Abstract
The complex of rapamycin with its intracellular receptor, FKBP12, interacts with RAFT1/FRAP/mTOR, the in vivo rapamycin-sensitive target and a member of the ataxia telangiectasia mutated (ATM)-related family of kinases that share homology with the catalytic domain of phosphatidylinositol 3-kinase. The function of RAFT1 in the rapamycin-sensitive pathway and its connection to downstream components of the pathway, such as p70 S6 kinase and 4E-BP1, are poorly understood. Here, we show that RAFT1 directly phosphorylates p70S6k, 4E-BP1, and 4E-BP2 and that serum stimulates RAFT1 kinase activity with kinetics similar to those of p70S6k and 4E-BP1 phosphorylation. RAFT1 phosphorylates p70S6k on Thr-389, a residue whose phosphorylation is rapamycin-sensitive in vivo and necessary for S6 kinase activity. RAFT1 phosphorylation of 4E-BP1 on Thr-36 and Thr-45 blocks its association with the cap-binding protein, eIF-4E, in vitro, and phosphorylation of Thr-45 seems to be the major regulator of the 4E-BP1–eIF-4E interaction in vivo. RAFT1 phosphorylates p70S6k much more effectively than 4E-BP1, and the phosphorylation sites on the two proteins show little homology. This raises the possibility that, in vivo, an unidentified kinase analogous to p70S6k is activated by RAFT1 phosphorylation and acts at the rapamycin-sensitive phosphorylation sites of 4E-BP1.
Increases in the translation of certain mRNAs are an important response to mitogen stimulation (1–3), but little is known about the signaling pathways that link growth stimuli to the activation of the protein synthesis machinery. Studies with rapamycin (4), a potent immunosuppressant, have uncovered a signaling pathway that modulates protein synthesis in yeast (5) and animal cells (6, 7). In vivo treatment with rapamycin affects the phosphorylation of several regulators of translation, including the ribosomal S6 protein and its specific kinase, p70S6k (8, 9); the eIF-4E binding proteins, 4E-BP1 (6, 7) and 4E-BP2 (10); and elongation factor 2 (11).
Ribosomal S6 and 4E-BP1 regulate the initiation of translation of distinct classes of mRNAs. Phosphorylation by p70S6k of the S6 protein of the small ribosomal subunit permits, through unknown mechanisms, efficient translation of mRNAs containing oligopyrimidine tracts in their 5′ untranslated regions (12, 13). Phosphorylation of 4E-BP1 controls cap-dependent translation of mRNAs with extensive secondary structure (3). Initiation factor 4F complexes with these mRNAs through the interaction of its eIF-4G subunit with eIF-4E, the cap-binding protein that recognizes the N7-methyl-GpppN structure of the 5′ end of all nonorganellar mRNAs. In quiescent cells, 4E-BP1 competes with eIF-4G for binding to eIF-4E and represses translation by displacing the initiation factor 4F from the mRNA. Growth stimuli activate phosphorylation of 4E-BP1, which decreases its affinity for eIF-4E and releases the block on cap-dependent translation (3). By preventing the phosphorylation of specific residues on p70S6k and 4E-BP1, rapamycin inhibits mitogen-stimulated activation of p70S6k and phosphorylation of S6 (8, 9), and disassociation of 4E-BP1 from eIF-4E (6, 7).
The FKBP12–rapamycin complex interacts with (14–17) and perturbs a function of RAFT1 (18, 19), a member of the ATM-related family (20). The precise role of RAFT1 in the rapamycin-sensitive signaling pathway and its connection to the downstream regulators p70S6k and 4E-BP1 are not well understood. In this paper, we show that RAFT1 directly phosphorylates p70S6k on Thr-389, a residue whose phosphorylation is rapamycin-sensitive in vivo and necessary for S6 kinase activity (21, 22). In vitro, RAFT1 phosphorylation of 4E-BP1 on Thr-36 and Thr-45 prevents its interaction with eIF-4E, and in vivo Thr-45 phosphorylation is a major regulator of the 4E-BP1–eIF-4E association.
MATERIALS AND METHODS
Plasmids and Fusion Proteins.
cDNAs for rat p70S6k or regions comprising residues 332–506, 332–415, 415–502, and 66–235 of p70S6k were amplified from p85S6k in Pmt2 using PCR with appropriate primers. The amplified products were cloned into the SalI and NotI sites of HA-prk5, myc-prk5, pGEX-4T (Pharmacia), or Pet32c (Novagen). The human 4E-BP1, mouse 4E-BP2, and mouse eIF-4E cDNAs were amplified from I.M.A.G.E. clones 526490, 475519, and 539369, respectively, using PCR with primers to the 5′ and 3′ ends of the respective ORFs (23, 24). The amplified products were cloned into the SalI and NotI sites of pGEX-4T, myc-pRK5, or Pet 32c. All cDNAs prepared with PCR were verified by DNA sequencing. Mutations were introduced into the RAFT1, p70S6k, and 4E-BP1 cDNAs via PCR mutagenesis (25) and confirmed by DNA sequencing. Glutathione S-transferase (GST)-p70S6k and GST-4EBP1/2 fusion proteins were purified as described (15). The 6xHis-TRX fusion proteins of eIF-4E, p70-S6-kinase, and residues 66–235 of p70S6k were purified under denaturing conditions according to the protocol provided by the manufacturer (Novagen). Fusion proteins of p70S6k or RAFT1 fused to GST at the N or C terminus in HA-prk5 were expressed in HEK293 cells and purified as described (26).
