by flow cytometry. The graph in Fig. 3 shows that ≈50% of the cells display a fluorescent signal significantly above background, indicative of PNA uptake.
Figure 3.
Cellular uptake of PNAs. Cells were permeabilized with SLO in the presence of fluorescein-conjugated PN-10 (Table 1) at a concentration of 1 μM. Fifteen minutes later, the cells were fixed in paraformaldehyde and examined by using a fluorescent microscope at ×40 power (A), with the 4′,6-diamidino-2-phenylindole (DAPI) staining shown in B. Cells treated with PNA but without SLO permeabilization are shown in C, with the associated DAPI stain shown in D. Cells were also examined with a laser scanning confocal imaging system (Bio-Rad MRC600) (E). The confocal images were captured by using a ×63 C-apochromat oil phase lens (Zeiss Axiovert 10 microscope). The percentage of cells taking up the fluorescein-tagged PNA was further quantified by flow cytometry (F). The solid line indicates the fluorescence profile obtained when cells were treated with the fluorescein-tagged PNA following permeabilization with SLO. The dotted line shows the profile of cells treated with the PNA in the absence of permeabilization.
Chromosomal Targeting of the supFG1 Gene.
After treatment of 3340 cells with the sequence-specific and control PNAs, genomic DNA was isolated for shuttle vector rescue and analysis via λ in vitro packaging (22, 28). Mutations in the λsupFG1 gene were identified by the presence of colorless plaques in contrast to the blue plaques produced by phage carrying wild-type supFG1. The specific oligomers that bind to the target site on the supFG1 gene, PN-8 and PN-10, induced mutations in supFG1 at frequencies almost 10-fold above those seen in cells treated with SLO alone (Table 2). The compiled results of four experiments are shown. When the cells were treated with the mismatched PNAs, RP-10, and CP-10, the mutation frequencies observed were similar to the background level (Table 2). These data indicate that the PNAs can be delivered into cells and bind to a chromosomal target site in a sequence-specific manner, resulting in mutations at the site.
Table 2.
PNA-targeted mutagenesis of a chromosomal gene in mouse cells
| Treatment | supFG1 gene, mutants/total | Frequency, ×10−5 | cII gene, mutants/total | Frequency, ×10−5 |
|---|---|---|---|---|
| No oligo | 7/74,093 | 9 | 2/51,868 | 4 |
| PN-8 | 52/62,359 | 83 | ND | ND |
| PN-10 | 62/71,437 | 86 | 4/45,288 | 9 |
| CP-10 | 10/73,185 | 14 | 4/49,846 | 8 |
| RP-10 | 14/67,551 | 21 | ND | ND |
The PNAs were introduced into mouse 3340 cells via SLO-mediated permeabilization. After 3–4 days, the cells were harvested for DNA preparation and rescue of the chromosomal shuttle vector. In each case, the frequency of mutant phage carrying mutations in the supFG1 and cII reporter genes was calculated. ND, not determined.
cII Gene Mutagenesis.
To assay for nonspecific PNA effects, potentially resulting in mutations at sites other than the target site, we looked for mutations within another gene in the λ vector, the cII gene, in cells treated with the PNAs (Table 2). The level of the cII gene product controls whether the rescued phage lyses or lysogenizes the bacterial host cell after infection. Mutations in the cII gene can be detected by plating the packaged phage on an hfl− E. coli strain; only the cII mutant phage will form plaques (29). As shown (Table 2), no mutations above background were induced in the cII gene by any of the PNAs, indicating that the PNA-induced mutagenesis is sequence-dependent and is not caused by a generalized disruption of cellular genomic integrity.
Sequence Analysis of supFG1 Mutations.
The DNA sequences of 28 mutations induced in the supFG1 gene by PN-8 and PN-10 are presented in Fig. 4. These mutations were generated in four independent experiments. All the mutations were point mutations, with 79% (22/28) located within the stretch of eight G:C base pairs from position 172 to 179. This sequence overlaps the PN-8 and PN-10 binding sites. Of the mutations in this region, 4 were single base deletions and 12 were single base insertions within the G run, indicative of replication slippage events. The six G:C to T:A transversions at base pair 177 may also represent slippage events. They could have arisen in an event in which a slipped helix is realigned so that the A or T in bp 180 is used as a template to extend the opposite strand at position 177. Such a conformation could theoretically be stabilized by the large number of consecutive G:C base pairs and by the possibility of the A or T from the A:T base pair at 183 binding to the complementary base at position 180.
Figure 4.
