Abstract
Background
Puccinia striiformis is a plant pathogenic fungus causing stripe rust, one of the most important diseases on cereal crops and grasses worldwide. However, little is know about its genome and genes involved in the biology and pathogenicity of the pathogen. We initiated the functional genomic research of the fungus by constructing a full-length cDNA and determined functions of the first group of genes by sequence comparison of cDNA clones to genes reported in other fungi.
Results
A full-length cDNA library, consisting of 42,240 clones with an average cDNA insert of 1.9 kb, was constructed using urediniospores of race PST-78 of P. striiformis f. sp. tritici. From 196 sequenced cDNA clones, we determined functions of 73 clones (37.2%). In addition, 36 clones (18.4%) had significant homology to hypothetical proteins, 37 clones (18.9%) had some homology to genes in other fungi, and the remaining 50 clones (25.5%) did not produce any hits. From the 73 clones with functions, we identified 51 different genes encoding protein products that are involved in amino acid metabolism, cell defense, cell cycle, cell signaling, cell structure and growth, energy cycle, lipid and nucleotide metabolism, protein modification, ribosomal protein complex, sugar metabolism, transcription factor, transport metabolism, and virulence/infection.
Conclusion
The full-length cDNA library is useful in identifying functional genes of P. striiformis.
Background
Puccinia striiformis Westend., a fungus in Pucciniacea, Uredinales, Basidiomycotina, Eumycota, causes stripe (yellow) rust. Based on specific pathogenicity on cereal crops and grasses, the fungal species consists of various formae speciales, such as P. striiformis f. sp. tritici on wheat (Triticum aestivum), P. striiformis f. sp. hordei on barley (Hordeum vulgare), P. striiformis f. sp. poae on bluegrass (Poa pratensis) and P. striiformis f. sp. dactylidis on orchard grass (Dactylis glomerata) [9,32]. Among the various formae speciales, the wheat and barley stripe rust pathogens are most economically important. Wheat stripe rust has been reported in more than 60 countries and all continents except Antarctica [6]. Devastating epidemics of wheat stripe rust often occur in many countries in Africa, Asia, Australia, Europe, North America and South America [6,32]. In the U. S., stripe rust of wheat has existed for more than 100 years [19,25]. The disease had been primarily a major problem in western US before 2000, but has become increasingly important in the south central and the Great Plains since 2000 [6,11,25]. Barley stripe rust is a relatively new disease in the west hemisphere. It has caused severe damage in some locations since it was introduced to Colombia in 1975 from Europe [14], and spread to Mexico in 1987 [1] and the U. S. in 1991 [5,9,29]. In spite of its importance, very little is known about the molecular biology and the genomics of the stripe rust fungus.
The life cycle of the stripe rust fungus consists of the dikaryotic uredial and diploid telial stages in the nature [24,32]. Teliospores can germinate to form haploid basidiniospores. Unlike the stem rust (P. graminis) and leaf rust (P. triticina) pathogens, the stripe rust pathogen does not have known alternate hosts for basidiniospores to infect, and thus, it does not have known sexual pycnial and aecial stages. Therefore, isolates of the fungus cannot be crossed through sexual hybridization, which makes it impossible to study the fungal genes through classic genetic approaches. The fungus reproduces and spreads through urediniospores and survives as mycelium in living host plants. Because urediniospores cannot keep their viability for very long, living plants (volunteers of wheat and barley crops and grasses, or crops and grasses in cool regions in the summer and in warm regions in the winter) are essential to keep the fungus alive from season to season. Although the pathogen does not have known sexual reproduction, there is a high degree of variation in virulence and DNA polymorphism in the natural populations of the stripe rust pathogens [5,6,8,9,11,25]. More than 100 races of P. striiformis f. sp. tritici and more than 70 races of P. striiformis f. sp. hordei have been identified in the U. S. [5,6] based on virulence/avirulence patterns produced on differential cultivars by isolates of the pathogens. The avirulence or virulence phenotypes have not been associated with any specific genes or DNA sequences due to the factors that the pathogen can not be studied by conventional analyses.
The expressed sequence tag (EST) technology is an approach to identify genes in organisms that are difficult to study using classic genetic approaches and gene mutation by insertional mutagenesis. Liu et al. [26] analyzed abundant and stage-specific mRNA from P. graminis. Lin et al. [23] isolated and studied the expression of a host response gene family encoding thaumatin-like proteins in incompatible oat-stem rust fungus interactions. Recently, EST libraries have been constructed for various fungal species including P. triticina [18], the probably most closely related fungal species to P. striiformis. ESTs provide valuable putative gene sequence information for genomic studies of targeted organisms. However, EST data has its own limitations such as incomplete cDNA sequence. Because ESTs are typically generated from the 3' end sequences of cDNA clones, EST libraries tend to be incomplete at the 5' end of the transcripts. The cDNA libraries constructed by conventional methods [17] normally contain a high percentage of 5' truncated clones due to the premature stop of reverse transcription (RT) of the template mRNA, particularly for cDNA clones derived from large mRNA molecules and those with the potential to form secondary structures. The size bias against large fragments commonly exists in conventional cDNA cloning procedures. Certain limitations also apply to the end products of the automatic EST assemblies, which may be composed of ESTs generated from different tissues or different developmental stages and may not reflect the accurate transcripts.
Several methods have been developed to construct cDNA libraries that are enriched for full-length cDNAs, including RNA oligo ligation to the 5' end of mRNA [21,33], 5' cap affinity selection via eukaryotic initiation factor [15], or 5' cap biotinylation followed by biotin affinity selection [2]. These methods can be used to improve the full-length cDNA clone content of the cDNA library, but they are all very laborious and involve several enzymatic steps that must be performed on mRNA. Therefore, they are prone to quality loss through RNA degradation. Furthermore, they all require high amounts of starting mRNA at μg level for reverse transcription and cloning processes. Comprehensive sets of accurate, full-length cDNA sequences would address many of the current limitations of the EST data. Genome-scale collections of full-length cDNA become important for analyses of the structures and functions of expressed genes and their products [31]. Full-length cDNA library is a powerful tool for functional genomics and is widely used as physical resources for identifying genes [36].
A full-length cDNA library should be an important resource for studying important genes of the P. striiformis pathogen, for sequencing the whole genome, and for determining its interaction with host plants. The objectives of the present study were to construct a full-length cDNA library for P. striiformis f. sp. tritici and characterize selected cDNA sequences in the library to identify putative functional genes of P. striiformis f. sp. tritici.
Results
Full-length cDNA library generation and characterization
Total RNA was extracted from 30 mg urediniospores of race PST-78 of P. striiformis f. sp. tritici and yielded approximately 7.5 μg total RNA of high purity. Full-length cDNA was synthesized by reverse transcription and enriched by subsequent long distance PCR (LD PCR). Only non-truncated first strand cDNAs were tagged by the SMART IV oligonucleotide sequence : 5'-AAGCAGTGGTATCAACGCAGAGTGGCCATTACGGCCGGG-3' during the initial reverse transcription. The PCR amplification products were digested with restriction enzyme sfiI to generate directional cloning ends. The agarose gel analysis of the digestion showed a significant amount of double stranded cDNA that appeared as a smear ranging from 300 bp to 12 kb. The sfiI-digested double strand cDNA was obtained from 5 fractionated gel zones. The gel zones containing smaller cDNA fragments (ranging from 500 bp to 4 kb) yielded approximately 800 ng to 1 μg of cDNA while the gel zones containing large cDNA fragments (ranging from 5 kb to 10 kb) had relatively lower cDNA yields in the 50 – 100 ng range. Although the large cDNA fragment output was relatively low, it was adequate for the subsequent ligation reaction for cloning.
Fractionated cDNA was cloned into the sfiI sites of the pDNR-LIB cloning vector and transformed into DH10B competent cells. One microliter of ligation yielded a range of 1,000 to 2,000 recombinant clones for cDNA inserts within the large fractionated gel zone. More than 3,000 recombinant clones were obtained for cDNA inserts from the medium and smaller fractionated gel zones. The clone evaluation of random samples revealed cDNA insert length ranging from 200 bp up to 9 kb across all the fractionation inserts. In general, most of the inserts were in the length range of 500 bp to 4 kb. Large scale transformation was conducted using ligation reactions from each of the fractions, and clones were picked in a mixed fashion using an automated robotic clone picker. A total of 42,240 cDNA clones were arrayed in 112 micro-plates of 384-wells each. An additional copy of the cDNA library was generated by manual duplication.
