Abstract
An RNA-dependent RNA polymerase is packaged within the virions of purified vesicular stomatitis virus, a nonsegmented negative-strand RNA virus, which carries out transcription of the genome RNA into mRNAs both in vitro and in vivo. The RNA polymerase is composed of two virally encoded polypeptides: a large protein L (240 kDa) and a phosphoprotein P (29 kDa). Recently, we obtained biologically active L protein from insect cells following infection by a recombinant baculovirus expressing L gene. During purification of the L protein from Sf21 cells, we obtained in addition to an active L fraction an inactive fraction that required uninfected insect cell extract to restore its activity. The cellular factors have now been purified, characterized, and shown to be β and γ subunits of the protein synthesis elongation factor EF-1. We also demonstrate that the α subunit of EF-1 remains tightly bound to the L protein in the inactive fraction and βγ subunits associate with the L(α) complex. Further purification of L(α) from the inactive fraction revealed that the complex is partially active and is significantly stimulated by the addition of βγ subunits purified from Sf21 cells. A putative inhibitor(s) appears to co-elute in the inactive fraction that blocked the L(α) activity. The purified virions also package all three subunits of EF-1. These findings have a striking similarity with Qβ RNA phage, which also associates with the bacterial homologue of EF-1 for its replicase function, implicating a possible evolutionary relationship between these host proteins and the RNA-dependent RNA polymerase of RNA viruses.
Vesicular stomatitis virus (VSV), a prototype of nonsegmented negative-strand RNA viruses, has long been a paradigm for studying gene expression of this class of RNA viruses that infect vertebrates, invertebrates, and plants (1). Some of the most common human pathogens that belong to this category are rabies, measles, mumps, and human parainfluenza. A hallmark of all negative strand RNA viruses is the obligate packaging of an RNA-dependent RNA polymerase within the mature virions (2) that transcribes the genome RNA into mRNAs both in vitro and in vivo (3). For VSV, the virion-associated RNA polymerase is generally thought to consist of two virally encoded protein subunits, L (240 kDa) and P (29 kDa), which remain tightly complexed within the virion (3). Studies on the structure and function of VSV RNA polymerase have been greatly aided by the ability to isolate the polymerase subunits from the virions in a relatively pure form (4, 5). Active reconstitution of transcription is achieved by mixing the genome RNA enwrapped with the nucleocapsid protein (N) (referred to as N-RNA template) and purified L and P proteins (4, 5). From a large body of evidence, it appears that the L protein possesses the catalytic activity for RNA synthesis and the P protein is a transcription factor essential for L function (3); no cellular protein(s) has so far been shown to be required for the RNA polymerase activity. Only recently, by using the P protein expressed in Escherichia coli in an unphosphorylated form (6), has the role of phosphorylation in P protein function by a cellular protein kinase, identified as casein kinase II, been established (7). On the other hand, a detailed study on the structure and function of the RNA polymerase (L) is lacking because of its unavailability in large amounts. We have recently reported overexpression of L protein in Sf21 insect cells in a biologically active form by infection with recombinant baculovirus (8). During purification of the L protein from insect cells, in addition to a transcriptionally active L fraction (pool I), an inactive L fraction (pool II) was obtained that needed uninfected cell extract to restore RNA transcription activity in vitro (8). These findings prompted us to identify the putative cellular component(s) from insect cells needed for L activity. Here, we report that the novel cellular protein, the translation elongation factor EF-1, specifically associates with the RNA polymerase L for its activity.
MATERIALS AND METHODS
Cell Cultures and Virus.
Sf21 and HeLa cells were grown as described previously (8). VSV (Indiana serotype, Mudd–Summers strain) was grown in Sf21 cells following an infection at a multiplicity of infection of 1.0, and the released virus was purified (6). The recombinant L expressed in Sf21 cells was purified as detailed elsewhere (8).
Purification of Cellular Factor(s).
