Abstract
Cationic lipids are being widely used for cell transfection in vitro. The lipid/DNA complexes, however, tend to aggregate into large and polydisperse particle mixtures; this hampers their use in vivo. Cationic detergents, on the contrary, do not mediate cell transfection per se, yet are capable of condensing individual DNA molecules into discrete entities. We have taken (only) the interesting features of both types of amphiphiles for the two-step formation of stable core particles reminiscent of viruses. Individual anionic plasmid molecules were cooperatively collapsed with a carefully tailored cationic cysteine-based detergent. The resulting 23-nm particles were then simply “frozen” by spontaneous aerobic dimerization of the cysteine-detergent into a cystine-lipid on the template DNA. The population of spherical particles is monodisperse and stable over days, in physiological conditions. Together with a negative surface potential, these properties should ensure good tissue dissemination and escape from the blood stream after i.v. injection.
Recombinant DNA viruses contain a single genome copy that can efficiently carry a foreign gene into eukaryotic cells in vivo (1). In contrast, DNA counterion-condensation by cationic polymers (2) or cationic lipids (3) used for gene transfer generally leads to large multimolecular and polydisperse aggregates. These complexes are efficient at transfecting cells in culture, yet are very poor at diffusing within a tissue or escaping from blood vessels.
Cationic detergents too have been shown to condense DNA into discrete particles (4) that, interestingly, can consist of a single nucleic acid molecule (5). However detergents have a much higher water solubility than lipids and, upon addition to cells, their fast release into the environment induces DNA decondensation and detergent-related cytotoxicity. As a consequence, this class of amphiphiles is unable to transfect cells per se (6).
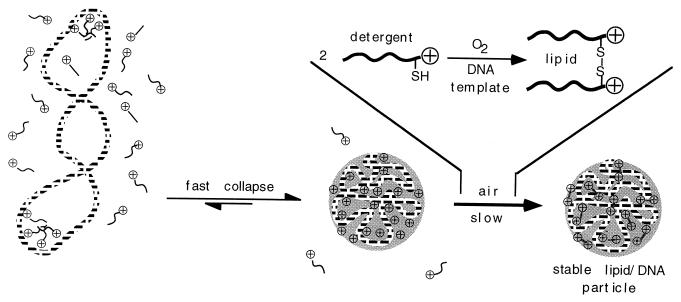
Using the basics of supramolecular chemistry (7, 8), we have thought taking the essential features of both types of surfactants toward the formation of stable core particles reminiscent of viruses (Fig. 1). First, anionic DNA molecules were individually condensed by addition of an appropriately designed cationic detergent. The resulting particles were then “frozen” by chemically converting the detergent into a lipid on the template DNA, via simple air-induced dimerization.
Figure 1.
Schematic representation of cationic detergent-induced monomolecular collapse of plasmid DNA. Particles are subsequently stabilized by detergent dimerization into a lipid.
EXPERIMENTAL PROCEDURES
Reagents.
Chemicals were purchased from Fluka or Aldrich, Janssen, and Lancaster Synthesis. pCMV-luc was propagated and purified as described (9).
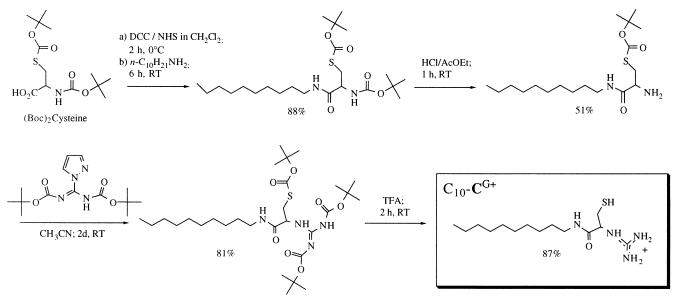
Synthesis of Guanidinocysteine N-Decylamide (C10-CG+).