Transient Transfections and Cell Treatments.
HEK293 cells plated on 10-cm dishes were transfected with the calcium phosphate precipitate method (27). When p70S6k or 4E-BP1 were transfected alone, 100 ng of p70S6k in HA-prk5 or 100 ng of 4E-BP1 in myc-prk5 and 9.9 μg of empty vector were used.
To make cells quiescent after transfection, plates were incubated overnight, rinsed once with PBS, and then placed in media without fetal bovine serum for 30–48 h. Cells were treated with 10 nM rapamycin (Calbiochem) or ethanol vehicle for 30 min before stimulation with 10% fetal bovine serum for 1 h or the indicated times. After appropriate treatments, cells on 10-cm dishes were rinsed once with PBS and lysed in 1 ml of ice cold buffer A (50 mM Hepes-KOH, pH 7.4/40 mM NaCl/1 mM EDTA/0.5% Triton X-100/1.5 mM Na3VO4/50 mM NaF/10 mM sodium pyrophosphate/10 mM sodium β-glycerophosphate) with 1 mM phenylmethylsulfonyl fluoride, 5 mg/ml aprotinin, 1 mg/ml antipain, 1 mg/ml leupeptin, 6 mg/ml chymostatin, and 0.7 mg/ml pepstatin A.
In vivo 32P labeling of transfected cells was performed as described (28).
In Vitro Kinase Assays.
HEK293 cells were transfected with 10 μg of RAFT1 cDNA in myc-prk5. After a 20-h incubation, cells were rinsed once with PBS and lysed in 1 ml of ice-cold buffer A. After clearing, the supernatant was used to prepare mycRAFT1 immunoprecipitates with 5 μg of anti-myc antibody and 50 μl of a 50% slurry of protein G agarose (Calbiochem). Immunoprecipitates were washed once with buffer W (50 mM Hepes-KOH, pH 7.4/40 mM NaCl/1 mM EDTA) containing 1% Triton X-100 and 0.05% SDS, twice with buffer W containing 0.5% Triton X-100 and 0.5 M LiCl, twice with buffer W containing 0.5 M LiCl, and once with 50 mM Hepes 7.4/150 mM NaCl/1 mM DTT (29). Kinase assays were performed in a volume of 22 μl at 30°C for 20 min and contained 1/5 the washed immunoprecipitates from one 10-cm dish, 1.5 μg of GST-4E-BP1 or GST-4E-BP2 or 200 ng of p70S6k fusion proteins, 2 μCi of [γ-32P]ATP (NEN), 25 mM Hepes-KOH, pH 7.4, 50 mM KCl, 20% glycerol, 10 mM MgCl2, 4 mM MnCl2, 1 mM DTT, and 50 μM unlabeled ATP. Reactions were stopped by the addition of 5 μl of 5 × gel sample buffer. After boiling the samples for 3 min, proteins were resolved by 4–12% SDS-PAGE and transferred to poly(vinylidene difluoride); phosphorylated proteins were then visualized with autoradiography.
Mitogen-activated protein (MAP) kinase and S6 kinase assays were performed as described (21).
Peptide Maps and Phosphoamino Acid Analysis.
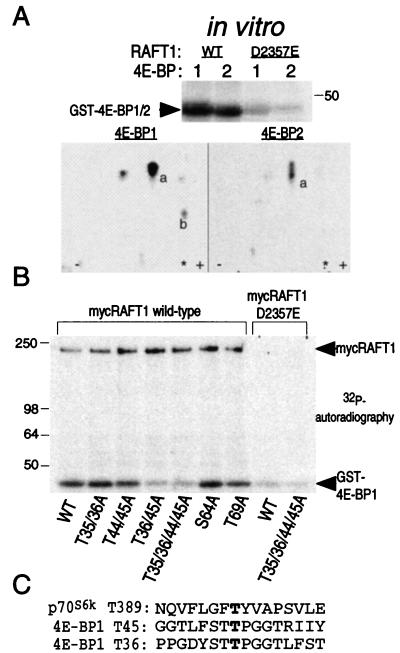
Phosphoamino acid analyses were as described (30). Peptide maps were prepared using pH 1.9 electrophoresis buffer (30). To identify the RAFT1 phosphorylation sites on 4E-BP1, we compared tryptic peptide maps of RAFT1-phosphorylated 4E-BP1 and 4E-BP2. We speculated that the 4E-BP2-derived peptide (marked “a” in Fig. 4A) is highly homologous to the one from 4E-BP1 but also contains a cysteine whose several oxidation states might give rise to peptides of similar but not identical mobilities. The six threonines between residues 20 and 50 of 4E-BP1 were chosen as candidate RAFT1 phosphorylation sites because they fulfilled these criteria. Peptide maps prepared from mutants with single or double alanine substitutions at the potential phosphorylation sites are not markedly different from those for wild-type 4E-BP1. However, separation by ascending chromatography of chymotryptic digests of peptide “a” purified from each TLC plate reveals that RAFT1 phosphorylates two homologous sites, threonines 35/36 and 44/45, that lie in the same tryptic peptide.