Sequences of supFG1 mutations induced by PNA treatment of mouse fibroblasts. Mutations induced by both PN-10 and PN-8 compiled from four separate experiments are shown. Three classes of mutations were observed, including single base substitutions, single base pair deletions and insertions, and multiple, simultaneous point mutations. The base substitutions are listed above the corresponding supFG1 gene sequence and represent changes with respect to the upper strand. Single base pair deletions and insertions are indicated by the “−” and “+” signs, respectively, above the affected site. The multiple point mutations are indicated by the underlining. Within the supFG1 sequence, the PNA binding site is underlined.
DISCUSSION
The results presented here demonstrate the potential use of peptide nucleic acids to target chromosomal DNA within mammalian cells and generate mutations at the target site. By using dimeric PNAs that can strand invade duplex DNA and form three-stranded PNA/DNA/PNA clamps, we observed up to a 10-fold induction of mutagenesis at the targeted chromosomal site in mouse cells. That these PNAs recognize and bind to the target site in a sequence-dependent manner was demonstrated in gel mobility shift assays (Fig. 2). Control PNAs with recognition sequence mismatches bound poorly to the duplex target and were ineffective at mutation induction.
In previous work, we have shown that DNA oligonucleotides capable of high affinity triplex formation can induce mutations in the supFG1 reporter gene when carried in an extrachromosomal simian virus 40-based vector in monkey and human cells (3). This effect was found to be repair-dependent, and we have hypothesized that the triplex constitutes a DNA lesion that is mutagenic at a low level. We have recently found that triplex-forming DNA oligomers can also target mutations to the supFG1 gene in the 3340 mouse cells used here (unpublished data). The results presented here suggest that the PNA clamp also represents an alteration of DNA structure that disrupts normal DNA metabolism and can lead to mutation.
Evidence exists that PNA binding can block restriction enzyme activity (19), and so it is conceivable that a PNA clamp could also constitute a partial block to DNA repair. In this regard, we had previously shown that a DNA triplex can inhibit repair endonuclease activity (32). In addition, the PNA clamp may constitute a hindrance to the progress of the DNA polymerase during replication. Inhibition or stalling of a DNA polymerase, either in the context of replication or during DNA repair synthesis, may promote template dislocation and strand slippage events (33). Such events could account for the mutation pattern observed, which was characterized by a predominance of single base insertions and deletions within the G:C base pair run overlapping the PNA binding site. In this process, the poly G:C target site may be important for two reasons. One, a polypurine/polypyrimidine site is needed for PNA clamp formation, because one segment of the clamp binds to the DNA via Hoogsteen bond formation, a process which is highly favored at polypurine sites. Although PNAs can bind to single-stranded DNA at any sequence, the triplex portion of the clamp has the same requirement for polypurine DNA as all DNA triplexes. Second, the poly G run may be especially prone to strand slippage by facilitating stabilization of the dislocated and realigned helix (33, 34). Hence, we might expect that PNAs would be less effective in targeting mutations to sequences lacking a homopurine repeat motif.
In an alternative model, the PNA clamp may actually undergo repair by the nucleotide excision repair pathway (35), so that a patch of DNA from the strand containing the PNA clamp is removed. This is conceivable, because the typical patch size for excision repair in mammalian cells is 28–30 nt (36), which would easily encompass the 10-nt PNA binding site. This would also be consistent with our previous observation that a DNA triple helix formed at a 10-bp site is repaired, whereas a longer DNA triplex covering a 30-bp site is resistant to repair (32). With the PNA clamp removed, the resulting gap would be filled in as usual for this repair pathway. However, the presence of the mononucleotide repeat sequence within the template for the repair synthesis might promote slippage errors during gap fill-in, producing the observed pattern of mutations. Hence, in this model, too, the mutagenesis would be enhanced by the G:C base pair repeat sequence within the PNA binding site. The results presented here do not distinguish between these models, and so determining the mechanism of the PNA-induced mutagenesis will require further investigation.
The uptake of oligonucleotides and related molecules into cells is of great importance for anti-sense and anti-gene approaches. Previous studies have shown that the oligonucleotides are taken up passively by cells to varying extents, but much of the reagent is sequestered in endosomes (37). Cellular delivery can be enhanced by using various liposome formulations (38). However, because PNAs have an uncharged backbone, these lipids cannot be used for delivering PNAs. We therefore used SLO to permeabilize the cells for PNA uptake (25). Fluorescein-conjugated PNAs were observed to be taken up by the cells in both the nuclear and cytoplasmic spaces following SLO treatment as evidenced by standard fluorescent and confocal microscopy. However, a significant portion of the PNAs appeared to be distributed in the cytoplasm, and so only a portion of the reagent was internalized into the nucleus (Fig. 3). With alternative methods, it may be possible to improve nuclear delivery and to further enhance the anti-gene effects of PNAs within mammalian cells. Also, advances in PNA chemistry, such as the generation of DNA–PNA chimeric oligomers, may help to overcome the present shortcomings of PNAs in this regard.