N,S-bis-ter-butoxycarbonyl-l-cysteine (10) was activated as the N-hydroxysuccinimide ester (1.2 equ. N-hydroxysuccinimide plus 1.1 equ. dicyclohexylcarbodiimide for 2 h at 0°C in CH2Cl2/tetrahydrofuran) and treated with decylamine (1.1 equ.) for 6 h. The cysteinamide was obtained in 88% yield after silica gel column chromatography. This compound was hemi-deprotected in 2 N HCl/AcOEt (6 equ. HCl) for 1 h, yielding the free amine (51% after silica gel chromatography) and coupled with 1-H-pyrazole-1-(N, N′-bis(tert-butoxycarbonyl))carboxamidine (11) (0.9 equ.) for 2 days (81% yield after silica gel chromatography). Mass spectrum of the Tris-ter-butoxycarbonyl-(C10-CG+): MS (FAB+), m/z: 603.3 (60%, [MH+]), 547.2 (9%, [MH+ - t-butyl]), 503.3 (14%, [MH+ - Boc]), 447.2 (13%, [MH+ - t-butyl - Boc]), 403.2 (4%, [MH+ - 2Boc]), 347.1 (100%, [MH+ - t-butyl - 2Boc]), 303.2 (10%, [MH+ - 3Boc]). The stable protected compound was kept as a solid at −20°C.
Final deprotection of an aliquot was performed in neat trifluoroacetic acid (2 h, 87% yield as judged by free thiol titration). Three hundred megaHertz 1H NMR, δ(CD3CD2OD): 0.9 [t, (CH3)]; 1.25–1.4 [m, (CH2)7]; 1.5–1.6 (m, CH2-CNCO); 2.9–3.03 (m, CH2-S); 3.15–3.35 (m, CH2-NCO); 4.35–4.4 (m, CH-N+). This 150 mM stock solution was kept under argon at −80°C and checked by NMR for degradation and/or oxidation prior to each series of measurements.
Determination of Critical Micelle Concentrations (c.m.c.).
A Langmuir balance (KSV2200 Surface Barostat, KSV Chemicals) was used to measure the surface tension of a solution of increasing detergent concentration in 50 ml Mes buffer 20 mM, pH 6, containing 10 mM DTT to avoid detergent oxidation. A plot of the surface tension (mN/m) vs. the logarithm of the concentration showed a sharp break toward a slope close to zero. The corresponding concentration was taken as the c.m.c.
Kinetics of the Detergent Oxidation.
The stock buffer solution (5 ml of 15 mM Tris⋅HCl, pH 8.4) was saturated with oxygen by three vacuum/oxygen atmosphere cycles. C10-CG+ was injected from a concentrated stock solution (6 mM in ethanol) to a final concentration of 90 μM. For each time point, a 0.5-ml aliquot was removed and mixed with 0.5 ml 2× Ellman’s reagent. The remaining free thiol was quantitated spectrophotometrically according to Riddles et al. (12). For the template experiment, pCMV-luc plasmid DNA was added to a final concentration of 60 μM phosphate before addition of the detergent.
Light Scattering Measurements of Particle Size and Zeta Potential.
The sample solution was prepared by adding the desired amount of detergent from a 6 mM stock solution to 10 μg plasmid DNA contained in 1 ml of Tris⋅HCl buffer 15 mM, pH 8.4. After 24 h, the solution was concentrated twice with a Speedvac and particle size was determined by light scattering using a Zetamaster 3000 (Malvern Instruments, Paris) with the following specifications: sampling time, 90 s; three to nine measurements per sample; medium viscosity, 1.054 cP; medium refractive index, 1.34; particle refractive index, 1.45 (typical liposome RI); scattering angle, 90°; temperature, 20°C. Zeta potentials were measured with the following specifications: sampling time, 30 s; three measurements per sample; medium viscosity, 1.054 cP; medium dielectric constant, 80; temperature, 20°C, beam mode F(Ka) = 1.50 (Smoluchowsky).
Transmission Electron Microscopy.
The sample solution was prepared by adding the detergent (40 μM final concentration) to a 30 μM phosphate pCMV-luc DNA solution in Hepes (15 mM, pH 7.4) or Tris⋅HCl (15 mM, pH 8.4) buffer and leaving the solution in aerobic condition overnight. Carbon films were prepared by sublimation on freshly cleaved mica and recovered by flotation on Cu/Rh grids (300 mesh, Touzard & Matignon, Courtaboeuf, France). After drying overnight, grids were kept on blotting paper in a Petri dish. Immediately before sample addition, grids were glow-discharged (110 mV, 25 s). A drop (5 μl) of sample solution was left on the grid for 1 min. Complexes were negatively stained with 30 μl aqueous uranylacetate (1% wt/wt) for 20 s, and excess liquid was removed with blotting paper. Observations were performed at 80 kV with a Philips EM 410 transmission electron microscope. Similar results were obtained with complexes observed in 0.15 M NaCl.
Cell Transfection.