The L-activating factor was purified from uninfected Sf21 cells by using cell pellet obtained from a 200-ml culture (2 × 109). The pellet was suspended in 5 ml of lysis buffer A containing 25 mM Tris⋅HCl (pH 7.5), 1 mM DTT, 2 mM EDTA, aprotinin (2 μg/ml), leupeptin (2 μg/ml), E64 (1 μg/ml), and pepstatin (1 μg/ml) and disrupted in a Dounce homogenizer followed by centrifugation at 15,000 × g for 20 min. The supernatant was collected and diluted to 20 ml with buffer B containing 25 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 1 mM DTT and adjusted to 40% saturation with ammonium sulfate [(NH4)2SO4] followed by centrifugation at 15,000 × g for 30 min. The supernatant was brought to 80% saturation with (NH4)2SO4, and the pellet fraction was dissolved in buffer C containing buffer B plus 10% glycerol and dialyzed overnight against the same buffer. The dialyzed fraction was loaded onto a 5-ml DEAE-cellulose (DE-52) column previously equilibrated with buffer C, and the bound proteins were eluted by a linear gradient (50–500 mM NaCl) in the same buffer. Fractions were monitored for their ability to activate transcription by inactive L protein in pool II fraction as described before (8). The activity peak (at ≈0.2 M NaCl) was dialyzed in buffer C and loaded onto a 1.5-ml phosphocellulose column preequilibrated with the same buffer. The proteins were eluted by a linear gradient of 50–700 mM NaCl in buffer C. The active fractions were pooled and dialyzed against buffer D containing 25 mM potassium phosphate (KP) (pH 7.5), 1 mM DTT, and 10% glycerol. The pooled fraction was then loaded onto a 1-ml hydroxylapatite (HAP) column preequilibrated with buffer D followed by a wash with 1 M NaCl in the same buffer. The proteins were eluted stepwise in buffer D containing various KP concentrations.
Microsequencing.
Protein sample (1 M KP eluate) was electrophoresed in a 10% Laemmli gel at constant power. The protein bands were transferred to the poly(vinylidene difluoride) membrane as described elsewhere (9), cut out after staining with Coomassie blue R-250, and subjected to trypsin digestion. The oligopeptides were purified by HPLC, and the individual peptides were microsequenced in an Applied Biosystems microsequencer, model 470A attached to an on-line model 120A phenylthiohydantoin analyzer (10).
Western Blot and Immunoprecipitation.
Western blot analysis was performed essentially as described (8) with minor modifications. In the case of anti-EF-1 antibodies, the blocking and wash buffers contained 0.3 M NaCl and 1% Nonidet P-40. Anti-Artemia EF-1 antibodies against different subunits were raised individually in rabbit (11). The immunoprecipitation was carried out in a buffer containing 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, and 0.25% Nonidet P-40, according to the method described elsewhere (12).
RESULTS
Characterization of Cellular Factor(s).
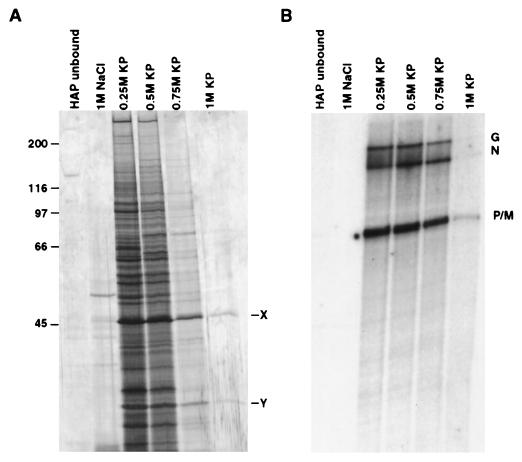
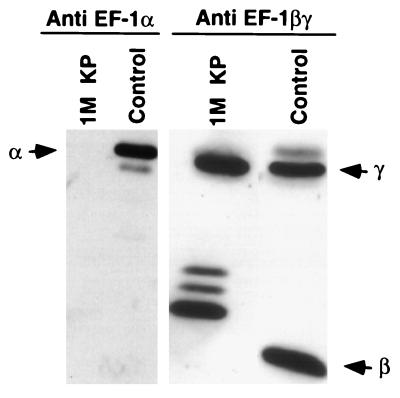
To purify the putative cellular factor(s) needed for the restoration of L activity in the inactive pool II fraction, the cytoplasmic extract prepared from uninfected Sf21 cells was subjected to a series of column chromatographic separation procedures as detailed under Materials and Methods. At the final purification step in HAP column chromatography, only two protein bands (46 kDa, X, and 32 kDa, Y) were visible in 1 M KP eluate, which restored transcription activity when added to the inactive pool II fraction (Fig. 1 A and B). Based on the calculated specific activity of the 0.25 M KP fraction, a 10-fold purification of the activity was achieved in the 1 M KP fraction. The specific activity of protein bands X and Y remained the same in each fraction. These protein bands (X and Y) were then blotted onto a poly(vinylidene difluoride) membrane (9) and