Figure 4.
RAFT1 phosphorylates 4E-BP1 on Thr-36 and Thr-45. (A) Phosphoamino acid analysis reveals that RAFT1 phosphorylates 4E-BP1 on threonine (data not shown). Tryptic peptide maps (Lower) of RAFT-phosphorylated 4E-BP1 and 4E-BP2 (Upper) share a major peptide (marked “a” in Lower) that migrates as a single spot in the 4E-BP1 map but as three closely spaced spots in the 4E-BP2 map. The asterisk indicates where the sample was applied. Spot “b” is seen in maps of 4E-BP1 phosphorylated by both wild-type and kinase-dead RAFT1 and represents the site of phosphorylation of a contaminating kinase. Spots visible to the left and right of spot “a” in the 4E-BP1 map are partial digestion products of the peptide in spot “a” (data not shown). (B) Thr-36 and Thr-45 are the major sites of phosphorylation by RAFT1 on 4E-BP1. The indicated GST-4E-BP1 mutants were phosphorylated in vitro by RAFT1 as in Fig. 1A. Phosphorylation of mutants S64A and T69A are shown for comparison. (C) Comparison of RAFT1 phosphorylation sites on p70S6k and 4E-BP1.
Antibodies and Immunoblots.
Anti-myc 9E-10 antibody (Calbiochem) was used to detect myc4E-BP1 and immunoprecipitate myc-RAFT1, anti-hemagglutinin (HA) antibody (Babco, Richmond, CA) was used to detect and immunoprecipitate HA-p70S6k, and anti-GST antibody (Santa Cruz) was used to detect GST-4E-BP1. Gel samples were resolved by SDS-PAGE, transferred to poly(vinylidene difluoride) in CAPS-methanol, pH 11, and probed with primary antibodies. Western blots were developed with anti-mouse or anti-rabbit antibodies (Amersham) and enhanced chemiluminescence (NEN).
7-Methyl-GTP and eIF-4E Affinity Chromatography.
To purify endogenous eIF-4E, 30 μl of a 50% slurry of 7-methyl-GTP Sepharose (Pharmacia) were added to the cleared cellular lysate and incubated for 30 min at room temperature. After washing the resin twice with 50 mM Hepes, pH 7.4/40 mM NaCl/2 mM EDTA/0.1% Triton X-100, bound proteins were eluted with 1.25× gel sample buffer and resolved with SDS/17% PAGE, and 4E-BP1 was visualized with the anti-myc antibody.
For in vitro binding reactions between 4E-BP1 and eIF-4E, 1 μg of TRX-eIF-4E fusion protein was bound to 20 μl of packed 7-methyl-GTP Sepharose for 30 min at room temperature, washed twice with buffer B (25 mM Hepes, 7.4/100 mM KCl/1 mM EDTA/1 mM DTT), and resuspended in 40 μl of the same buffer. 20 μl of the reaction products of kinase assays containing 100 ng of GST-4E-BP1 were added, incubated for 1 h on ice, and washed twice with buffer B; bound proteins were then eluted with gel sample buffer. After Western blotting, 4E-BP1 was detected with anti-GST antibody.
RESULTS
In Vitro RAFT1 Phosphorylates p70 S6 Kinase and 4E-BP1.
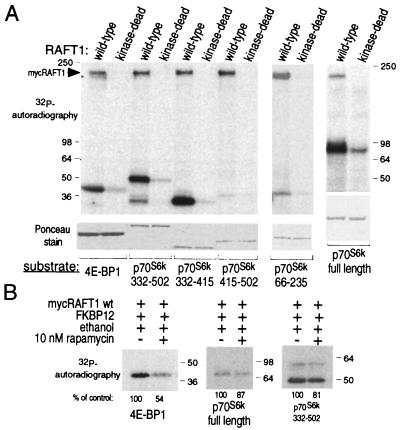
Because RAFT1 autophosphorylates (18, 31) and is a member of the ATM-related protein kinase family, we asked whether RAFT1 might directly phosphorylate p70S6k and 4E-BP1. In vitro, wild-type, but not kinase-dead (D2357E), RAFT1 expressed in HEK293 cells and purified by immunoprecipitation phosphorylates bacterially produced p70S6k and 4E-BP1 (Fig. 1A). RAFT1 strongly prefers p70S6k as a substrate, as over 10-fold more 4E-BP1 (2,000 ng) than p70S6k (150 ng) is necessary to obtain about equal levels of phosphorylation by RAFT1 (Fig. 1A, lower panel). The major RAFT1 phosphorylation site(s) in p70S6k is C-terminal to the catalytic domain within residues 332–415. This region contains Thr-389 and Ser-404, whose phosphorylation is known to be inhibited by rapamycin in vivo (21). In vitro, RAFT1 does not phosphorylate the N-terminal half of p70S6k (residues 66–235), which contains Thr-229, the third in vivo rapamycin-sensitive phosphorylation site (21).