BNL CL.2 murine hepatocytes were cultured and plated as described (9). Complexes were formed by adding at once the desired amount of C10-CG+ from the concentrated stock solution (6 mM in ethanol) to the plasmid solution (60 μM phosphate) in oxygen-saturated Tris⋅HCl 15 mM, pH 8.4. The solution was left under oxygen for 24 h. Complexes (100 μl solution corresponding to 2 μg plasmid per well) were added to the cells maintained in serum-free medium. Fetal calf serum was added to a final concentration of 1% after 3 h, then to 10% (4 h). After 6 h incubation, the solution was replaced by fresh 10% serum-containing cell culture medium. Polyethylenimine (25 kDa) was used as a positive control.
Luciferase gene expression and protein content were determined after 24 h using conventional procedures and commercial kits (9).
RESULTS AND DISCUSSION
As described in the introduction, single DNA molecule condensation into particles withstanding physiological conditions was based on the conversion of a cationic detergent into a cationic lipid according to Fig. 1. The detergent molecule was designed de novo. A preference for mild chemistry and for the use of endogeneous-like compounds led us to thiol as a dimerizable function, hence to cysteine as the nontoxic recipient backbone for constructing the polyfunctional molecule. Once having carried DNA inside the endosomal compartment of cells, lipophilic cystine ester or amide molecules could, in principle, be enzymatically converted back to cysteine plus an aliphatic alcohol or amine with concomitant DNA decondensation and possible endosome lysis.
Clean monomolecular DNA collapse was expected to occur only for detergent concentrations below the c.m.c. Indeed, micelles could provide a link for detergent/DNA complexes to merge into larger structures. A series of n-alkyl esters of cysteine were therefore synthesized by DMAP-catalyzed dicyclohexylcarbodiimide dehydration. To assess the optimal length of the hydrocarbon chain, their c.m.c.s were determined with a Langmuir balance, giving the following results (n-alkyl chain length/c.m.c.): dodecane/15 μM; undecane/80 μM; decane/320 μM; octane/> 2,000 μM. Typical working plasmid concentrations for gene delivery experiments being in the 50-μM range, the decane chain seemed a safe choice. Unfortunately, the weakly basic nature of the cysteine ester primary amine function (pKa = 6.8) precluded simultaneous DNA binding (which requires a protonated detergent) and reasonable thiol oxidation rates in slightly alkaline medium. The amine function was therefore converted into a strongly basic guanidine (pKa ≈ 13) that is known to interact strongly with DNA (C10-CG+, Fig. 2).
Figure 2.
The four-step synthesis of the dimerizable cationic amphiphile guanidinocysteine N-decylamide C10-CG+.
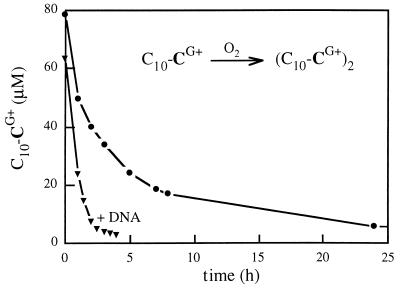
Oxidative detergent dimerization was expected to be prone to a template effect of DNA (in other words, the local concentration of thiol groups in the particles may be orders of magnitude higher than in solution, Fig. 1). Fast intra-complex reaction is a key aspect to formation of small and stable particles since extensive detergent dimerization into lipid prior to DNA binding would obviously have brought our system back to the classical cationic lipid/DNA complex formation/aggregation process. Enhanced oxidation rates were indeed observed in the presence of DNA, whether in an oxygen atmosphere (Fig. 3) or in air or with added FeIII catalyst (not shown).
Figure 3.
Oxidation of the cysteine detergent C10-CG+ into a cystine lipid (C10-CG+)2 occurs faster in the presence of template DNA.
Below the c.m.c., the major driving force for complex formation is of electrostatic nature. Anionic DNA binds isolated cationic amphiphile molecules that tend to sequester due to their higher local concentration and to the hydrophobic effect (Fig. 1). Therefore the cooperative DNA collapse that follows leads to particles that cannot be cationic even with some excess detergent remaining in solution. As a preliminary evidence for complex formation, plasmid DNA (60 μM phosphate in 15 mM Tris⋅HCl, pH 8.4) was mixed with C10-CG+ (to 90 μM, i.e., a cation/anion charge ratio of 1.5) and left in aerobic conditions for 24 h. After the instantaneous complex formation step, the typical UV absorption spectrum of DNA centered at 260 nm was increased by 15%; no further change (once corrected for disulfide absorption) and no turbidity was observed during oxidation. The spectrum remained time-independent over a period of 24 h. The most stringent test of stability, however, was that performed in physiologically relevant ionic concentrations, where cationic lipid/DNA complexes are known to aggregate (3, 13). Remarkably, the UV spectrum of (C10-CG+)2/DNA complexes remained unchanged after 3 days in 150 mM NaCl. Spectroscopy thus gave indirect evidence for the formation of small and stable amphiphile/DNA complexes.