subjected to microsequence analyses (10) followed by a homology search from a protein sequence database with the blast program. As shown in Table 1, the three peptide fragments derived from protein band X were 72–88% identical in amino acid sequence to the protein synthesis elongation factor EF-1γ of brine shrimp, Artemia salina (13). On the other hand, the two peptide fragments derived from protein band Y bear 82 and 88% amino acid sequence identity with the protein synthesis elongation factor EF-1β of silkworm (14). These results strongly suggested that the β and γ subunits of EF-1, which normally remain as a complex (11), were directly involved in activating the L protein in pool II fraction. To establish that the protein bands X and Y were indeed the γ and β subunits of EF-1, respectively, we carried out Western blot analyses of the 1 M KP eluate fraction by using antibodies raised against α, β, and γ subunits of EF-1 of A. salina (11), which are highly homologous to the insect cell EF-1 subunits (14). As shown in Fig. 2, the 1 M KP fraction cross-reacted with anti-EF-1βγ antibody but not with anti-EF-1α antibody, indicating that the fraction contained only β and γ subunits of EF-1. It should be noted that the β subunit of Sf21 routinely displayed multiple bands that migrated more slowly than the Artemia β subunit. The reason for these differences is presently unknown. Thus, based on microsequence and Western blot analyses it seems that protein bands X and Y correspond to γ and β subunits, respectively, of EF-1, which are involved in the activation of L protein. This conclusion is further supported by the fact that the βγ complex purified from A. salina (11) activated pool II L protein (see Fig. 7).
Figure 1.
Purification of cellular factor(s). Cytoplasmic extract prepared from uninfected Sf21 cells was fractionated through a column chromatography series as detailed under Materials and Methods. (A) An equal volume (30 μl) of each fraction from the HAP column was electrophoresed and stained with silver reagent. (B) In vitro transcription reconstitution of each fraction was carried out with 0.5 μg of N-RNA template, 0.1 μg of recombinant P, 0.5 μg of pool II L, and 7 μl of each HAP fraction. [32P]UMP-labeled RNA products were analyzed on a 5% urea/polyacrylamide gel, and the transcripts (G, N, P, and M) were visualized by autoradiography. X and Y represent protein bands eluted at 1 M KP fraction. Molecular weight markers are indicated.
Table 1.
Microsequence analysis of protein bands X and Y
| Protein band | Microsequence of peptides | Protein obtained through search | Percent identity |
|---|---|---|---|
| X, ≈46 kDa | (i) VAPNFVFGETNK | Artemia EF-1γ | 75 |
| (ii) VPAYESADG | 88 | ||
| (iii) TFLVTER | 72 | ||
| Y, ≈32 kDa | (i) QADFQVFQQVG | Silkworm EF-1β | 82 |
| (ii)NYVSGYTP | 88 |
The 1 M KP eluate from HAP column (Fig. 1A) was concentrated and electrophoresed in a 10% SDS/polyacrylamide gel. The protein bands (X and Y) were then transferred onto a poly(vinylidene difluoride) (PVDF) membrane and subjected to peptide digestion followed by microsequencing analyses. The peptide sequences obtained from each protein band were used to search for similar sequences in the protein databank by using the blast program. The amino acid residues in boldface indicate the exact match, and the underlined residues correspond to conservative changes.
Figure 2.
Immunoblot analysis of protein bands X and Y in 1 M KP fraction. An equal volume (30 μl) of the fraction was electrophoresed in a 10% SDS/polyacrylamide gel and transferred to nitrocellulose membrane for Western blot analysis with subunit-specific antibodies against Artemia EF-1, and protein bands were visualized by ECL. In control lanes, purified subunits from Artemia were used. Positions of Artemia α, β, and γ subunits are shown by arrows.