Figure 1.
RAFT1 phosphorylates 4E-BP1 and the C-terminal half of p70S6k in vitro. (A) Wild-type and kinase-dead RAFT1 expressed in HEK293 cells were purified by immunoprecipitation and incubated in a kinase assay with [γ-32P]ATP and the indicated fusion proteins of 4E-BP1 or p70S6k. The phosphorylated proteins are revealed by autoradiography (Upper), and a Ponceau stain reveals the relative amounts of each fusion protein in the assays (Lower). (B) FKBP12-rapamycin partially blocks RAFT1 phosphorylation of 4E-BP1 and p70S6k. Kinase assays using the indicated substrates were performed as in A except that the immunoprecipitates were first incubated on ice for 1 h with 100 nM FKBP12 with or without 10 nM rapamycin. These results are representative of five similar experiments.
Preincubation of RAFT1 with FKBP12-rapamycin decreases phosphorylation of 4E-BP1 and p70S6k to about 50% and 85%, respectively, of controls with FKBP12 alone (Fig. 1B). The incomplete inhibition by FKBP12-rapamycin is not unexpected because RAFT1 complexed to a GST-FKBP12-rapamycin affinity resin can still robustly autophosphorylate (31). Moreover, rapamycin treatment does not completely eliminate the putative kinase activity of the Tor2 protein, a yeast homologue of RAFT1, as rapamycin treatment of yeast does not reproduce all the effects observed when replacing the wild-type Tor2 with a kinase-dead version of the protein (32).
Time Course of RAFT1 Activation and 4E-BP1 and p70 S6 Kinase Phosphorylation.
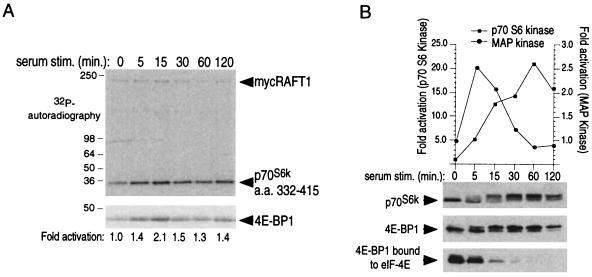
As one might expect of an upstream kinase, serum-induced RAFT1 activation precedes p70S6k and 4E-BP1 phosphorylation but follows a similar pattern of activation and deactivation. The RAFT1 activation phase is fast and peaks at 15 min, but deactivation is slow and activity remains elevated at 2 h after serum stimulation (Fig. 2A). RAFT1 activation is not inhibited by a 30-min treatment with 10 nM rapamycin before serum stimulation (data not shown). Phosphorylation of p70S6k and 4E-BP1, as reflected by increases in their apparent molecular weights, begins within 5 min after serum addition, rises significantly by 15 min, peaks at 1 h, and then decays slowly (Fig. 2B). Activation of the S6 kinase and disruption of the 4E-BP1–eIF-4E complex follow the same time course, which differs from the kinetics of MAP kinase activation and deactivation in response to serum (Fig. 2B). Because RAFT1 has significant activity in quiescent cells (Fig. 2A) in which p70S6k and 4E-BP1 are dephosphorylated, the specific activity of RAFT1 cannot be the sole determinant of the p70S6k and 4E-BP1 phosphorylation states. Thus, in addition to RAFT1 activity, other factors, such as the accessibility of p70S6k and 4E-BP1 to the RAFT1 kinase domain and/or the activity of phosphatases that target them, must also play an important role in controlling the extent of their phosphorylation.
Figure 2.
Serum stimulates RAFT1 kinase activity toward p70S6k and 4E-BP1. (A) Wild-type RAFT1 purified from quiescent HEK293 cells stimulated with 10% serum for the indicated times and purified by immunoprecipitation was incubated in a kinase assay with indicated fusion proteins and [γ-32P]ATP. After SDS-PAGE, transfer to poly(vinylidene difluoride) and Ponceau staining, the bands corresponding to p70S6k 332–415 and 4E-BP1 were excised, and the amount of radioactivity incorporated into each was determined by scintillation counting. The increases in 32P incorporation were similar for p70S6k 332–415 and 4E-BP1, and the fold activation for p70S6k is shown below the autoradiograph. The results shown are representative of three similar experiments. (B) Time course of p70S6k phosphorylation (Top) and activation (graph) and 4E-BP1 phosphorylation and release from eIF-4E (Middle and Bottom). p70S6k and 4E-BP1 were detected with immunoblotting. Kinetics of endogenous MAP kinase activation is shown for comparison (graph).