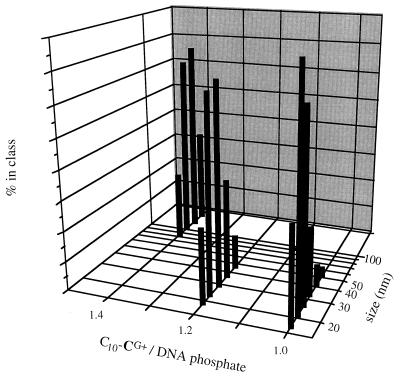
The size and surface charge of the particles were measured by laser light scattering. The detergent/plasmid particles did not diffuse sufficient light intensity for accurate measurements to be made. After 24 h oxidation into lipid/DNA complexes at equimolar ionic charge ratio, the particle mean size could be estimated in the 20–30 nm range (Fig. 4). Increasing the initial detergent concentration led to larger particles. The zeta potential of particles formed with initial cationic detergent/DNA phosphate ionic ratio of 1 to 1.5 was found surprisingly negative (−45 to −40 mV). This can be rationalized taking into account that hydrophobic side chain-mediated detergent aggregation cannot occur at the particle surface, hence DNA must be constituting the interface between the particle and the aqueous medium. For a small particle, the amount of exposed nucleic acid can be considerable: a straightforward calculation shows that up to 60% of the nucleic acid is contained in the 3.4 nm-thick (i.e., the double helix diameter) shell of a 25-nm particle.
Figure 4.
Dynamic laser light scattering measurement of the size of (C10-CG+)2/DNA complexes obtained from various initial C10-CG+ detergent/plasmid DNA ratio.
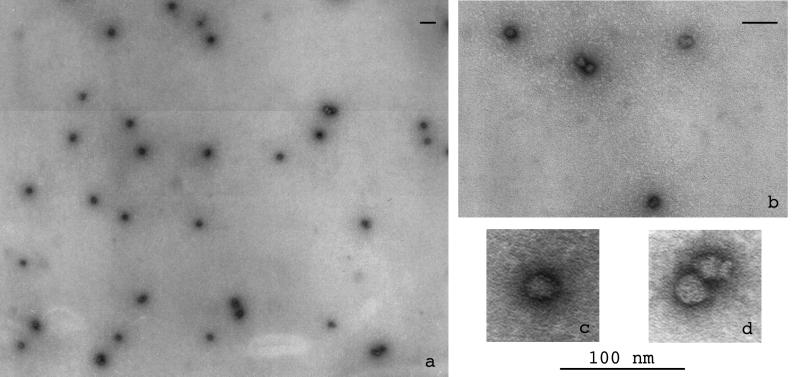
Transmission electron microscopy of an unfiltered solution of the stable oxidized complexes showed a unexpectedly homogeneous population of nearly spherical objects (71% monomers, n = 109), with a few larger structures (Fig. 5a). Moreover, the larger structures were clearly side-by-side spheres giving dimers (Fig. 5 b and d) or oligomers that may have arisen incidentally and that showed no tendency to merge into more intimately fused aggregates as cationic lipids do (3). At the highest magnification (Fig. 5c) the particle size was measured to be 23 ± 4 nm. A rough calculation using typical DNA and amphiphile molecular dimensions led to a sphere of 28 nm diameter when adding the volumes of a 5.5-kb plasmid to that of 5,500-lipid molecules. Transmission electron microscopy thus provides strong evidence for monomolecular DNA collapse. The topology of DNA should not influence the particles morphology. Yet, their size varying as the cubic root of the DNA size, interesting extrapolations can be made. For instance, a particle of the size of an adenovirus would carry ≈400 kb DNA; similarly, along possible gene therapy applications, condensation of a mammalian artificial chromosome would lead to a particle still smaller than 200 nm.
Figure 5.