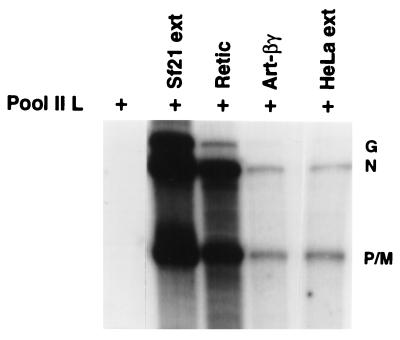
Figure 7.
Transcriptional activation of pool II L by mammalian cell extract. The in vitro VSV transcription reaction was carried out as described in Fig. 1. The cytoplasmic extract of Sf21 or HeLa cells (10 μg), rabbit reticulocyte lysate (5 μl), and βγ purified from A. salina (25 ng) were added to the transcription reactions. The transcription products were analyzed as in Fig. 1.
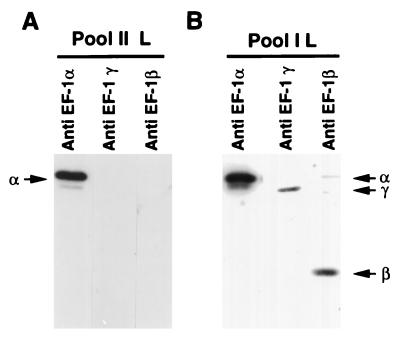
Because the L protein in pool II fraction required a βγ subunit for transcription, it was important to show that this fraction is indeed devoid of these subunits. By Western blot analyses, it was apparent that the βγ subunits were absent in pool II fraction, but interestingly, this fraction contained the other subunit EF-1α, indicating that the α subunit is possibly bound to L protein or co-purified with it during purification (Fig. 3A). The transcriptionally active pool I fraction (8) contained all three subunits (Fig. 3B), strongly suggesting that the L protein which remained complexed with αβγ subunits of EF-1 in pool I must have lost the βγ subunits and eluted in the pool II fraction as an inactive L(α) complex.
Figure 3.
Western blot analyses of pool I and pool II L. Twenty microliters of pool II L (A) or 25 μl of pool I L (B) were electrophoresed in a 10% SDS/polyacrylamide gel and transferred to a nitrocellulose membrane. The membranes were probed with subunit-specific antibodies against Artemia EF-1, and protein bands were visualized by ECL. Positions of Sf21 α, β, and γ subunits are shown by arrows.
Specific Association of EF-1 with the L Protein.
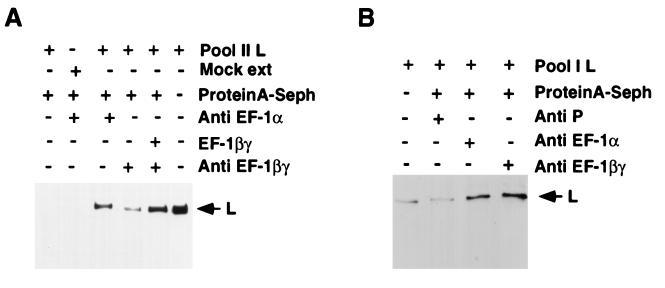
To directly test whether EF-1α is indeed bound to the L protein, we treated pool II fraction with anti-EF-1α antibody, and the immune complex was analyzed for L protein by Western blot analyses by using anti-L antibody. As shown in Fig. 4A, anti-EF-1α precipitated L protein (>50%), indicating that the α subunit is bound to a major fraction of the L protein. In a control experiment, anti-EF-1βγ antibody precipitated a trace amount of L protein; however, the amount of L protein substantially increased when immunoprecipitation was carried out after the addition of purified βγ fraction (1 M KP eluate). Thus, we conclude that βγ subunits of EF-1 bind to the L protein either directly or via the α subunit to become an integral part of the RNA polymerase L. As expected, similar analyses on active pool I fraction indicated that all three subunits were also bound to the L protein (Fig. 4B). It seems that the L protein has a high affinity for the α subunit, which remains tightly associated with the L protein even at high ionic strength (>0.7 M NaCl), whereas the β and γ subunits are dissociated from the L(α) complex at this salt concentration. Additional co-immunoprecipitation studies clearly demonstrated that P protein was able to bind L(α) in pool II fraction (data not shown), suggesting that the P protein can interact with L(α) in the absence of βγ complex. Taken together, these results strongly suggest that the active RNA polymerase of VSV is composed of L protein associated with three cellular proteins, EF-1αβγ, and forms the active holoenzyme with P protein. An approximately equimolar amount of the αβγ subunits was found to be complexed with the L protein at optimal RNA polymerase activity, as determined by quantitation of the associated subunits by Western blot analyses with a known amount of the subunit proteins as control (data not shown).