RAFT1 Phosphorylates p70 S6 Kinase on Thr-389.
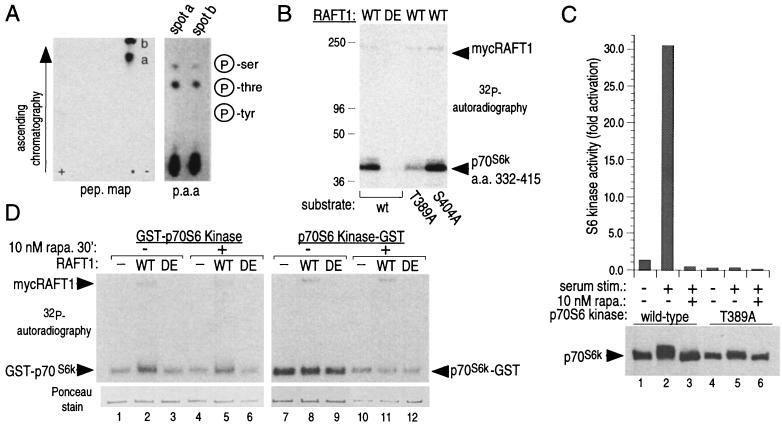
We determined the sites of RAFT1 phosphorylation on p70S6k and 4E-BP1 to see if their mutation to nonphosphorylatable amino acids would mimic the effects of rapamycin. Residues 332–415 of p70S6k contain the major site of RAFT1 phosphorylation (Fig. 1A). Chymotryptic digestion of the RAFT1-phosphorylated fusion protein yields hydrophobic peptides that contain phosphothreonine (Fig. 3A). Thr-389 is the major site of RAFT1 phosphorylation in vitro (Fig. 3B), as an alanine substitution of Thr-389 and not Ser-404 eliminates the majority of RAFT1 phosphorylation. No other kinase has been reported to phosphorylate Thr-389 and it is one of three residues (Thr-229, Thr-389, and Ser-404) whose phosphorylation is completely blocked by rapamycin treatment in vivo (21). Under our in vitro experimental conditions, we do not find phosphorylation of Thr-229 or Ser-404 of p70S6k, although they have significant homology to Thr-389 (21). Further work will be required to determine if RAFT1 cannot phosphorylate these sites because they are inaccessible in the fusion proteins tested or if they do not conform to the preferred RAFT1 phosphorylation motif. The importance of Thr-389 phosphorylation for activation of p70S6k is well documented, and the phosphorylation status of this residue correlates with S6 kinase activity (21, 22). We find that compared with wild-type, a T389A mutant of p70S6k has less than 1% of the S6 kinase activity, does not respond to serum stimulation, and displays decreased laddering on SDS-PAGE (Fig. 3C). Rapamycin has similar effects on wild-type p70S6k (Fig. 3C).
Figure 3.
RAFT1 phosphorylates p70S6k on Thr-389. (A) Chymotryptic peptide map of RAFT1-phosphorylated p70S6k 332–415 and phosphoamino acid analysis of the purified peptides. Peptides “a” is an incomplete digest of peptide “b,” and tryptic peptide maps reveal only one spot (data not shown). (B) Substitution of a T389A mutant for wild-type p70S6k 332–415 eliminates the majority of RAFT1 phosphorylation. (C) T389A p70S6k expressed in HEK293 cells is not activated by serum stimulation (graph) and displays decreased laddering on SDS-PAGE. (D) Wild-type (WT) but not kinase-dead (DE) RAFT1 phosphorylates HEK293-purified p70S6k fused to GST at the N terminus (Left) but not at the C terminus (Right). All the p70S6k fusion proteins are capable of autophosphorylating (−). The amount of added p70S6k is roughly equal except for those with p70S6k-GST from rapamycin-treated cells, which contain about one-third of the amount of the others (Lower).