The average cDNA insert size and their distribution were analyzed by random sampling of cDNA clones from randomly selected plates. A total of 320 cDNA clones were double-digested by HindIII/EcoRI. The average cDNA insert size was 1.9 kb. Approximately, 96% of the clones had inserts longer than 500 bp, 54% of the cDNA clones had inserts longer than 1.5 kb, and 15% of the clones contained inserts longer than 3 kb. Only 3% of the clones had inserts smaller than 500 bp (Fig. 1). Therefore, the size fractionation procedure used in this library construction was effective for obtaining cDNA inserts of different lengths.
Figure 1.
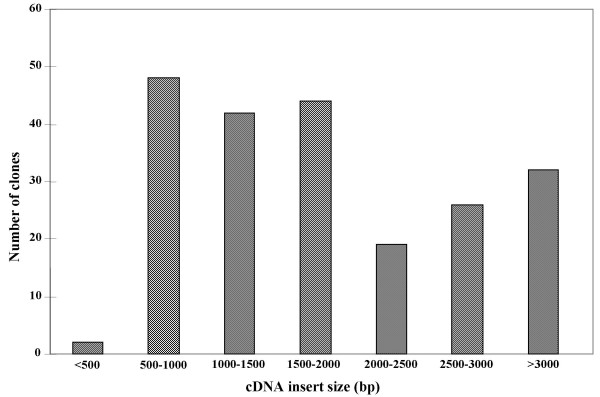
The insert size distribution of urediniospore cDNA clones of Puccinia striiformis f. sp. tritici. The insert sizes of 320 randomly picked cDNA clones were determined by HindIII/EcoRI double digestion.
cDNA sequence analysis
A total of 198 cDNA clones were sequenced with a single pass reading from both ends of the cloning sites. Sequence reads of 800 – 1,000 bp were achieved for most of the clones. For each sampled cDNA clone, two sequence reads from both ends were aligned and were comparatively edited to generate a consensus sequence contig. Of the 196 clones, we obtained a completed cDNA sequence for 149 clones. The remaining 47 cDNA clones had two partial sequences because they had insert sizes that exceeded the single pass sequencing capability. The 243 single sequences were deposited in the EST sequence database of the GenBank (Accession numbers EG374272 – EG374514).
All edited sequence contigs were searched against the NCBI fungal gene databases and the all-organism gene databases with their translated amino acid sequences. We consider that if a cDNA clone of P. striiformis f. sp. trtici and a gene in the fungal database share homology significant at an e-value of <1.00E-5, they likely belong to the same gene family and should share a similar broad sense function. A total of 73 cDNA clones (36.9%) met this requirement, and therefore, were considered with functions identified, of which 50 clones had completed sequences, 13 clones had partial sequences that hit the same or similar genes, and 10 clones had one partial sequence hitting a characterized gene (Table 1). These genes represented 51 different protein products that are involved in amino acid metabolism, cell defense, cell cycle, cell signaling, cell structure and growth, energy cycle, lipid and nucleotide metabolism, protein modification, ribosomal protein complex, sugar metabolism, transcription factor, transport metabolism and virulence/infection. Examples of these genes are glycine hydroxymethyltransferase, saccharopine dehydropine, mitogen-activated protein kinase (MAPK), serine/threonine kinase, β-tubulin, deacetylase, mitochondrial ATPase alpha-subunit, fatty acid oxidoreductase, phosphatidyl synthase, endopeptidase, elongation factor, ribosomal RNA unit, glucose-repressible protein, transaldolase, TATA-box binding protein, cell wall glucanase and pectin lyase. Thirty-seven clones (18.9%) had certain levels of homology to genes in other fungi, but the significance levels were not adequate for considering the functions identified (Table 2). Sequences of 36 clones (18.4%) were homologous to fungal genes with functions unclassified and the most of them were hypothetical proteins. Although many of the hypothetical protein genes had e-value < 1.00E-05, they are listed in Table 2 because of their unclear functions. Some of the hypothetical protein genes were homologous to genes in other plant pathogens, such as Ustilago maydis, Gibberella zeae and Magnapothe grisea. These genes could be related to plant infection. Many of the cDNA clones had homology of various levels to genes from plants (12%), other eukaryotes (34%), or to proteins of bacterial origin (11%) (data not shown). There were 50 clones (25.5%) with full-length sequences resulting in no-hit, indicating that they had no homology to any sequence available in the current NCBI databases (Table 3). These genes could be unique to P. striiformis f. sp. tritici. Alternatively, similar genes in other fungi have not been identified or desposited into the databases.
Table 1.
Putative genes idenitified in cDNA clones of Puccinia striiformis f. sp. tritici based on their sequence comparison with other fungal genes through Blastx search of the NCBI databases
| Category & clone no. | GenBank accession | Size (bp) | Full length or partiala | Best hit in the NCBI fungal databases | |||
| Protein | Accession | Organism | e-value | ||||
| 1. Amino acid metabolism | |||||||
| 65N4 | EG374380 | 2044 | F | Glycine hydroxymethyltransferase | gb|AAW45780.1 | Cryptococcus neoformans | 1.00E-156 |
| 60J18a | EG374421 | 1142 | P | Potential kynurenine 3-monooxygenase | gb|EAK98864.1 | Candida albicans | 2.00E-06 |
| 60J18b | EG374422 | 1220 | P | Potential kynurenine 3-monooxygenase | gb|EAK98864.1 | Candida albicans | 1.00E-12 |
| 58D15a | EG374299 | 897 | P | Saccharopine dehydrogenase | gi|70993695 | Aspergillus fumigatus | 2.00E-55 |
| 58D15b | EG374300 | 780 | P | Spermidine synthase | emb|CAD71251.1 | Neurospora crassa | 3.00E-78 |
| 2. Cell Defense | |||||||
| 35A16 | EG374447 | 1351 | F | Related to stress response protein | emb|CAD21425.1 | Neurospora crassa | 2.00E-23 |
| 3. Cell division/cycle | |||||||
| 80F12 | EG374389 | 1560 | F | Cell division control protein | gb|AAB69764.1 | Candida albicans | 2.00E-28 |
| 65O23 | EG374383 | 2037 | F | Cyclin c homolog 1 | ref|NP_596149.1 | Schizosaccharomyces pombe | 3.00E-07 |
| 4. Cell signaling/cell communication | |||||||
| 40D3 | EG374466 | 1534 | F | Autophagy-related protein | gb|AAW43831.1 | Cryptococcus neoformans | 6.00E-45 |
| 70C17a | EG374441 | 1206 | P | Fasciclin I family protein | gi|44890027 | Aspergillus fumigatus | 3.00E-06 |
| 58J15b | EG374311 | 807 | P | GTPase activating protein | gb|AAW43777.1 | Cryptococcus neoformans | 2.00E-09 |
| 55B10a | EG374277 | 861 | P | MAP kinase 1 | gb|AAO61669.1 | Cryptococcus neoformans | 3.00E-19 |
| 55B10b | EG374278 | 932 | P | MAP kinase | gb|AAU11317.1 | Alternaria brassicicola | 7.00E-74 |
| 65M20 | EG374379 | 1098 | F | Nucleoside-diphosphate kinase | emb|CAD37041.1 | Neurospora crassa | 9.00E-53 |
| 70E5 | EG374404 | 1766 | F | Serine/threonine kinase | gi|58262703 | Cryptococcus neoformans | 3.00E-61 |
| 10D13a | EG374414 | 1122 | P | Serine palmitoyl transferase subunit | gb|AAP47107.1 | Aspergillus nidulans | 4.00E-27 |
| 10D13b | EG374416 | 1170 | P | Serine palmitoyl transferase subunit | gb|AAP47107.1 | Aspergillus nidulans | 2.00E-18 |
| 30G12 | EG374337 | 1131 | F | Signal peptidase 18 KD subunit | emb|CAE76335.1 | Neurospora crassa | 3.00E-10 |
| 5. Cell Structure and growth | |||||||
| 58H22a | EG374306 | 920 | P | Beta-tubulin | emb|CAC83953.1 | Uromyces viciae-fabae | 3.00E-72 |
| 58H22b | EG374307 | 859 | P | Beta-tubulin | emb|CAC83953.1 | Uromyces viciae-fabae | 5.00E-68 |
| 10I12 | EG374325 | 1105 | F | Conidiation protein 6 | emb|CAD70456.1 | Neurospora crassa | 2.00E-10 |
| 30J9 | EG374343 | 1302 | F | Deacetylase | emb|CAD10036.1 | Cryptococcus neoformans | 2.00E-43 |
| 60C15 | EG374348 | 1456 | F | Deacetylase | gb|AAW47023.1 | Cryptococcus neoformans | 6.00E-35 |
| 65D17 | EG374372 | 1449 | F | Deacetylase | emb|CAD10036.1 | Cryptococcus neoformans | 4.00E-36 |