Figure 4.
Association of EF-1 with pool I and pool II. (A) Twenty microliters of pool II L either alone or after incubating with 30 μl of 1 M KP eluate (purified βγ) was immunoprecipitated with subunit-specific antibodies against Artemia EF-1 and protein A-Sepharose beads. (B) Immunoprecipitation of a similar volume of pool I L was carried out as described above. The immunoprecipitates were analyzed on a 10% SDS/polyacrylamide gel followed by immunoblotting and probing with an L-specific peptide antibody. The blot was detected by ECL. Mock extract, protein A-Sepharose, and anti-P antibody (B) were used as controls. The migration position of L protein is shown by the arrow.
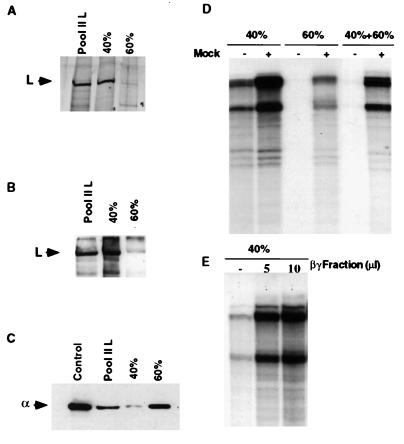
Purification of L(α) Complex from Pool II Fraction.
The above results indicate that the L(α) complex in pool II is transcriptionally inactive but requires βγ complex to regain activity. Alternatively, a possibility exists that L(α) by itself is active, but a putative inhibitor present in this fraction blocks its activity, and the βγ subunit restores that by sequestering the inhibitor with concomitant complex formation with L(α). Accordingly, we further purified the L(α) complex by (NH4)2SO4 fractionation of pool II. More than 90% of L protein was recovered from pool II in 40% saturated (NH4)2SO4 fraction, as determined by silver staining (Fig. 5A) and by Western blot analyses (Fig. 5B). However, a major portion (more than 80%) of α subunit in pool II was recovered in 40–60% (NH4)2SO4, although a small but significant amount of α remained associated with the L protein in 40% (NH4)2SO4 fraction (Fig. 5C) with an approximate molar ratio of 1:1. Interestingly, the L(α) complex in 40% (NH4)2SO4 fraction was found to be active in transcription, albeit partially (Fig. 5D); however, addition of mock extract (Fig. 5D) or purified βγ fraction (Fig. 5E) significantly stimulated transcription by approximately 5-fold. The 40–60% (NH4)2SO4 fraction, on the other hand, resembled the inactive pool II fraction, i.e., transcription was dependent on the addition of mock extract (Fig. 5D). Moreover, addition of 40–60% fraction to active 40% fraction strongly inhibited transcription, indicating that the former fraction contained a potent inhibitor of RNA polymerase L (Fig. 5D). Thus, it seems that L(α) by itself is partially active but remains inactive in pool II because of the presence of a putative inhibitor co-eluting with the L protein in pool II. The βγ subunits possibly sequester the inhibitor as well as complex with the L(α) (Fig. 4) to form the stable holoenzyme L(αβγ) for optimal transcription.
Figure 5.
Fractionation of pool II L. The pool II L was treated with (NH4)2SO4 to 40% saturation, and the precipitate was recovered by centrifugation. The supernatant fraction was brought to 60% saturation and the precipitate was recovered. The precipitates from both 0–40% and 40–60% fractions (indicated as 40% and 60%, respectively) were dissolved in 0.5 ml of buffer C and dialyzed against the same buffer as described under Materials and Methods. (A) 20 μl of each fraction was electrophoresed in a 10% polyacrylamide gel followed by staining with silver reagent. Immunoblots of the gel (A) with either anti-L antibody (B) or anti-EF-1α antibody (C) are shown. (D) In vitro transcription was carried out with 0.5 μg of N-RNA template, 0.1 μg of recombinant P, and 10 μl of each fraction in the presence of either mock extract (3 μg of total protein) or purified βγ fraction (E). RNA products were analyzed as described in Fig. 1. The position of the Artemia α subunit is shown in a control lane.