Activation of p70S6k in vivo requires phosphorylation at multiple sites in addition to Thr-389, such as Ser-367 (33). It is not surprising then, that bacterially produced 6xHis-Trx-p70S6k fusion protein is inactive toward S6 either before or after RAFT1 phosphorylation (data not shown). We reasoned that a p70S6k that is phosphorylated at all the necessary residues except Thr-389 should have the potential to be activated by RAFT1 phosphorylation in vitro. To obtain such a protein, we purified recombinant p70S6k fusion proteins from HEK293 cells that had been treated with rapamycin for only 15 min, conditions that result in the selective dephosphorylation of Thr-389 (21). p70S6k fused to GST at its N terminus is completely inactive toward S6 when purified from control or rapamycin-treated cells (data not shown), indicating that the GST moiety at the N terminus prevents an activating input that is necessary for S6 kinase activity or disturbs the normal folding of the protein. That input cannot be phosphorylation of Thr-389, as in vitro phosphorylation by immunoprecipitated RAFT1 (Fig. 3D) does not activate the protein (data not shown). A p70S6k variant with GST attached to the C terminus is stimulated by serum, in a rapamycin-sensitive fashion, to 25% of wild-type levels (data not shown). Surprisingly, however, this protein, when purified from either control or rapamycin-treated cells, is not a RAFT substrate in vitro (Fig. 3D). Furthermore, a soluble RAFT1 fusion protein purified from HEK293 cells cannot phosphorylate mycp70S6k immunoprecipitated from control or rapamycin-treated cells (data not shown). The addition to the kinase reaction of a variety of cellular extracts or lipids, including 3′ and 4′ phosphorylated phosphatidylinositols, does not restore the capacity of soluble RAFT1 to phosphorylate immunoprecipitated mycp70S6k or of immunoprecipitated RAFT1 to phosphorylate soluble p70S6k-GST (data not shown). These experiments indicate that Thr-389 is hidden from RAFT1 in potentially active versions of p70S6k, and it is likely that access to this residue requires a conformational change in the protein induced by cofactor(s) present in vivo but not in vitro. The inactive p70S6k variants, such as bacterially made p70S6k or HEK239-produced GST-p70S6k, have (perhaps because they are misfolded) accessible Thr-389 but are likely missing other modifications necessary for S6 kinase activity.
RAFT1 Phosphorylation of 4E-BP1 on Thr-36 and Thr-45 Blocks Its Ability to Interact with eIF-4E.
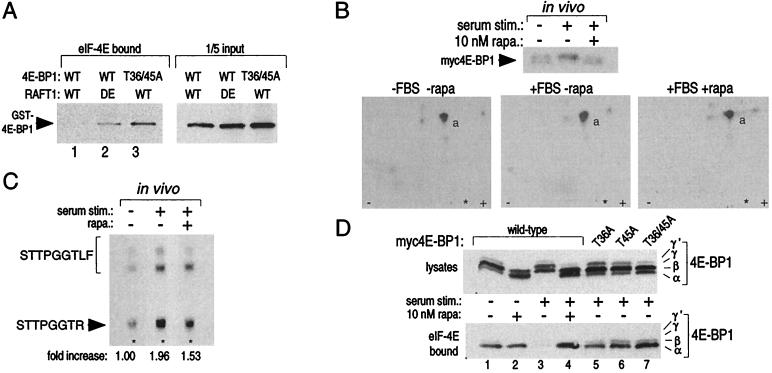
Using comparative tryptic peptide mapping of RAFT1-phosphorylated 4E-BP1 and 4E-BP2 to aid us, we identified Thr-36 and Thr-45 as the RAFT1 phosphorylation sites on 4E-BP1 (Fig. 4 A and B; see Materials and Methods for details). MAP kinase phosphorylation of 4E-BP1 occurs on Ser-64 (34) and, with longer kinase reactions, also at Thr-36 and Thr-45 (35). Protein alignment of Thr-36, Thr-45, and Thr-389 and their surrounding residues reveals that the RAFT1 phosphorylation sites on 4E-BP1 and p70S6k share little amino acid homology; large hydrophobic amino acids flank Thr-389 in p70S6k, whereas Thr-36 and Thr-45 of 4E-BP1 are in S/TP motifs (Fig. 4C). RAFT1 phosphorylation of 4E-BP1 in vitro decreases its capacity to interact with eIF-4E, and substitution of wild-type 4E-BP1 with the T36A/45A double mutant or of wild-type RAFT1 with the kinase-dead variant eliminates the ability of RAFT1 to decrease the interaction (Fig. 5A). Given the dissimilarity between the RAFT1 phosphorylation sites on 4E-BP1 and p70S6k, the preference of RAFT1 for p70S6k as a substrate, and the finding that MAP kinase also phosphorylates 4E-BP1 and affects its binding to eIF-4E (34), one must be careful in accepting this in vitro result as enough evidence to declare RAFT1 to be the in vivo 4E-BP1 kinase. We find that Thr-36 and Thr-45 are phosphorylated in 4E-BP1 isolated from 32P-labeled cells (Fig. 5B) and that rapamycin blocks about 50% of the serum-stimulated increase in phosphate at these sites (Fig. 5C). A recent report (35) finds that rapamycin partially blocks the insulin-stimulated phosphorylation of several S/TP sites in 4E-BP1, including Thr-36 and Thr-45 but also Ser-64, Thr-69, and Ser-82. To further evaluate the role of Thr-36 and Thr-45 phosphorylation in vivo, we expressed in quiescent HEK293 cells wild-type, T36A, T45A, or T36/45A 4E-BP1 and examined the effect of serum stimulation on their function. Mutation of Thr-36 to alanine substantially increases the amount of 4E-BP1 that remains bound to eIF-4E after serum stimulation, and the T45A and T36/45A mutations have more pronounced effects (Fig. 5D). Even after serum stimulation, T45A containing mutants bind to eIF-4E to the same degree as wild-type 4E-BP1 does in the absence of serum stimulation or after serum stimulation in the presence of rapamycin (Fig. 5D, compare lanes 1 and 4 to 7). These results indicate that the threonines phosphorylated by RAFT1 in vitro, particularly Thr-45, are important mediators of the interaction between 4E-BP1 and eIF-4E in vivo.