| 40F18 | EG374469 | 1117 | F | Deacetylase | emb|CAD10036.1 | Cryptococcus neoformans | 2.00E-31 |
| 55D17 | EG374475 | 1619 | F | Deacetylase | emb|CAD10036.1 | Cryptococcus neoformans | 5.00E-18 |
| 35C19b | EG374494 | 836 | P | Deacetylase | emb|CAD10036.1 | Cryptococcus neoformans | 6.00E-18 |
| 10C3 | EG374321 | 1479 | F | Deacetylase | gb|AAW47023.1 | Cryptococcus neoformans | 6.00E-26 |
| 35N24 | EG374461 | 783 | F | Hydrophobin | emb|CAD42710.1 | Davidiella tassiana | 5.00E-34 |
| 32H21a | EG374436 | 1176 | P | Intraorganellar peroxisomal translocation component Pay32p (PAY32) gene | gi|5821763 | Yarrowia lipolytica | 4.00E-32 |
| 40B22 | EG374465 | 1708 | F | Nuclear filament-containing protein | emb|CAA93293.1| | Schizosaccharomyces pombe | 5.00E-16 |
| 35G11a | EG374497 | 819 | P | Pria_lened pria protein | emb|CAA43289.1 | Lentinula edodes | 2.00E-12 |
| 65M2 | EG374413 | 2097 | F | UDP-glucose dehydrogenase | gb|AAS20528.1 | Cryptococcus neoformans | 1.00E-145 |
| 6. Energy/TCA cycle | |||||||
| 35D23b | EG374496 | 629 | P | 64 kDa mitochondrial NADH dehydrogenase | gb|AAW44492.1 | Cryptococcus neoformans | 1.00E-07 |
| 40H12 | EG374471 | 1249 | F | Iron-sulfur cluster Isu1-like protein | gb|AAQ98966.1 | Cryptococcus neoformans | 8.00E-56 |
| 55E23a | EG374279 | 957 | P | Mitochondrial ATPase alpha-subunit | gb|AAA33560.1 | Neurospora crassa | 6.00E-78 |
| 55E23b | EG374280 | 870 | P | Mitochondrial ATPase alpha-subunit | gb|AAA33560.1 | Neurospora crassa | 1.00E-101 |
| 90M15 | EG374409 | 1570 | F | Mitochondrial carrier family protein | gb|EAK95613.1 | Candida albicans | 1.00E-46 |
| 30N15a | EG374419 | 1078 | P | Succinate dehydrogenase flavoprotein subunit precursor | gb|AAW45324.1 | Cryptococcus neoformans | 1.00E-63 |
| 30N15b | EG374420 | 1143 | P | Succinate dehydrogenase flavoprotein subunit precursor | gb|AAW45324.1 | Cryptococcus neoformans | 1.00E-136 |
| 10A2 | EG374481 | 1114 | F | V-type ATPase subunit G | gb|AAB41886.1| | Neurospora crassa | 6.00E-15 |
| 7. Lipid metabolism | |||||||
| 65D3 | EG374370 | 1809 | F | Diacylglycerol O-acyltransferase | gi|58268157 | Cryptococcus neoformans | 1.00E-84 |
| 65G21a | EG374424 | 1078 | P | Fatty acid oxidoreductase | gb|AAW46114.1 | Cryptococcus neoformans | 2.00E-05 |
| 65G21b | EG374425 | 1149 | P | Fatty acid oxidoreductase | gb|AAW46114.1 | Cryptococcus neoformans | 3.00E-32 |
| 58J11b | EG374309 | 732 | P | Phosphatidyl synthase | gi|70999337 | Aspergillus fumigatus | 2.00E-20 |
| 8. Nucleotide metabolism | |||||||
| 58C19a | EG374297 | 827 | P | Uracil DNA N-glycosylase | gb|AAW41098.1 | Cryptococcus neoformans | 7.00E-16 |
| 58C19b | EG374298 | 857 | P | Uracil DNA N-glycosylase | gb|AAW41098.1 | Cryptococcus neoformans | 1.00E-19 |
| 9. Protein modification | |||||||
| 65B1 | EG374366 | 1847 | F | Carboxypeptidase | gi|19115337 | Schizosaccharomyces pombe | 7.00E-06 |
| 66B11a | EG374437 | 1145 | P | Endopeptidase | gb|AAW41068.1 | Cryptococcus neoformans | 2.00E-69 |
| 66B11b | EG374438 | 1200 | P | Endopeptidase | gb|AAW41068.1 | Cryptococcus neoformans | 1.00E-48 |
| 80N15 | EG374397 | 1944 | F | Translation elongation factor eEF-1 alpha chain | pir||S57200 | Puccinia graminis | 0.00E+00 |
| 10. Protein translational modification | |||||||
| 55N13 | EG374483 | 833 | F | Ubiquitin-conjugating enzyme | ref|NP_594859.1 | Schizosaccharomyces pombe | 7.00E-21 |
| 11. Ribosomal protein complex | |||||||
| 55B4 | EG374472 | 770 | F | 16S small subunit ribosomal RNA | gi|52699765 | Xanthoria elegans | 2.00E-08 |
| 35O22 | EG374462 | 938 | F | 18S ribosomal RNA | gi|21702995 | Gymnosporangium libocedri | 1.00E-154 |
| 60E22 | EG374352 | 1117 | F | 18S ribosomal RNA | gi|34493860 | Puccinia graminis f. sp.tritici | 3.00E-142 |
| 65C12 | EG374368 | 1136 | F | 18S ribosomal RNA | gi|34493860 | Puccinia graminis f. sp.tritici | 2.00E-66 |
| 90D5a | EG374432 | 1119 | P | 18S ribosomal RNA | gi|21724233 | Puccinia striiformis f. sp.tritici | 6.00E-102 |
| 90D5b | EG374431 | 1147 | P | ITS1, ITS2 and 5.8S ribosomal RNA | gi|3668067 | Tricholoma matsutake | 9.00E-54 |
| 58E11b | EG374302 | 831 | P | 25S ribosomal RNA | gi|169606 | Puccinia graminis f. sp. dactylis | 1.00E-09 |
| 23H10b | EG374283 | 1921 | F | 28S ribosomal RNA | gi|37703614 | Puccinia allii | 1.00E-83 |
| 35M12a | EG374458 | 763 | F | 28S ribosomal RNA | gi|21724230 | Puccinia graminis f. sp. tritici | 2.00E-14 |
| 35N2 | EG374460 | 917 | F | 28S ribosomal RNA | gi|46810582 | Fuscoporia viticola | 4.00E-06 |
| 35P13 | EG374463 | 888 | F | 28S ribosomal RNA | gi|86160913 | Melampsora epitea | 2.00E-16 |
| 40A4 | EG374464 | 951 | F | 28S ribosomal RNA | gi|58532805 | Puccinia carthami | 4.00E-05 |
| 55J11 | EG374479 | 957 | F | 28S ribosomal RNA | gi|21724233 | Puccinia striiformis f. sp. tritici | 2.00E-26 |
| 35I10b | EG374502 | 422 | P | 28S ribosomal RNA | gi|21914221 | Puccinia graminis | 5.00E-77 |
| 35I22a | EG374505 | 716 | P | 28S ribosomal RNA | gi|21914221 | Puccinia graminis | 2.00E-70 |
| 35I22b | EG374504 | 878 | P | ITS1, ITS2 and 5.8S ribosomal RNA | gi|21724233 | Puccinia striiformis f. sp.tritici | 5.00E-134 |
| 10G18 | EG374323 | 1108 | F | 28S ribosomal RNA | gi|84452427 | Cladosporium cladosporioides | 1.00E-59 |
| 30C19 | EG374333 | 1117 | F | 28S ribosomal RNA | gi|62005831 | Puccinia ferruginosa | 2.00E-13 |
| 30H3 | EG374340 | 1052 | F | 28S ribosomal RNA | gi|21724233 | Puccinia striiformis f. sp. tritici | 3.00E-71 |
| 30I12 | EG374341 | 1067 | F | 28S ribosomal RNA | gi|21724233 | Puccinia striiformis f. sp. tritici | 2.00E-39 |
| 30M20 | EG374347 | 1008 | F | 28S ribosomal RNA | gi|21914221 | Puccinia graminis | 1.00E-93 |
| 60J23 | EG374357 | 2112 | F | calnexin | gb|AAS68033.1 | Aspergillus fumigatus | 1.00E-133 |
| 12. Sugar/glycolysis metabolism | |||||||
| 30I15b | EG374418 | 617 | P | Glucose-repressible protein | emb|CAC28672.1 | Neurospora crassa | 2.00E-14 |
| 90C20 | EG374401 | 1130 | F | Glucose-repressible protein | gi|70996962 | Aspergillus fumigatus | 7.00E-18 |
| 55J22b | EG374287 | 887 | P | Glyoxal oxidase precursor | gb|AAW44259.1 | Cryptococcus neoformans | 2.00E-90 |
| 55J22a | EG374286 | 764 | P | Glyoxal oxidase precursor | gb|AAW41343.1 | Cryptococcus neoformans | 3.00E-30 |
| 90H16 | EG374405 | 1753 | F | Phosphopyruvate hydratase | gi|1086120 | Cladosporium herbarum | 1.00E-139 |
| 30K8 | EG374344 | 1547 | F | Transaldolase | gb|AAW46393.1 | Cryptococcus neoformans | 3.00E-95 |
| 13. Transcription factor | |||||||
| 58E6 | EG374485 | 1310 | F | TATA-box binding protein | gb|AAB57876.1 | Emericella nidulans | 7.00E-63 |
| 14. Transport metabolism | |||||||
| 65M6 | EG374378 | 1119 | F | Cation transport-related protein | gb|AAW42114.1 | Cryptococcus neoformans | 3.00E-13 |
| 15. virulence/infection related protein | |||||||
| 70I2 | EG374433 | 1952 | F | Cell wall glucanase | gi|70998053 | Aspergillus fumigatus | 2.00E-25 |
| 30M9 | EG374345 | 1162 | F | Differentiation-related/infection protein | gb|AAD38996.1 | Uromyces appendiculatus | 7.00E-11 |
| 80C7 | EG374385 | 1180 | F | Differentiation-related/infection protein | gb|AAD38996.1 | Uromyces appendiculatus | 1.00E-10 |
| 60E18 | EG374351 | 2147 | F | Pectin lyase | gb|AAA21817.1 | Glomerella cingulata | 2.00E-06 |
a F = full-length sequence and P = partial sequence.