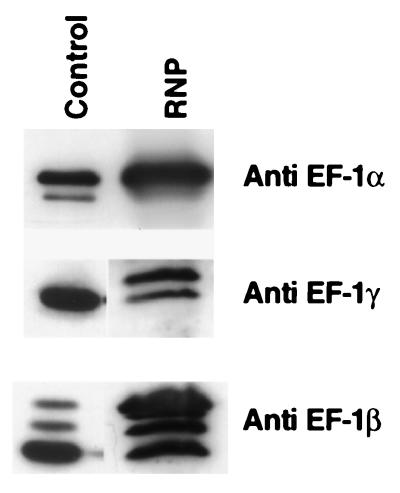
Packaging of EF-1 in the Purified Virion.
Because purified ribonucleoprotein (RNP) complex of VSV is transcriptionally active (15), we next tested whether the subunits of EF-1 are also bound to the L protein during virus maturation and subsequently packaged within the transcribing RNP. VSV was grown in Sf21 cells, and the recovered virus was purified. As shown in Fig. 6, the RNPs from the virions contained all three subunits of EF-1 by Western blot analyses. Although multiple bands of β were seen as observed before (Fig. 2), two bands were detected for the γ subunit in RNP. It is presently not known whether these multiple bands correspond to different isoforms of the subunits. It is important to mention that the α subunit is present in a larger amount in different preparations of RNP compared with the other two subunits, confirming the tight association of α subunit with the viral L protein as observed for recombinant L purified from insect cells.
Figure 6.
Packaging of EF-1 in VSV virion. The Sf21 cells were infected with VSV at a multiplicity of infection of 1.0, and the virus was purified and RNP prepared as described earlier (5). Purified RNP (40 μg), pool II L serving as a control for Sf21 α, and HAP fraction (1 M KP eluate) serving as a control for Sf21 βγ were subjected to SDS/PAGE followed by immunoblot analysis with subunit-specific antibodies against Artemia EF-1. The bands were visualized by ECL.
Effect of Mammalian Extracts in L Activity.
Finally, we investigated whether mammalian extracts (the natural hosts of VSV) containing the homologous EF-1 also activate L protein. We used reticulocyte lysate (rich in translation elongation factors) (16) and HeLa cell extract in restoring L activity in pool II fraction. As shown in Fig. 7, reticulocyte lysate stimulated transcription almost to the same level as the Sf21 extract, whereas HeLa cell extract and βγ complex purified from A. salina stimulated to the same extent, albeit at a lower level. These results suggest that reticulocyte lysate or HeLa cells presumably contain the elongation factor EF-1 β and γ subunits and effectively form an active complex with the insect cell α subunit, which is in complex with the L protein in pool II fraction. In support of this contention, we found that EF-1βγ partially purified from rabbit reticulocyte lysate also effectively complemented the insect cell-derived L(α) complex (pool II) in stimulating transcription in vitro (data not shown).
DISCUSSION
Overexpression of the RNA-dependent RNA polymerase (L) of VSV in recombinant baculovirus-infected insect cells in a biologically active form (8) provides us an opportunity to study structure and function as they relate to the transcription and replication of the viral genome RNA. During purification of recombinant L protein from the insect cells, we previously made an observation that a fraction (pool II) containing L protein was inactive unless uninfected cell extracts were added to it (8). In this communication, we purified the putative cellular proteins and identified them as the protein synthesis elongation factor βγ subunits (Figs. 1 and 2). Interestingly, the inactive L protein in pool II contained the other major subunit, α, tightly complexed with the L protein (8), leading us to conclude that inactivity of the L protein was because of removal of the βγ complex from the active complex L(αβγ). This conclusion was further supported by the findings that the active pool I contained L protein associated with αβγ subunits (Figs. 3 and 4). However, subsequent purification of L(α) from the inactive pool II fraction revealed that a putative inhibitor(s) co-eluted in this fraction and inhibited L(α), which is otherwise partially active in transcription (Fig. 5). Thus, the restoration of L(α) activity in pool II by the addition of βγ subunit appears to be because of possible sequestration of the putative inhibitor(s) by the βγ subunits. However, the observed association of purified βγ with L(α) (Fig. 4A) as well as complex formation of all three subunits with L in active pool I fraction (Fig. 4B) suggest that the βγ subunits are probably required to restore full activity of L(α). Thus, it can be postulated that L(α) is the RNA polymerase core enzyme and the βγ subunits help form a fully active and stable L(αβγ) holoenzyme. Further studies will be needed to establish the precise role of EF-1 subunits in L activity. The nature of the putative inhibitor is currently unknown. It appears to be present in uninfected insect cells (data not shown), and a detailed study to characterize the inhibitor is now under way.