Figure 5.
Thr-36 and Thr-45 are phosphorylated in vivo, and mutations of these residues to alanine create a 4E-BP1 variant that binds constitutively to eIF-4E in vitro and in vivo. (A) 4E-BP1 phosphorylated in vitro by wild-type (WT) RAFT1, but not by kinase-dead (DE) RAFT, cannot bind to eIF-4E (Left, lanes 1 and 2). Even after an incubation with wild-type RAFT1 in a kinase assay, the T36/45A mutant still binds to eIF-4E (Left, lane 3). Equal amounts of GST-4E-BP1 were offered for binding in each condition (Right). Kinase assays of 30-min duration were performed as in Fig. 1A except that only 100 ng of fusion protein was used. The reaction products were then offered for binding to recombinant eIF-4E prebound to 7-methyl-GTP Sepharose. Bound 4E-BP1 was detected by immunoblotting with an anti-GST antibody. (B) Tryptic peptide maps (Lower) of 4E-BP1 isolated form cells labeled in vivo with 32P (Upper) and treated with the indicated conditions. Peptide “a” is eliminated in maps prepared with a T36/45A mutant (data not shown). (C) Serum stimulation increases phosphate content at Thr-45 2-fold, and rapamycin blocks 50% of the increase. Peptide “a” was purified from the TLC plates of B and analyzed as described in Materials and Methods. The spots were quantitated with densitometry. (D) Alanine mutations at Thr-36 and Thr-45 prevent serum-induced release of 4E-BP1 from eIF-4E and mimic the effects of rapamycin. HEK293 cells were transfected with plasmids encoding wild-type or the indicated mutant 4E-BP1s, treated with rapamycin and serum, and the migration pattern of 4E-BP1 (Upper) or the amount of 4E-BP1 bound to eIF-4E (Lower) was determined by immunoblotting.
DISCUSSION
In this study, we provide links between three components of the rapamycin-sensitive signaling pathway, an important mediator of growth factor-induced increases in protein synthesis. We demonstrate that RAFT1 is the elusive hydrophobic-directed Thr-389 kinase of p70S6k. The rapamycin sensitivity and importance for S6 kinase activation of Thr-389 phosphorylation in vivo is well established (21, 22). Furthermore, RAFT1 phosphorylates 4E-BP1 on sites that regulate its interaction with the cap-binding protein eIF-4E in vitro and in vivo. p70S6k and 4E-BP1 (19) are the first known exogenous substrates for RAFT1, and their phosphorylation states are key switches for regulating protein translation (3).
The finding that RAFT1 phosphorylates 4E-BP1 on residues that control its function is provocative and makes the 4E-BP1 pathway dissimilar from that leading to S6 phosphorylation, where an intermediate kinase, p70S6k, transduces and amplifies the initial signal. Although the work presented here supports RAFT1 as a physiological 4E-BP1 kinase, we cannot exclude the possibility that an unidentified 4E-BP1 kinase exists that is analogous to p70S6k, recognizes all the rapamycin-sensitive S/TP sites of 4E-BP1, and contains a p70S6k-like RAFT1-phosphorylation motif whose phosphorylation controls its activity.
We find that phosphorylation of p70S6k and 4E-BP1 is only partially inhibited by FKBP12-rapamycin binding to RAFT1. This contrasts with a recent report on RAFT1 (mTOR) phosphorylation of 4E-BP1 (PHAS-1) in which almost complete inhibition of 4E-BP1 phosphorylation is observed (19). A major difference between our studies is the concentration of rapamycin employed. Whereas we use 10 nM rapamycin in our in vitro kinase reaction, Brunn et al. (19) employed 10 μM rapamycin, which is over 1000-fold the concentration needed to inhibit 4E-BP1 phosphorylation in vivo. Exactly how the drug–receptor complex affects RAFT1 in vivo remains unknown, but it is likely that the end result of this interaction is an inhibition of RAFT1 kinase activity only toward specific protein substrates, such as p70S6k. FKBP12-rapamycin could do this through several potential mechanisms, such as sterically blocking access to the RAFT1 kinase domain, preventing RAFT1 activation by secondary modifications or interacting proteins, or altering its subcellular localization. On the other hand, the decrease in its kinase activity toward target proteins may be enough to prevent the phosphorylation of proteins that are under the constant control of competing kinases and phosphatases.
Acknowledgments
We thank Joseph Avruch for the generous gift of the p85 S6 kinase cDNA and Joseph Hurt for valuable input. This work was supported by the U.S. Public Health Service (Grant MH-18501), Research Scientist Award DA-00074 (to S.H.S.), and Training Grant GM-07309 (to D.M.S. and P.E.B.).