Table 2.
cDNA clones showing homology to genes with characterized or unclassified proteins through Blastx search of the NCBI fungal databases
| Category & clone no. | GenBank accession | Size (bp) | Full length or partiala | Best hit in the NCBI databases | |||
| Protein | Accession | Organism | e-value | ||||
| 1. Amino acid metabolism | |||||||
| 35I14 | EG374455 | 766 | F | Cystathionine beta-lyase | gi|6636350 | Botryotinia fuckeliana | 5.70E+00 |
| 2. Cell Defense | |||||||
| 66C24a | EG374440 | 1175 | P | 88 kDa immunoreactive mannoprotein MP88 | gb|AAL87197.1 | Cryptococcus neoformans | 1.00E-03 |
| 3. Cell Division/cycle | |||||||
| 10F19 | EG374412 | 1877 | F | g1/s-specific cyclin pcl1 (cyclin hcs26) | gb|AAW44590.1 | Cryptococcus neoformans | 2.00E-04 |
| 4. Cell signaling/cell communication | |||||||
| 65G15 | EG374514 | 1106 | P | Protein kinase | gi|15072451 | Cryphonectria parasitica | 1.20E+00 |
| 30E21 | EG374336 | 1128 | F | Serine/threonine kinase | gi|22531808 | Ustilago maydis | 3.90E-01 |
| 65C6 | EG374367 | 1649 | F | Serine/threonine phosphatase | gi|33087517 | Hypocrea jecorina | 3.90E-01 |
| 80G5b | EG374428 | 1230 | P | Mitogen-activated protein kinase | gi|57227328 | Cryptococcus neoformans | 1.70E-00 |
| 5. Cell Structure and growth | |||||||
| 58G9 | EG374486 | 1714 | F | Beta tubulin | gi|47834278 | Penicillium flavigenum | 6.40E-00 |
| 40G6b | EG374274 | 888 | P | Cell wall protein | gi|68471254 | Candida albicans | 4.60E-01 |
| 58C4b | EG374296 | 819 | P | Cell surface protein | gi|70983232 | Aspergillus fumigatus | 2.60E-02 |
| 10D19 | EG374322 | 1212 | F | Cell wall mannoprotein | ref|NP_012685.1 | Saccharomyces cerevisiae | 1.00E-03 |
| 90I19 | EG374406 | 1240 | F | Cell wall mannoprotein | gi|6322611 | Saccharomyces cerevisiae | 1.50E-02 |
| 90C22 | EG374402 | 1641 | F | Cytoplasm protein | gb|AAW42379.1 | Cryptococcus neoformans | 1.00E-04 |
| 10I15 | EG374326 | 1088 | F | Mitochondrial outer membrane beta-barrel protein | gi|45758780 | Neurospora crassa | 1.70E-01 |
| 60H1 | EG374354 | 1035 | F | Nuclear pore complex subunit | gi|46437749 | Candida albicans | 5.00E-00 |
| 70I19a | EG374443 | 1132 | P | Nucleoskeletal-like protein | gi|172053 | Saccharomyces cerevisiae | 1.30E-01 |
| 6. Differentiation- related protein | |||||||
| 70A18 | EG374371 | 1207 | F | Differentiation-related protein | gb|AAD38996.1 | Uromyces appendiculatus | 6.00E-03 |
| 7. Mating type | |||||||
| 30M10 | EG374346 | 1025 | F | Mating type alpha locus | gi|73914085 | Cryptococcus gattii | 6.80E+00 |
| 30C22 | EG374334 | 1110 | F | Mating type alpha locus | gi|73914085 | Cryptococcus gattii | 7.50E+00 |
| 8. Nucleotide metabolism | |||||||
| 35K8 | EG374456 | 1572 | F | Ribonuclease H2 subunit | gi|6320485 | Saccharomyces cerevisiae | 9.00E+00 |
| 9. Protein translational modification | |||||||
| 100C10 | EG374490 | 1179 | F | Non-ribosomal peptide synthetase | gi|62006079 | Hypocrea virens | 1.20E+00 |
| 10. Ribosomal protein complex | |||||||
| 35L17 | EG374457 | 585 | F | 18S ribosomal RNA | gi|51102377 | Microbotryum dianthorum | 4.20E-02 |
| 40C19a | EG374512 | 706 | P | 18S ribosomal RNA | gi|28412377 | Leotiomycete sp. | 5.40E-01 |
| 35H2b | EG374500 | 786 | P | 26S large subunit ribosomal RNA | gi|30313824 | Pichia guilliermondii AjvM13 | 1.00E-03 |
| 35E4 | EG374451 | 897 | F | 28S ribosomal RNA | gi|46810582 | Fuscoporia viticola | 5.00E-03 |
| 35P11a | EG374506 | 667 | P | 28S ribosomal RNA | gi|62005826 | Puccinia artemisiae-keiskeanae | 1.00E-04 |
| 55B15 | EG374473 | 954 | F | 28S ribosomal RNA | gi|84794517 | Puccinia striiformoides | 3.60E-01 |
| 58B3 | EG374484 | 884 | F | 28S ribosomal RNA | gi|46810582 | Fuscoporia viticola | 3.30E-01 |
| 58N22 | EG374488 | 996 | F | 28S ribosomal RNA | gi|20452324 | Rhodotorula pilati | 3.30E-01 |
| 66I12 | EG374338 | 1167 | F | 28S ribosomal RNA | gi|46810582 | Fuscoporia viticola | 3.00E-04 |
| 80G5a | EG374427 | 1106 | P | Calnexin | gi|45551624 | Aspergillus fumigatus | 2.30E-00 |
| 11. Sugar/glycolysis metabolism | |||||||
| 58G18b | EG374304 | 796 | P | Pyruvate decarboxylase | gi|68480982 | Candida albicans | 1.40E+00 |
| 10N6 | EG374330 | 1029 | F | Pyruvate kinase | gi|168073 | Aspergillus nidulans | 6.00E+00 |
| 12. Transport metabolism | |||||||
| 30G15 | EG374339 | 1087 | F | Membrane zinc transporter | gi|47156070 | Aspergillus fumigatus | 5.70E-01 |
| 40H8a | EG374275 | 656 | P | amino acid transporter | gi|70985369 | Aspergillus fumigatus | 3.10E+00 |
| 80K19 | EG374395 | 1728 | F | Na+-ATPase | gi|1777377 | Zygosaccharomyces rouxii | 2.00E-04 |
| 55L18b | EG374289 | 845 | P | Peptide transporter | gi|70982509 | Aspergillus fumigatus | 5.30E-01 |
| 13. Unclassified | |||||||
| 80G10 | EG374391 | 1132 | F | Genomic sequence | gi|48056381 | Phakopsora pachyrhizi | 7.00E-53 |
| 04F9 | EG374470 | 1127 | F | Hypothetical protein | gi|71006713 | Ustilago maydis | 1.00E-06 |
| 10N10 | EG374331 | 1106 | F | Hypothetical protein | gi|58258450 | Cryptococcus neoformans | 6.00E-22 |
| 30I21 | EG374342 | 1906 | F | Hypothetical protein | gi|71023234 | Ustilago maydis | 1.00E-21 |
| 35B6 | EG374449 | 1060 | F | Hypothetical protein | gb|EAA67250.1 | Gibberella zeae | 1.00E-03 |
| 35C10 | EG374450 | 1465 | F | Hypothetical protein | gi|71004383 | Ustilago maydis 521 | 2.00E-08 |
| 35G21 | EG374454 | 1332 | F | Hypothetical protein | gb|EAK81105.1 | Ustilago maydis | 5.00E-09 |
| 35H2a | EG374499 | 758 | P | Hypothetical protein | gi|71021872 | Ustilago maydis | 1.80E+00 |
| 40B2a | EG374508 | 603 | P | Hypothetical protein | gi|85114517 | Neurospora crassa | 3.00E-05 |
| 40C12a | EG374510 | 792 | P | Hypothetical protein | gi|71019552 | Ustilago maydis | 4.00E-01 |
| 55L8 | EG374491 | 1417 | F | Hypothetical protein | gi|71004813 | Ustilago maydis | 1.50E-01 |
| 58C4a | EG374296 | 764 | P | Hypothetical protein | MGG_09875.5b | Magnaporthe grisea | 6.00E-12 |
| 60D4 | EG374350 | 1123 | F | Hypothetical protein | gi|50259357 | Cryptococcus neoformans | 7.00E-04 |
| 60I14 | EG374356 | 1565 | F | Hypothetical protein | gi|58263159 | Cryptococcus neoformans | 2.00E-09 |