Because PNAs bind to duplex DNA via strand invasion, which requires some opening of the duplex, moderate salt concentrations, as exist within cells, can inhibit PNA binding to duplex DNA. As a result, concerns exist regarding the ability of PNAs to carry out strand invasion under physiologic conditions. However, specific inhibition of transcription in mammalian cells has been achieved with PNAs (21, 39), and strand invasion in intact chromatin has been demonstrated (40). In addition, the development of dimeric PNAs, which can form clamps, as used here, was found to enhance the strand invasion capabilities of these reagents (14, 31). Other work has shown that DNA topology can influence PNA strand invasion into duplex DNA, with negative supercoiling favoring PNA binding, even at physiologic salt concentrations (41). Hence, PNA strand invasion may be more efficient inside cells than expected because DNA in vivo is predominantly negatively supercoiled (41). In this regard, transcriptional activity is associated with negative supercoiling, and this may further promote PNA accessibility (41). In vitro transcription studies on a linear template have shown that PNA binding correlates with transcriptional activity, suggesting that the transcription bubble may also facilitate PNA invasion (42).
Our results suggest that a chromosomal target is accessible to PNA binding, at least under certain conditions. It may be, however, that the activity of PNAs is highest in periods when the DNA helix is transiently open, as is the case in transcription or replication. In our vector construct, the supFG1 gene does not have a mammalian promoter, but immediately downstream of the gene is the simian virus 40 promoter driving the expression of the neo gene. The cells were grown in the presence of the neomycin analog, G418, to select for cells with transcriptional activity at this site. In experiments with DNA triplex-forming oligonucleotides, we found that this adjacent transcriptional activity was necessary to enable triplex-forming oligonucleotide targeting to the supFG1 gene (unpublished data). In addition, other strategies, such as synchronizing the cells to be in S-phase or inducing transcription of the target gene, might further facilitate strand invasion and enhance PNA-induced mutagenesis, but these approaches remain to be tested. In any case, the data presented here show that PNA clamps can be used to induce site-specific mutagenesis that can knock out a chromosomal gene in mammalian cells. This capability may prove useful as a tool in research applications and may eventually lead to the use of PNAs in gene or anti-viral therapy.
Acknowledgments
We thank G. Wang, D. Brown, M. Seidman, L. Narayanan, L. Lacroix, R. Franklin, S. J. Baserga, and L. Cabral for their help. We also thank R. Carbone of the Yale Cancer Center Flow Cytometry Shared Resource (CA16359). This work was supported by the Leukemia Society of America and the National Institutes of Health (CA64186).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: PNA, peptide nucleic acid; SLO, streptolysin-O.
References
- 1.Helene C. Anti-Cancer Drug Design. 1991;6:569–584. [PubMed] [Google Scholar]
- 2.Wang G, Levy D D, Seidman M M, Glazer P M. Mol Cell Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang G, Seidman M M, Glazer P M. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 4.Postel E H, Flint S J, Kessler D J, Hogan M E. Proc Natl Acad Sci USA. 1991;88:8227–8231. doi: 10.1073/pnas.88.18.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maher L J d, Wold B, Dervan P B. Science. 1989;245:725–730. doi: 10.1126/science.2549631. [DOI] [PubMed] [Google Scholar]
- 6.Faruqi A F, Seidman M M, Segal D J, Carroll D, Glazer P M. Mol Cell Biol. 1996;16:6820–6828. doi: 10.1128/mcb.16.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottesfeld J M, Neely L, Trauger J W, Baird E E, Dervan P B. Nature (London) 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 8.Geierstanger B H, Mrksich M, Dervan P B, Wemmer D E. Science. 1994;266:646–650. doi: 10.1126/science.7939719. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen P E, Egholm M, Berg R H, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 10.Egholm M, Buchardt O, Nielsen P E, Berg R H. J Am Chem Soc. 1992;114:1895–1897. [Google Scholar]
- 11.Egholm M, Buchardt O, Christensen L, Behrens C, Freier S M, et al. Nature (London) 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 12.Peffer N J, Hanvey J C, Bisi J E, Thomson S A, Hassman C F, Noble S A, Babiss L E. Proc Natl Acad Sci USA. 1993;90:10648–10652. doi: 10.1073/pnas.90.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wittung P, Nielsen P, Norden B. J Am Chem Soc. 1996;118:7049–7054. [Google Scholar]