Although the L protein in both fractions (pool I and II) can be immunoprecipitated by antibody against αβγ, it is unclear at the present time whether all L molecules are complexed with the EF-1 subunits. So far we have not been able to completely dissociate the α subunit from the L protein, leading us to conclude that it has a strong affinity for the L protein. Thus, it is difficult to precisely determine the molar ratio of the subunits in the active L fraction, although by a rough approximation it appears to be equimolar. Once each of the EF-1 subunits is separately purified and L protein devoid of EF-1 is obtained, reconstitution experiments should reveal the precise composition of the holoenzyme. It is important to note that we have routinely observed the presence of δ, the remaining subunit of EF-1 (11, 14, 16) in the active pool I fraction (data not shown), suggesting its possible involvement in L function.
The fact that the EF-1 subunits are packaged within the purified virions (Fig. 6) strongly suggests that they are essential for L function and play an important role in the VSV transcription process. Moreover, stimulation of transcription of L(α) in pool II by the elongation factors present in rabbit reticulocyte lysate (Fig. 7) or purified from it (data not shown) also indicates that the mammalian counterpart of these cellular components is important for L protein activity. It is noteworthy that antibodies raised against the elongation factor subunits purified from rabbit reticulocyte lysate poorly cross-react with the corresponding insect cell subunits (unpublished observation), due primarily to the lack of homology between the corresponding subunits of the two species. Use of antibodies against mammalian elongation factor subunits would certainly help us understand their roles in mammalian cell context.
The involvement of the three subunits of EF-1 in RNA polymerase activity of VSV bears a striking similarity with phage Qβ replicase, which also requires multiple host-encoded proteins as subunits (17). Two of them are the bacterial homologues of the translation elongation factors Ts and Tu, and the other is ribosomal protein S1. These host proteins were shown to be essential for the replicase to recognize the template, initiate RNA synthesis, and play a fundamental structural role in the replicase function (18, 19). Our findings that the RNA polymerase of a distantly evolved animal RNA virus may also require functionally similar host proteins for its activity provide an evolutionary insight into this unique host protein–virus interaction. It is tempting to speculate that a variety of RNA-dependent RNA polymerases may have evolved from a common ancestor, and an intimate interaction with their respective host EF-1 subunits, perhaps around the active site, is fundamental to the preservation of the RNA polymerase activity.
Finally, the association of α, β, and γ subunits of EF-1 with the L protein for its activity may now help us explain some earlier unexplained observations in the VSV transcription reaction. For example, inconsistent transcription activity demonstrated by the L protein purified from different batches of the virion may have been because of removal of one or more of the associated EF-1 subunits from the L protein packaged within the virion. The EF-1 subunits associated with the L protein in the RNP remained undetected in earlier studies, possibly because of their co-migration with viral nucleocapsid protein N (49 kDa) and matrix protein M (30 kDa). The GTP/GDP binding property of the α subunit may explain the mechanism of the unique 5′-capping reaction involving GDP (20, 21). Additionally, the observed protein kinase activity of βγ complex (13, 22) may provide clues to the origin of the elusive L-associated kinase activity (23, 24). The demonstrated strong affinity of the βγ subunit for tubulin (11) may also provide insight into the mechanism of interaction of the L protein with the other RNA polymerase subunit, the P protein, via the N-terminal acidic domain (25). Thus, a detailed study along these lines would certainly help us understand the structure and function of the VSV RNA polymerase.