| 60L15 | EG374359 | 2073 | F | Hypothetical protein | gb|EAA47832.1 | Magnaporthe grisea | 7.00E-10 |
| 60N2 | EG374363 | 1109 | F | Hypothetical protein | gi|46096746 | Ustilago maydis | 7.00E-03 |
| 60N6 | EG374364 | 1071 | F | Hypothetical protein | gi|49642978 | Kluyveromyces lactis | 8.00E-17 |
| 65H5 | EG374374 | 1390 | F | Hypothetical protein | gi|85095053 | Neurospora crassa | 1.40E+00 |
| 65I3 | EG374375 | 1870 | F | Hypothetical protein | gb|EAK86140.1 | Ustilago maydis | 1.00E-129 |
| 65O15 | EG374381 | 1893 | F | Hypothetical protein | gi|71006255 | Ustilago maydis | 1.10E+00 |
| 66B6 | EG374316 | 1263 | F | Hypothetical protein | gb|EAK81690.1 | Ustilago maydis | 1.00E-03 |
| 66B11a | EG374437 | 1145 | P | Hypothetical protein | AN2903.3b | Aspergillus nidulans | 3.00E-57 |
| 66B11b | EG374438 | 1200 | P | Hypothetical protein | FG10782.1b | Fusarium graminearum | 5.00E-49 |
| 66C18 | EG374327 | 2043 | F | Hypothetical protein | gb|EAA59593.1 | Aspergillus nidulans | 2.00E-12 |
| 70A3 | EG374360 | 1835 | F | Hypothetical protein | SS1G_14513.1b | Sclerotinia sclerotiorum | 8.00E-18 |
| 70C17b | EG374442 | 1191 | P | Hypothetical protein | AN0768.3b | Aspergillus nidulans | 1.00E-07 |
| 70H16 | EG374426 | 1121 | F | Hypothetical protein | gi|38100779 | Magnaporthe grisea | 2.60E+00 |
| 70I19b | EG374443 | 1190 | P | Hypothetical protein | NCU02808.2b | Neurospora crassa | 2.00E-08 |
| 70K15b | EG374320 | 933 | P | Hypothetical protein | gi|58261561 | Cryptococcus neoformans | 1.00E-07 |
| 70L24b | EG374446 | 1168 | P | Hypothetical protein | gb|EAA28928.1 | Neurospora crassa | 3.00E-23 |
| 80I9 | EG374394 | 1060 | F | Hypothetical protein | gi|58259618 | Cryptococcus neoformans | 1.50E+00 |
| 90O3 | EG374410 | 1725 | F | Hypothetical protein | gi|85119288 | Neurospora crassa | 1.20E-02 |
| 90O18 | EG374411 | 1973 | F | Hypothetical protein | CHG04543.1b | Chaetomium globosum | 4.00E-07 |
| 66C24b | EG374440 | 1271 | P | Macrofage activating glycoprotein | gi|15722495 | Cryptococcus neoformans | 3.00E-08 |
| 30E3 | EG374335 | 1406 | F | Probable gEgh 16 protein | emb|CAE85538.1 | Neurospora crassa | 8.00E-07 |
| 60I8 | EG374355 | 1039 | F | Related to ars binding protein 2 | gi|18376044 | Neurospora crassa | 6.60E+00 |
| 55J15b | EG374285 | 896 | P | Telomeric sequence DNA | gi|173051 | Saccharomyces cerevisiae | 2.00E-05 |
| 55E7 | EG374477 | 1253 | F | Unknown protein in chromosome E | gi|49654999 | Debaryomyces hansenii | 3.00E-06 |
| 55F15a | EG374281 | 461 | P | Unknown protein in chromosome G | gi|50427978 | Debaryomyces hansenii | 2.00E-03 |
| 60L20 | EG374361 | 1646 | F | Unknown protein in chromosome VI | gi|39975020 | Magnaporthe grisea | 3.00E-18 |
| 60N1 | EG374362 | 2024 | F | Unknown protein in chromosome 1 | gi|46110618 | Gibberella zeae | 2.00E-09 |
| 70F20 | EG374415 | 1818 | F | Unknown protein in chromosome III | gi|58270250 | Magnaporthe grisea | 1.60E+00 |
| 80M4 | EG374396 | 1985 | F | Unknown protein in chromosome G | gi|49657202 | Debaryomyces hansenii | 1.00E-03 |
| 80N10 | EG374430 | 563 | P | Phytochrome | gi|57337632 | Emericella nidulans | 4.30E-00 |
| 90B8 | EG374400 | 2011 | F | Unknown protein in chromosome G | gi|49657202 | Debaryomyces hansenii | 4.90E-02 |
| 90L21 | EG374408 | 2002 | F | Unknown protein in chromosome A | gi|49524079 | Candida glabrata | 1.20E+00 |
a F = full-length sequence and P = partial sequence.
b Data generated from Blastx search of the fungal database of the Broad Institute [34].
Table 3.
cDNA clones that produced no hit in the Blastx search of the NCBI fungal databases
| Category & Clone no. | GenBank accession | Size (bp) | Full length or partiala | Category & clone no. | GenBank accession | Size (bp) | Full length or partiala |
| 04A1 | EG374448 | 1188 | F | 55N9 | EG374482 | 1171 | F |
| 04C13 | EG374459 | 1423 | F | 55B9a | EG374292 | 585 | P |
| 04P11 | EG374434 | 1133 | F | 55B9b | EG374293 | 930 | P |
| 100B17 | EG374489 | 1137 | F | 58E11a | EG374301 | 542 | P |
| 10B5 | EG374492 | 1161 | F | 58G18a | EG374303 | 791 | P |
| 10C11 | EG374503 | 1235 | F | 58J11a | EG374308 | 672 | P |
| 10I7 | EG374324 | 1112 | F | 58J15a | EG374310 | 921 | P |
| 10K3 | EG374328 | 1687 | F | 58L3 | EG374487 | 959 | F |
| 10L3 | EG374272 | 1099 | F | 58M15a | EG374314 | 719 | P |
| 10N5 | EG374329 | 1090 | F | 58M15b | EG374315 | 718 | P |
| 10O19 | EG374332 | 1359 | F | 58M7a | EG374312 | 788 | P |
| 30I15a | EG374417 | 1032 | P | 58M7b | EG374313 | 934 | P |
| 32B15 | EG374294 | 1296 | F | 58N10a | EG374317 | 287 | P |
| 32H21b | EG374436 | 1249 | P | 58N10b | EG374318 | 837 | P |
| 35C19a | EG374493 | 739 | P | 60F10 | EG374353 | 1131 | F |
| 35D23a | EG374495 | 775 | P | 60L12 | EG374358 | 1239 | F |
| 35F14 | EG374453 | 971 | F | 60O23 | EG374365 | 1084 | F |
| 35F7 | EG374452 | 1086 | F | 65C23 | EG374369 | 2047 | F |
| 35G11b | EG374498 | 757 | P | 65G1 | EG374373 | 1631 | F |
| 35I10a | EG374501 | 807 | P | 65G15b | EG374514 | 1158 | P |
| 35P11b | EG374507 | 682 | P | 65I10 | EG374376 | 1010 | F |
| 40B2b | EG374509 | 860 | P | 65K18 | EG374377 | 1230 | F |
| 40C12b | EG374511 | 921 | P | 65P1 | EG374384 | 1814 | F |
| 40C19b | EG374513 | 857 | P | 66M21 | EG374349 | 1437 | F |
| 40E10 | EG374467 | 713 | F | 70C4 | EG374382 | 1518 | F |
| 40E23 | EG374468 | 734 | F | 70D12 | EG374393 | 1285 | F |
| 40G6a | EG374273 | 779 | P | 70K15a | EG374319 | 722 | P |
| 40H8b | EG374276 | 811 | P | 70L24a | EG374445 | 1104 | P |
| 50M2 | EG374305 | 1182 | F | 80D10 | EG374386 | 1147 | F |
| 55C20 | EG374474 | 868 | F | 80E22 | EG374388 | 2064 | F |
| 55E2 | EG374476 | 1272 | F | 80E4 | EG374387 | 1173 | F |
| 55F12 | EG374478 | 935 | F | 80F15 | EG374390 | 2129 | F |
| 55F15b | EG374282 | 865 | P | 80G19 | EG374392 | 1124 | F |
| 55J15a | EG374284 | 660 | P | 80N10a | EG374429 | 1091 | P |
| 55L18a | EG374288 | 930 | P | 80O12 | EG374398 | 1517 | F |
| 55M5 | EG374480 | 942 | F | 80O24 | EG374399 | 2098 | F |
| 55N22a | EG374290 | 813 | P | 90H10 | EG374403 | 1748 | F |
| 55N22a | EG374291 | 282 | P | 90K17 | EG374407 | 1896 | F |
a F = full-length sequence and P = partial sequence.