- 14.Egholm M, Christensen L, Dueholm K L, Buchardt O, Coull J, Nielsen P E. Nucleic Acids Res. 1995;23:217–222. doi: 10.1093/nar/23.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demidov V V, Potaman V N, Frank-Kamenetskii M D, Egholm M, Buchardt O, Sonnichsen S H, Nielsen P E. Biochem Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 16.Hanvey J C, Peffer N J, Bisi J E, Thomson S A, Cadilla R, et al. Science. 1992;258:1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- 17.Koppelhus U, Zachar V, Nielsen P E, Liu X, Eugen-Olsen J, Ebbesen P. Nucleic Acids Res. 1997;25:2167–2173. doi: 10.1093/nar/25.11.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Praseuth D, Grigoriev M, Guieysse A L, Pritchard L L, Harelbellan A, Nielsen P E, Helene C. Biochim Biophys Acta. 1996;1309:226–238. doi: 10.1016/s0167-4781(96)00146-7. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen P E, Egholm M, Berg R H, Buchardt O. Nucleic Acids Res. 1993;21:197–200. doi: 10.1093/nar/21.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mollegaard N E, Buchardt O, Egholm M, Nielsen P E. Proc Natl Acad Sci USA. 1994;91:3892–3895. doi: 10.1073/pnas.91.9.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boffa L C, Morris P L, Carpaneto E M, Louissaint M, Allfrey V G. J Biol Chem. 1996;271:13228–13233. doi: 10.1074/jbc.271.22.13228. [DOI] [PubMed] [Google Scholar]
- 22.Narayanan L, Fritzell J A, Baker S M, Liskay R M, Glazer P M. Proc Natl Acad Sci USA. 1997;94:3122–3127. doi: 10.1073/pnas.94.7.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leach E G, Gunther E J, Yeasky T M, Gibson L H, Yang-Feng T L, Glazer P M. Mutagenesis. 1996;11:49–56. doi: 10.1093/mutage/11.1.49. [DOI] [PubMed] [Google Scholar]
- 24.Faruqi A F, Krawczyk S H, Matteucci M D, Glazer P M. Nucleic Acids Res. 1997;25:633–640. doi: 10.1093/nar/25.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barry E L R, Gesek F A, Friedman P A. BioTechniques. 1993;15:1018–1020. [PubMed] [Google Scholar]
- 26.Gunther E J, Yeasky T M, Gasparro F P, Glazer P M. Cancer Res. 1995;55:1283–8. [PubMed] [Google Scholar]
- 27.Gunther E J, Murray N E, Glazer P M. Nucleic Acids Res. 1993;21:3903–3904. doi: 10.1093/nar/21.16.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glazer P M, Sarkar S N, Summers W C. Proc Natl Acad Sci USA. 1986;83:1041–1044. doi: 10.1073/pnas.83.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakubczak J L, Merlino G, French J E, Muller W J, Paul B, Adhya S, Garges S. Proc Natl Acad Sci USA. 1996;93:9073–9078. doi: 10.1073/pnas.93.17.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Havre P A, Gunther E J, Gasparro F P, Glazer P M. Proc Natl Acad Sci USA. 1993;90:7879–7883. doi: 10.1073/pnas.90.16.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffith M C, Risen L M, Greig M J, Lesnik E A, Sprankle K G, Griffey R H, Kiely J S, Freier S M. J Am Chem Soc. 1995;117:831–832. [Google Scholar]
- 32.Wang G, Glazer P M. J Biol Chem. 1995;270:22595–22601. doi: 10.1074/jbc.270.38.22595. [DOI] [PubMed] [Google Scholar]
- 33.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzah E, Inoye M. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Kunkel T A, Soni A. J Biol Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- 35.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 36.Huang J C, Svoboda D L, Reardon J T, Sancar A. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gewirtz A M, Stein C A, Glazer P M. Proc Natl Acad Sci USA. 1996;93:3161–3163. doi: 10.1073/pnas.93.8.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis J G, Lin K-Y, Kothavale A, Flanagan W M, Matteucci M D, DePrince R B, Mook R A, Jr, Hendren R W, Wagner R W. Proc Natl Acad Sci USA. 1996;93:3176–3181. doi: 10.1073/pnas.93.8.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boffa L C, Carpaneto E M, Mariani M R, Louissaint M, Allfrey V G. Oncol Res. 1997;9:41–51. [PubMed] [Google Scholar]
- 40.Boffa L C, Carpaneto E M, Allfrey V G. Proc Natl Acad Sci USA. 1995;92:1901–1905. doi: 10.1073/pnas.92.6.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bentin T, Nielsen P E. Biochemistry. 1996;35:8863–8869. doi: 10.1021/bi960436k. [DOI] [PubMed] [Google Scholar]
- 42.Larsen H J, Nielsen P E. Nucleic Acids Res. 1996;24:458–463. doi: 10.1093/nar/24.3.458. [DOI] [PMC free article] [PubMed] [Google Scholar]