Transmission electron microscopy of an unfiltered solution of (C10-CG+)2 cationic lipid/DNA particles showing a very homogeneous population (a) of spheres (b) with a diameter of ≈23 nm (c, d). Bar = 100 nm. Similar results were obtained with complexes observed in 0.15 M NaCl.
Raising stepwise the detergent concentrations above the c.m.c. of C10-CG+ progressively led to less clearcut UV, light scattering, and transmission electron microscopy results due to slow aggregation (see Fig. 4), in agreement with the hypotheses underlying the concept.
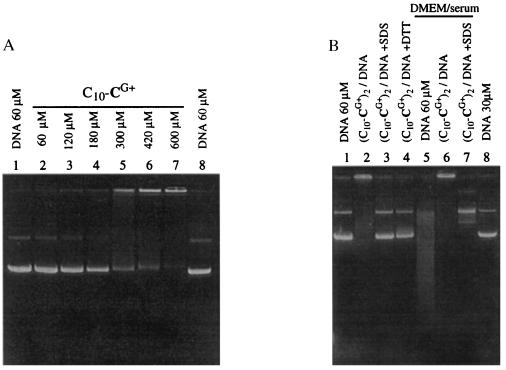
In a last series of experiments, the reversibility of complex formation as well as the stability of complexed DNA toward degradation were assessed by electrophoresis. C10-CG+ was unable to retard DNA migration (Fig. 6A), except for concentrations >300 μM, well above the c.m.c., where presumably precipitation occurred. In contrast, the oxidized 60 μM (C10-CG+)2/DNA particles did not migrate and were stable during electrophoresis (Fig. 6B, lane 2). The cationic lipid/DNA particles could either be destroyed with excess SDS detergent (SDS, lane 3) or by converting the lipid back to the original C10-CG+ detergent with excess reducing DTT (lane 4). Serum nucleases present in cell culture medium rapidly degrade supercoiled plasmid DNA (controls, Fig. 6B, lanes 1 and 8) into a smear of fragments with many nicks (lane 5). The lipid/DNA particles, however, were physically stable in this medium (lane 6) and protected the nucleic acid from degradation, as the plasmid was essentially converted into a slower migrating circular relaxed form (revealed after SDS treatment, lane 7). Protection was not complete, a finding that may be related to the highly negative zeta potential of the particles (see above), which both are indicative of surface-exposed DNA.
Figure 6.
Physical and chemical stability of the (C10-CG+)2/DNA complexes as revealed by agarose gel electrophoresis. (A) Twenty microliter samples containing 0.4 μg pCMV-luc plasmid DNA in Hepes buffer (15 mM, pH 7.4) and concentrations of C10-CG+ detergent as indicated were loaded onto the gel after 5 min and run. (B) The 20-μl samples contained 0.4 μg (lanes 1–4) or 0.2 μg (lanes 5–8) pCMV-luc plasmid DNA. Complexes were made by adding a 100-fold concentrated C10-CG+ detergent solution (90 μM final concentration) to a solution of plasmid (60 μM) in Hepes buffer (15 mM, pH 7.4). The solution was gently mixed once and left to oxidize for 24 h. Lane 1, 60 μM plasmid DNA. Lane 2, 60 μM oxidized (C10-CG+)2/DNA complexes. Lane 3, as lane 2 in 3 mM final SDS before loading. Lane 4, as lane 2 in 6 mM DTT before loading. Lane 5, 60 μM plasmid incubated for 1 h at 37°C with an equivalent volume of DMEM cell culture medium containing 10% serum. Lane 6, 60 μM oxidized (C10-CG+)2/DNA complexes incubated as in lane 5. Lane 7, as lane 6 in 3 mM final SDS before loading. Lane 8, 30 μM pCMV-luc plasmid DNA. The 1% agarose gel was run for 90 min at 8 V/cm in 40 mM Tris-acetate buffer, pH 8.
Being devoid of excess cationic surface charge, these particles are not expected to bind efficiently to cell-surface polyanions (3, 14). As a consequence they should not transfect cells. Preliminary experiments performed with a luciferase reporter gene and BNL CL.2 murine hepatocytes indeed confirmed this view. However larger cationic particles formed with a 3-fold excess detergent led to a moderate reporter gene expression (3.106 luciferase light units/mg total protein), showing that, once in the cell, the fate of the complexes followed that of a typical cationic vector.