Identification of open reading frames
Various lengths of open reading frames (ORFs) were identified from 167 cDNA clones using the Lasergene sequence analysis software (DNASTAR package, WI. USA). The quality of the cDNA libraries with respect to the full-length (intactness) of cDNA was evaluated using three parameters: 1) identification of the 5'-end sequence structures of the insert, 2) ATG start site at their 5'-end for complete ORF contents and 3) Blastx evaluation of pre-determined ORF with corresponding amino acid sequences in the GenBank. Multiple ORFs with different length were frequently identified in a given cDNA sequence. When methionine was found aligned (including gaps) with first amino acid of a completed sequence (within the longest ORFs) with the first ATG start codon at the 5' end, a cDNA sequence was determined as a full-length transcript. Most of the cDNA sequences retained the specific 5'-end priming sequences (5'-CGGCCGGG-3'). A total of 128 complete ORFs were identified with first translation initiation codon ATG. The longest ORF was 951 bp, and the shortest ORF was 93 bp. The longest ORF sequence was selected from each analyzed cDNA and validated with the corresponding amino acid sequences to determine the genuine ORF. Four cDNA sequences were identified which contain incomplete ORF sequences, indicating incomplete transcripts for those cDNA clones. Nearly 86% of the cDNA sequences were found containing completed ORFs with a translation initiation codon (ATG). Each of the validated ORFs was able to translate into a continuous protein sequence with a translation initiation codon. This finding indicated high percentage of cDNA clones containing full-length transcripts with various sizes of ORFs in the cDNA library.
Discussion
A cDNA library can provide molecular resources for analysis of genes involved in the biology of a plant pathogenic fungus, such as genes responsible for the development, survival, pathogenicity and virulence. In order to initiate studies on the basic genome structure and gene expression of P. striiformis with infective state, we constructed a full-length cDNA library and a BAC library from urediniospores of a predominant race of P. striiformis f. sp. tritici [10]. The full-length cDNA library can be used to study the normal transcription profiles for the uredinial state, the biologically and epidemiologically essential stage of the fungus. The current cDNA library will serve as a major genetic resource for identifying and isolating full-length genes and functional units from the P. striiformis genome. Because this cDNA library was constructed from urediniospores of the pathogen, it should include expressed genes unique to this spore stage. Therefore, the cDNA library should have avoided EST limitations that are commonly generated by automatic assemblies of transcripts from different tissues. Controlled greenhouse conditions and careful handling of the plants and spores minimized possibility of contaminations by other fungal spores. Powdery mildew or leaf rust, which sometimes contaminates stripe rust spores, were not observed on the stripe rust – sporulating plants. Therefore, genes or cDNA sequences identified in this study should be from urediniospores of P. striiformis f. sp. tritici. This also was confirmed in a separate study, in which primers of all 12 randomly picked cDNA clones were successfully amplified clones in the BAC library constructed with the same race of the pathogen (data not shown).
A urediniospore of P. striiformis is an infectious structure that is critical for the rust to initiate the infection process. Although the fungus produces other spores, teliospores and basidiospores, they do not result in infection of host plants because the fungus does not have alternate hosts for basidiospores to infect. Compared to mycelium, a urediniospore is relatively more resistant to adverse environmental conditions. Therefore, the urediniospore stage should contain most of the pathogen genes involved in the pathogen development, survival and pathogenicity. Thus, our first full-length cDNA library for P. striiformis was constructed using urediniospores. Such transcript (gene) collection should include the genes that are important for the unique physical properties and characters of the urediniospores of P. striiformis. These genes are essential to maintain their germination and infective abilities. Therefore, the current full-length cDNA library would be one of the useful genomic resources for the functional genomic study of this important agricultural pathogen. Our full-length cDNA library reported here is the first large scale transcript collection for P. striiformis. As expression of certain genes are stage-specific and genes involved in plant-pathogen interactions express in haustoria [4,13], currently, we are working together with Scot Hulbert's lab to construct a full-length cDNA library from haustoria of the same stripe rust race used in this study.
The technology used in this study for full-length cDNA enrichment is robust and only requires less than 1 μg of starting total RNA. By using the MMLV reverse transcriptase, only the 5'-end tagged cDNAs are not prematurely terminated and can be amplified into full-length by an RNA oligo-specific primer [35,37]. The size fractionation process was modified in this study to generate large directional full-length cDNA inserts, which enriched full-length cDNA clones to have an insert size up to 9 kb. The enrichment of the full-length cDNA was achieved by PCR amplification following the cDNA synthesis. Because selection bias could favor the smaller cDNA, we used fewer PCR cycles to minimize such bias as previously suggested [35]. The conventionally constructed cDNA libraries rarely carry cDNA inserts over 2 kb, because the longer transcripts are often easily truncated during cDNA synthesis process, causing size bias against the larger cDNA fragments in cloning process. In our study, up to 22 PCR amplification cycles were used to generate adequate amount cDNA for cloning. The evaluation of cDNA insert size and its distribution showed a low level of insert size bias in the final cDNA library. Most of the cDNA inserts ranged from 500 bp to 1,500 bp, and there were high number of cDNA clones harboring inserts over 3,000 bp. Such results indicate that the size fraction is an effective selection approach to ensure the full-length cDNA content level in the cDNA library. The high quality of the initial total RNA and the optimal LD PCR conditions also resulted in low size bias level for the insert size distribution in this library. High quality and adequate amount of the initial mRNA is the key for yielding sufficient amount of the first strand full-length cDNA by reverse transcription. To reduce the redundancy and to avoid underrepresentation of different transcript species, cDNA fragments with different fractionated sizes were balanced and subjected to library construction. A considerable number of clones with an insert over 3 kb were found in our cDNA library, such big insert size is rarely found in conventional cDNA libraries.
The sequences of 5'-end transcripts are important for finding the signals for initiation of transcription. Irrespective of the length of cDNA, identification of the specific 5'-end nucleotide sequences in cDNA is commonly used to determine the full-length cDNA content and quality. In many cases, the 5'-end nucleotide sequences are referred to as a 5' cap structure [3,15,20,27]. We also found that nearly 95% of the cDNA clones contained the known 5'-end sequence : 5'-CGGCCGGG-3' (DB Clontech. USA), where as (G)3 at 3'-end will bind to the intact reveres transcripts which has nucleotide priming site CCC at its 5'-end. Completed ORFs were identified in cDNA sequence having the 5'-end sequence structure (5'-CGGCCGGG-3'). Presence of the ATG initiation codon aligned with amino acid methionine also was used as an indicator for the quality of full-length cDNA.
Blastx was used to search the entire NCBI GenBank with e-value of 10-5, which revealed 37% of the cDNA clones with high homologies to genes with known functions in the database. The relative low match rate to homologous genes from the blastx search might be due to the lack of gene information in the database for fungi. During the search process, the longest ORFs in each given cDNA sequence was also evaluated with amino acid alignments. The results showed that 86% of the cDNA clones contain ORFs with the translation initiation codon and stop codon. In addition, the existence of multi-exonic structure within some ORFs is additional evidence that supports their biological reality of genes or transcripts. The Kozak rules were found not totally applicable in determining ORFs in this study. Perhaps the Kozak rules are more suitable for analysis of mammalian genomes [22].