ABBREVIATIONS
- GST
glutathione S-transferase
- HA
hemagglutinin
- MAP
mitogen-activated protein
- ATM
ataxia telangiectasia mutated
References
- 1.Nielsen F C, Ostergaard L, Nielsen J, Christiansen J. Nature (London) 1995;377:358–362. doi: 10.1038/377358a0. [DOI] [PubMed] [Google Scholar]
- 2.Brown E J, Schreiber S L. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 3.Sonenberg N. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 245–269. [Google Scholar]
- 4.Martel R R, Klicius J, Galet S. Can J Physiol Pharmacol. 1977;55:48–51. doi: 10.1139/y77-007. [DOI] [PubMed] [Google Scholar]
- 5.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Manteuffel S R, Gingras A C, Ming X F, Sonenberg N, Thomas G. Proc Natl Acad Sci USA. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 8.Chung J, Kuo C J, Crabtree G R, Blenis J. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 9.Kuo C J, Chung J, Fiorentino D F, Flanagan W M, Blenis J, Crabtree G R. Nature (London) 1992;358:70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- 10.Lin T A, Lawrence J C. J Biol Chem. 1996;271:30199–30204. doi: 10.1074/jbc.271.47.30199. [DOI] [PubMed] [Google Scholar]
- 11.Redpath N T, Foulstone E J, Proud C G. EMBO J. 1996;15:2291–2297. [PMC free article] [PubMed] [Google Scholar]
- 12.Jefferies H B, Reinhard C, Kozma S C, Thomas G. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jefferies H B, Fumagalli S, Dennis P B, Reinhard C, Pearson R B, Thomas G. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown E J, Albers M W, Shin T B, Ichikawa K, Keith C T, Lane W S, Schreiber S L. Nature (London) 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 15.Sabatini D M, Erdjument-Bromage H, Lui M, Tempst P, Snyder S H. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 16.Chiu M I, Katz H, Berlin V. Proc Natl Acad Sci USA. 1994;91:12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabers C J, Martin M M, Brunn G J, Williams J M, Dumont F J, Wiederrecht G, Abraham R T. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 18.Brown E J, Beal P A, Keith C T, Chen J, Shin T B, Schreiber S L. Nature (London) 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 19.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi H, Houghton P J, Lawrence J C, Jr, Abraham R T. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 20.Keith C T, Schreiber S L. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 21.Pearson R B, Dennis P B, Han J W, Williamson N A, Kozma S C, Wettenhall R E, Thomas G. EMBO J. 1995;14:5279–5287. doi: 10.1002/j.1460-2075.1995.tb00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennis P B, Pullen N, Kozma S C, Thomas G. Mol Cell Biol. 1996;16:6242–6251. doi: 10.1128/mcb.16.11.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pause A, Belsham G J, Gingras A C, Donze O, Lin T A, Lawrence J C, Sonenberg N. Nature (London) 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 24.Hu C, Pang S, Kong X, Velleca M, Lawrence J C., Jr Proc Natl Acad Sci USA. 1994;91:3730–3734. doi: 10.1073/pnas.91.9.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 26.Alessi D R, James S R, Downes C P, Holmes A B, Gaffney P R, Reese C B, Cohen P. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 27.Wigler M, Sweet R, Sim G K, Wold B, Pellicer A, Lacy E, Maniatis T, Silverstein S, Axel R. Cell. 1979;16:777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- 28.Cohen N A, Brenman J E, Snyder S H, Bredt D S. Neuron. 1996;17:759–767. doi: 10.1016/s0896-6273(00)80207-x. [DOI] [PubMed] [Google Scholar]
- 29.Sabatini D M, Pierchala B A, Barrow R K, Schell M J, Snyder S H. J Biol Chem. 1995;270:20875–20878. doi: 10.1074/jbc.270.36.20875. [DOI] [PubMed] [Google Scholar]
- 30.van der Geer P, Luo K, Sefton B M, Hunter T. In: Protein Phosphorylation. Hardie D G, editor. Oxford: Oxford Univ. Press; 1993. pp. 31–59. [Google Scholar]
- 31.Brunn G J, Williams J, Sabers C, Wiederrecht G, Lawrence J C, Jr, Abraham R T. EMBO J. 1996;15:5256–5267. [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng X F, Fiorentino D, Chen J, Crabtree G R, Schreiber S L. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 33.Moser B A, Dennis P B, Pullen N, Pearson R B, Williamson N A, Wettenhall R E H, Kozma S C, Thomas G. Mol Cell Biol. 1997;17:5648–5655. doi: 10.1128/mcb.17.9.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin T A, Kong X, Haystead T A, Pause A, Belsham G, Sonenberg N, Lawrence J C., Jr Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 35.Fadden P, Haystead T A J, Lawrence J C. J Biol Chem. 1997;272:10240–10247. doi: 10.1074/jbc.272.15.10240. [DOI] [PubMed] [Google Scholar]