PERSPECTIVES
The most promising future for these small negatively charged DNA-containing particles will be in vivo gene delivery. They should not raise the problems inherent to cationic lipid-mediated delivery, such as binding to circulating and extracellular matrix anionic proteoglycans (3) or complement activation (15). To penetrate into cells, however, our particles will have to be equipped with cell-surface targeting residues (16) able to trigger their endocytosis. Additional protection against nucleases and phagocytes may be brought about by insertion of polyethyleneglycol (17) or oligosaccharide residues as linkers. The size of the particles is on the lower limit of virus particles, hence good tissue dissemination and escape from the blood stream after i.v. injection can be predicted.
Fusogenic proteins provide viruses with efficient cell break-in mechanisms; on the other hand, along the way out of the cell, viral genomes are generally packaged into preformed nucleocapsids (18). Previous work (19) as well as the process described here show that similar functions can be obtained in a conceptually different manner, e.g., by proton sponge-mediated endosome swelling and cooperative detergent-induced DNA collapse, respectively. This, hopefully, will open the door for the development of fully artificial gene delivery devices devoid of the immunological problems raised by viruses.
Acknowledgments
This work was supported by grants from the Association Francaise de Lutte contre la Mucoviscidose, the Association Francaise contre les Myopathies, and the Association pour la Recherche sur le Cancer. We warmly thank Michel Terray (Malvern, France) for advice and access to the Zetasizer.
ABBREVIATIONS
- C10-CG+
guanidinocysteine N-decylamide
- c.m.c.
critical micelle concentrations
- DMAP
dimethylaminopyridine
- DTT
dithiothreitol
References
- 1.Mulligan R C. Science. 1993;260:926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 2.Tang M X, Szoka F C. Gene Ther. 1997;4:823–832. doi: 10.1038/sj.gt.3300454. [DOI] [PubMed] [Google Scholar]
- 3.Labatmoleur F, Steffan A M, Brisson C, Perron H, Feugeas O, Furstenberger P, Oberling F, Brambilla E, Behr J P. Gene Ther. 1996;3:1010–1017. [PubMed] [Google Scholar]
- 4.Behr J P. Tetrahedron Lett. 1986;27:5861–5864. [Google Scholar]
- 5.Melnikov S M, Sergeyev V G, Yoshikawa K. J Am Chem Soc. 1995;117:2401–2408. [Google Scholar]
- 6.Behr J P. Acc Chem Res. 1993;26:274–278. [Google Scholar]
- 7.Lehn J M. Angew Chem Int Ed Engl. 1988;27:89–112. [Google Scholar]
- 8.Ringsdorf H, Schlarb B, Venzmer J. Angew Chem Int Ed Engl. 1988;27:113–158. [Google Scholar]
- 9.Zanta M A, Boussif O, Adib A, Behr J P. Bioconjugate Chem. 1997;8:839–844. doi: 10.1021/bc970098f. [DOI] [PubMed] [Google Scholar]
- 10.Muraki M, Mizoguchi T. Chem Pharm Bull. 1971;19:1708–1713. [Google Scholar]
- 11.Bernatowicz M S, Wu Y, Matsueda G R. Tetrahedron Lett. 1993;34:3389–3392. [Google Scholar]
- 12.Riddles P W, Blakeley R L, Zerner B. Anal Biochem. 1979;94:75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- 13.Gustafsson J, Arvidson G, Karlsson G, Almgren M. Biochim Biophys Acta. 1995;1235:305–312. doi: 10.1016/0005-2736(95)80018-b. [DOI] [PubMed] [Google Scholar]
- 14.Mislick K A, Baldeschwieler J D. Proc Natl Acad Sci USA. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plank C, Mechtler K, Szoka F C, Wagner E. Hum Gene Ther. 1996;7:1437–1446. doi: 10.1089/hum.1996.7.12-1437. [DOI] [PubMed] [Google Scholar]
- 16.Remy J S, Kichler A, Mordvinov V, Schuber F, Behr J P. Proc Natl Acad Sci USA. 1995;92:1744–1748. doi: 10.1073/pnas.92.5.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong K L, Zheng W W, Baker A, Papahadjopoulos D. FEBS Lett. 1997;400:233–237. doi: 10.1016/s0014-5793(96)01397-x. [DOI] [PubMed] [Google Scholar]
- 18.Fields B N, Knipe D M. Virology. New York: Raven; 1990. [Google Scholar]
- 19.Boussif O, Lezoualch F, Zanta M A, Mergny M D, Scherman D, Demeneix B, Behr J P. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]