So far, there have been no other reports on the genome of P. striiformis in relation to function and biology of this important pathogen. In this study, we have identified genes encoding 51 different protein products involved in eleven aspects of the pathogen cell biology and plant infection. These genes are the first group of genes reported for the stripe rust pathogen. The genes identified for virulence/infection can be used in transient expression to confirm their function in pathogenicity. Although we sequenced only a small portion of the cDNA library, the study demonstrated the high efficiency of this procedure for the identification of putative genes of known function. As more and more genes with identified functions from other organisms are deposited into the databases, genes with important functions in P. striiformis should be more efficiently identified using our cDNA library. Even though sequences of only 196 clones were characterized in this study, we identified 19 cDNA clones encoding ribosomal RNA subunits, seven clones encoding deacetylase, and two clones encoding the glucose-repressible protein. The results may indicate the mRNA abundance of these genes. In this study, 10 cDNA clones had one of the two partial sequences with high homology (e-value ranging from 3E-06 to 5E-77) to genes identified in other fungi, but another partial sequence produced no hit. The results may indicate that these genes have very long sequences, and also may reflect that similar gene sequences in other fungi are mainly short EST sequences. When blastx search was conducted using other fungal genomic databases [34], seven cDNA clones, which produced no hit when blasted with the NCBI database, were identified to have some homology with unknown functions in various fungal species. In this study, we identified 37.2% of the clones with known genes, 18.4% encoding hypothetical proteins, and 25.5% no hit. These numbers are quite different from the 11%, 23%, and 66% of these categories, respectively, found in the urediniospore EST library of P. graminis f. sp. tritici, the wheat stem rust pathogen (L. Szabo, personal communication). The differences could be due to the clone sampling sizes of the studies and the different types of libraries (the full-length cDNA library for P. striiformis f. sp. tritici and conventional EST library for P. graminis f. sp. tritici). As more genes or ESTs from other Puccinia species infecting cereal crops become available, it will be more feasible to identify genes common to this group of the rust pathogens and also identify genes unique to particular species.
Conclusion
A full-length cDNA library was constructed using urediniospores of the wheat stripe rust pathogen. Using the library, we identified 51 genes involved in amino acid metabolism, cell defense, cell cycle, cell signaling, cell structure and growth, energy cycle, lipid and nucleotide metabolism, protein modification, ribosomal protein complex, sugar metabolism, transcription factor, transport metabolism, and virulence/infection. The results of function-identified genes demonstrated that the full-length library is useful in the study of functional genomics of the important plant pathogenic fungus. Research will be conducted to identify genes involved in the development, survival and pathogenicity of the pathogen using the cDNA library.
Methods
Total RNA isolation from urediniospores of P. striiformis f. sp. tritici
Urediniospores from race PST-78 of P. striiformis f. sp. tritici, a predominant race of the wheat stripe rust [11], were harvested from infected leaves 15 days after inoculation. The inoculation method and conditions for growing plants before and after inoculation were as described by Chen and Line [7]. For total RNA extraction, approximately 30 mg urediniospores were pre-chilled with liquid nitrogen in a glass vial. Spores were ground in liquid nitrogen with mortar and pestle, and then 10 mM Tris buffer (PH 8.0) was added. Ground frozen powder was transferred to an RNase-free microcentrifuge tube. The SV Total RNA Isolation kit (Pormega. Madison, WI. USA) was used to isolate total RNA from ground urediniospores. The extraction procedure recommended by the kit manufacturer was followed with slight modifications to adapt the use of fungal material. The quantity and purity of isolated total RNA was analyzed by 1% agarose gel electrophoresis and spectrophotometer.
Full-length cDNA synthesis and size fractionation
First-strand cDNA was synthesized from approximately 500 ng of total RNA using the Creator SMART cDNA Library Construction kit (DB Clontech. USA) following a slightly modified manufacturer's protocol. The first-strand cDNA mixture was used as template to synthesize double-stranded DNA with long distance (LD) PCR. PCR reactions were facilitated by 20 pmol of 5' end PCR primer containing sfiI A site (5'AAGCAGTGGTATCAACGCAGAGTGGCCATTACGGCCGGG-3'), and 20 pmol of CDSIII/3' end polyT PCR primers containing sfiI B site [5'-ATTCTAGAGGCCGAGGCGGCCGACATG-d(T)30N-1N-3']. In a 100 μL PCR reaction, 2 μL first-stranded cDNA were used as the template. The PCR reaction mixture contained 20 pmol of 10× PCR buffer, dNTP mix and 5 units of Taq polymerase. The LD PCR was performed in a GeneAmp 9600 thermal cycler (ABI Biosystem, USA) with the following program: denature at 95°C for 20 s followed by 22 cycles of 95°C for 5 s, 68°C for 6 min and 4°C soaking. The double stranded cDNA was then treated with proteinase K at 45°C for 20 min to inactivate the remaining DNA polymerase. The double stranded cDNA was then phenol-extracted and precipitated with 10 μL of 3 M sodium acetate, 1.3 μL of glycogen (20 μg/μL) and 2.5 volumes of 100% ethanol. Double stranded cDNA pellet was washed with 80% ethanol, air dried and suspended in 20 μL of water.
Double stranded cDNA was subjected to sfiI digestion, 100 μL sfiI digestion reaction containing 79 μL of cDNA, 10 μL 10× NE buffer 2 (New England Biolabs, USA) (10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol), 1 μL of 100× BSA (100 μg/ml) and 10 units of sfiI restriction enzyme (New England Biolabs, USA). Digestion was performed under 50°C for 2 h. Digested cDNA was size-fractionated on 1% agarose gel with 6 V/cm electrophoresis and the size fraction of 500 bp to 10 kb was excised. The excised gel slice was further divided into 5 zones (5 smaller gel slices) corresponding to a cDNA size ranging from 500 bp to 10 kb. Then cDNA in each gel slice was extracted and purified using the MinElute Gel Extraction kit (Qiagen, USA). The final cDNA concentration was adjusted to 5 ng/μl.
Construction of cDNA library
Approximately 30 ng sfiI-digested cDNA fragments were ligated to 100 ng of the pDNR-LIB cloning vector (DB Clontech, USA) using T4 DNA ligase (New England Biolabs, USA) under 16°C for 16 h. The ligation product was directly transformed into competent cell DH10B (Epicentre Technologies, USA) by electroporation. After 1 h SOC recovery incubation, transformed bacterial strain were grown on LB agar plates containing chloramphenicol (12.5 μg/ml), incubated at 37°C for 20 h. Since only the cDNA fragments with both sfiI A and sfiI B ends were allowed to be ligated into vector pDNR-LIB, only the recombinant clones were able to grow and were clearly identified as white colonies. The cDNA clones were randomly sampled and mini-prepared for a quality check using HindIII and EcoRI double-digestion to release inserts. The ligations with insert size larger than 500 bp were selected for large scale transformation. These colonies were subsequently picked and arrayed with a Q-Bot (Genetix, UK) into 384-well micro-titer plates. Each well on the culture plate contained 75 μl of LB freezing storage medium [360 mM K2HPO4, 132 mM KH2PO4, 17 mM Na citrate, 4 mM MgSO4, 68 mM (NH4)2SO4, 44% (v/v) glycerol, 12.5 μg/ml of chloramphenicol, LB]. Colonies were incubated at 37°C overnight, and then stored at -70°C.
Full-length cDNA library evaluation and cDNA clone sequence analysis
To evaluate the quality of the current full-length cDNA library, 400 individual cDNA clones were randomly picked from 12 storage plates, and grown in 5 ml of LB with 12.5 μg/ml of chloramphenicol under 37°C with 200 rpm shaking for 16 h. Plasmid DNA was isolated using the alkaline-lysis method [30] and digested with HindIII and EcoRI. The cDNA inserts were analyzed by 1% agarose gel electrophoresis with ethidium bromide staining. The average cDNA insert size and the cDNA length distribution profiles were obtained.
Two hundred cDNA clones were randomly selected for sequencing analysis. Prior to sequencing, all plasmids were isolated from cDNA bacterial clones by cellular lysis and purified in 96-well plates. Single pass sequencing was performed from both directions using two "in-house" sequencing primers. Phred software [16] was used for base calling. Each sequence was edited manually by removing vector sequences and the ambiguous reads. The overlapping sequences (from both 3' and 5' ends) were evaluated and aligned into full consensus sequence contigs using the DNA analyzing software DNA for Windows 2.2.1 [12]. The non-overlapping sequences were formatted and treated as two separated sequence contigs. All aligned sequence contigs were analyzed with the Lasergene 5.0 software (DNA STAR, Madison, WI, USA) for identifying ORFs. Consensus sequences were searched against the National Center for Biotechnology Information (NCBI) [28] fungal database and the all-organism database under E-value of 10-3 and 10-6, respectively. The genuine ORF fragments were cross validated by these two different scales of NCBI blast analysis.