Acknowledgments
We thank Dr. Kunio Misono for protein sequence analysis, Laura Tripepi for secretarial assistance, and Dorthy Herzberg for editorial help. This work was supported by National Institutes of Health Grant AI 26585 (to A.K.B.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: VSV, vesicular stomatitis virus; KP, potassium phosphate; HAP, hydroxylapatite; RNP, ribonucleoprotein.
References
- 1.Wagner R R, Rose J K. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 1. New York: Raven; 1996. pp. 1121–1135. [Google Scholar]
- 2.Baltimore D, Huang A S, Stampfer M. Proc Natl Acad Sci USA. 1970;66:572–576. doi: 10.1073/pnas.66.2.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee A K, Barik S. Virology. 1992;188:417–428. doi: 10.1016/0042-6822(92)90495-b. [DOI] [PubMed] [Google Scholar]
- 4.Emmerson S U, Yu Y-H. J Virol. 1975;15:1348–1356. doi: 10.1128/jvi.15.6.1348-1356.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De B P, Banerjee A K. Biochem Biophys Res Commun. 1985;126:40–49. doi: 10.1016/0006-291x(85)90568-6. [DOI] [PubMed] [Google Scholar]
- 6.Barik S, Banerjee A K. J Virol. 1991;65:1719–1726. doi: 10.1128/jvi.65.4.1719-1726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barik S, Banerjee A K. Proc Natl Acad Sci USA. 1992;89:6570–6574. doi: 10.1073/pnas.89.14.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathur M, Das T, Banerjee A K. J Virol. 1996;70:2252–2259. doi: 10.1128/jvi.70.4.2252-2259.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsudaira P J. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 10.Edman P, Begg G. Eur J Biochem. 1967;1:80–90. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- 11.Janssen G M C, Moller W. Eur J Biochem. 1988;171:119–129. doi: 10.1111/j.1432-1033.1988.tb13766.x. [DOI] [PubMed] [Google Scholar]
- 12.Das T, Schuster A, Schneider-Schaulies S, Banerjee A K. Virology. 1995;211:218–226. doi: 10.1006/viro.1995.1394. [DOI] [PubMed] [Google Scholar]
- 13.Maessen G D F, Amos R, Zeelen J P, Moller W. FEBS Lett. 1987;223:181–186. doi: 10.1016/0014-5793(87)80532-x. [DOI] [PubMed] [Google Scholar]
- 14.Taira H, Kamiie K, Kakuta A, Ooura H, Matsumoto S, Ejiri S, Katsumata T. Nucleic Acids Res. 1992;20:6734. doi: 10.1093/nar/20.24.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abraham G, Banerjee A K. Virology. 1976;71:230–241. doi: 10.1016/0042-6822(76)90108-2. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho J F, Carvalho M G C, Merrick W C. Arch Biochem Biophys. 1984;234:591–602. doi: 10.1016/0003-9861(84)90309-6. [DOI] [PubMed] [Google Scholar]
- 17.Blumenthal T, Carmichael G G. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 18.Blumenthal T, Hill D. J Biol Chem. 1980;255:11713–11716. [PubMed] [Google Scholar]
- 19.Blumenthal T. Proc R Soc London Ser B. 1980;210:321–335. doi: 10.1098/rspb.1980.0137. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee A K. Microbiol Rev. 1980;44:175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shuman S. Virology. 1997;227:1–6. doi: 10.1006/viro.1996.8305. [DOI] [PubMed] [Google Scholar]
- 22.Ejiri S, Honda H. Biochem Biophys Res Commun. 1985;128:53–60. doi: 10.1016/0006-291x(85)91643-2. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez D, De B P, Banerjee A K. J Gen Virol. 1985;66:1024–1036. [Google Scholar]
- 24.Hammond D C, Haley B E, Lesnaw J A. J Gen Virol. 1992;73:67–75. doi: 10.1099/0022-1317-73-1-67. [DOI] [PubMed] [Google Scholar]
- 25.Chattopadhyay D, Banerjee A K. Proc Natl Acad Sci USA. 1988;85:7977–7981. doi: 10.1073/pnas.85.21.7977. [DOI] [PMC free article] [PubMed] [Google Scholar]