Authors' contributions
PL constructed the full-length cDNA library, participated in the cDNA sequencing and analysis, and drafted the manuscript; MW contributed to cDNA sequencing, Blast-searching the databases, and drafted the manuscript; XC conceived and coordinated the study, contributed materials and resources, interpreted the data, and wrote the manuscript; KGC contributed resources and participated in planning the experiemnets. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This research was supported in part by the US Department of Agriculture (USDA), Agricultural Research Service (ARS), USDA-ARS Postdoctoral Program, and Washington Wheat Commission. PPNS No. 0440, Department of Plant Pathology, College of Agricultural, Human, and Natural Resource Sciences Research Center, Project No. 13C-3061-3923, Washington State University, Pullman, WA 99164-6430, USA. We thank the Sequencing Core Facility of Washington State University for the support of automated cDNA clone array, Dr. Pat Okubara for the assistance on the NCBI database blast search, Mr. Dat Q. Le for his technical assistance. We also are grateful to Dr. Lee Hadwiger and Dr. Weidong Chen for their critical review of the manuscript.
Contributor Information
Peng Ling, Email: pling@wsu.edu.
Meinan Wang, Email: meinan_wang@wsu.edu.
Xianming Chen, Email: xianming@wsu.edu.
Kimberly Garland Campbell, Email: kgcamp@wsu.edu.
References
- Calhoun DS, Arhana S, Vivar HE. Chemical control of barley stripe rust, a new disease for North America. Barley Newsl. 1988;32:109–112. [Google Scholar]
- Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, Muramatsu M, Hayashizaki Y, Schneider C. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;37:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Cono H, Okazaki Y, Muramatsu M, Hayashizaki Y. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapad discovery of new genes. Genome Res. 2000;10:1617–1630. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti AM, Dodds PN, Lawreance GJ, Ayliffe MA, Ellis JG. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell. 2000;18:243–256. doi: 10.1105/tpc.105.035980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XM. Epidemiology of barley stripe rust and races of Puccinia striiformis f. sp. hordei: the firstdecade in the United States. Cereal Rusts and Powdery Mildews Bulletin. 2004. http://www.crpmb.org/ 2004/1029chen.
- Chen XM. Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can J Plant Pathol. 2005;27:314–337. [Google Scholar]
- Chen XM, Line RF. Inheritance of stripe rust resistance inwheat cultivars used to differentiate races of Puccinia striiformis in North America. Phytopathology. 1992;82:633–637. [Google Scholar]
- Chen XM, Line RF, Leung H. Relationship between virulence variation and DNA polymorphism in Puccinia striiformis. Phytopathology. 1993;83:1489–1497. doi: 10.1094/Phyto-83-1489. [DOI] [Google Scholar]
- Chen XM, Line RF, Leung H. Virulence and polymorphic DNA relationships of Puccinia striiformis f. sp. hordei to other rusts. Phytopathology. 1995;85:1335–1342. doi: 10.1094/Phyto-85-1335. [DOI] [Google Scholar]
- Chen XM, Ling P. Towards cloning wheat genes for resistance to stripe rust and functional genomics of Puccinia striiformis f. sp. tritici. Proc of the 11th Intl Cereal Rusts and Powdery Mildew Conf, Norwich, England, 22–27 August 2004 Abstracts A210, Cereal Rusts and Powdery Mildews Bulletin.
- Chen XM, Moore M, Milus EA, Long DL, Line RF, Marshall D, Jackson L. Wheat stripe rust epidemics and races of Puccinia striiformis f. sp. tritici in the United States in 2000. Plant Dis. 2002;86:39–46. doi: 10.1094/PDIS.2002.86.1.39. [DOI] [PubMed] [Google Scholar]
- DNA for Windows http://www.dna-software.co.uk
- Dodds PN, Lawrence GJ, Catanzariti A, Ayliffe MA, Ellis JG. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell. 2004;16:755–768. doi: 10.1105/tpc.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin HJ, Stubbs RW. Epidemic spread of barley stripe rust in South America. Plant Dis. 1986;70:141–144. [Google Scholar]
- Edery I, Chu LL, Sonenberg N, Pelletier J. An efficient strategy to isolate full-length cDNAs based on an mRNA cap retention procedure (CAPture) Mol Cell Biol. 1995;15:3363–3371. doi: 10.1128/mcb.15.6.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Gubler U, Hoffman BJ. A simple and very efficient method for generating cDNA libraries. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hu GG, Linning R, Kamp A, Joseph C, McCallum B, Banks T, Cloutier S, Butterfield Y, Liu J, Kirkpatrick R, Stott J, Yang G, Smailus D, Jones S, Marra M, Schein J, Pei JM, Westwood T, Bakkeren G. Generation of a wheat leaf rust, Puccinia triticina, EST database and microarray from stage-specific cDNA libraries. Proc of the 11th Int Cereal Rusts and Powdery Mildew Conf, Norwich, England, 22–27 August 2004 Abstracts A147, Cereal Rusts and Powdery Mildews Bulletin.
- Humphrey HB, Hungerford CW, Johnson AG. Stripe rust (Puccinia glumarum) of cereals and grasses in the United States. J Agric Res. 1924;29:209–227. [Google Scholar]
- Kato S, Ohtoko K, Ohtake H, Kimura T. Vector-capping: a simple method for preparing a high-quality full-length cDNA library. DNA Res. 2005;12:53–62. doi: 10.1093/dnares/12.1.53. [DOI] [PubMed] [Google Scholar]
- Kato S, Sekine S, Oh SW, Kim NS, Umezawa Y, Abe N, Yokoyama KM, Aoki T. Construction of a human full-length cDNA bank. Gene. 1994;150:243–250. doi: 10.1016/0378-1119(94)90433-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mammalian Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- Lin KC, Bushnell WR, Szabo LJ, Smith AG. Isolation and expression of a host response gene family encoding thaumatin-like proteins in incompatible oat-stem rust fungus interactions. Mol Plant-Microbe Interact. 1996;9:511–522. doi: 10.1094/mpmi-9-0511. [DOI] [PubMed] [Google Scholar]
- Line RF. Stripe rust of wheat and barley in North America: a retrospective historical review. Ann Rev Phytopathol. 2002;40:75–118. doi: 10.1146/annurev.phyto.40.020102.111645. [DOI] [PubMed] [Google Scholar]
- Line RF, Qayoum A. Virulence, aggressiveness, evolution, and distribution of races of Puccinia striiformis (the cause of stripe rust of wheat) in North America, 1968–87. US Department of Agriculture Technical Bulletin. 1992;1788:44. [Google Scholar]
- Liu Z, Szabo LJ, Bushnell WR. Molecular cloning andanalysis of abundant and stage-specific mRNAs from Pucciniagraminis. Mol Plant Microbe Interact. 1993;6:84–91. doi: 10.1094/mpmi-6-084. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Sugano S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov
- Roelfs AP, Huerta-Espino J, Marshall D. Barley stripe rust in Texas. Plant Dis. 1992;76:538. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York; 1989. [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, Muramatsu M, Hayashizaki Y, Kawai J, Carninci P, Itoh M, Ishii Y, Arakawa T, Shibata K, Shinagawa A, Shinozaki K. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- Stubbs RW. Stripe rust. In: Roelfs AP, Bushnell WR, editor. The Cereal Rusts: Diseases, distribution, epidemiology and control. II. Academic Press, Orlando, FL; 1985. pp. 61–101. [Google Scholar]
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S. Construction and characterization of a full length-enriched and a 5'- end-enriched cDNA library. Gene. 1997;200:149–156. doi: 10.1016/S0378-1119(97)00411-3. [DOI] [PubMed] [Google Scholar]
- The Broad Institute http://www.broad.mit.edu
- Wellenreuther R, Schupp I, Poustka A, Wiemann S. SMART amplification combined with cDNA size fractionation in order to obtain large full-length clones. BMC Genomics. 2004;5:36. doi: 10.1186/1471-2164-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann S, Mehrle A, Bechtel S, Wellenreuther R, Pepperkok R, Poustka A. cDNAs for functional genomics and proteomics: the German cDNA Consortium. CR Biol. 2003;326:1003–1009. doi: 10.1016/j.crvi.2003.09.036. [DOI] [PubMed] [Google Scholar]
- Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]


