Abstract
We report an optimized synthesis of all canonical 2′-O-TOM protected ribonucleoside phosphoramidites and solid supports containing [13C5]-labeled ribose moieties, their sequence-specific introduction into very short RNA sequences and their use for the structure determination of two protein–RNA complexes. These specifically labeled sequences facilitate RNA resonance assignments and are essential to assign a high number of sugar–sugar and intermolecular NOEs, which ultimately improve the precision and accuracy of the resulting structures. This labeling strategy is particularly useful for the study of protein–RNA complexes with single-stranded RNA in solution, which is rapidly an increasingly relevant research area in biology.
INTRODUCTION
Unambiguous RNA resonance assignments and identification of a sufficient number of intermolecular NOEs is often one of the limiting factors preventing a rapid structure determination of protein–RNA complexes with NMR spectroscopy (1,2). Reasons for this limitation originate from the severe resonance overlap that is present in RNA compared to proteins especially within the sugar protons of the RNA that all resonate within one p.p.m. except for the anomeric proton (3). To overcome this problem, methods based on in vitro transcription have been developed to isotopically label RNA with 13C and 15N and to facilitate RNA resonance assignments and the structure determination of RNAs and protein–RNA complexes (4,5). Although, they have been successfully used to study RNA of 20 nt or more in size in complex with proteins (1,2), labeling methods to produce short RNA oligonucleotides (2 to 10 in size) by in vitro transcription failed (6). Yet, isotopic labeling of short RNAs would be very useful to confidently assign the resonances of short RNAs in complex with proteins and to improve the precision of the resulting structures. Recently, three structures of proteins in complex with short single-stranded RNAs have been determined with NMR spectroscopy, namely SF1 (7), TIS11d (6) and the Argonaute PAZ domain (8). All three were studied with unlabeled RNA making the resonance assignment of the RNA in complex very challenging, especially in the presence of an RNA with low sequence complexity like 5′-UUAUUUAUU-3′ used in the TIS11d–RNA complex (6).
Here, we report the synthesis of several short RNA sequences (4 to 7 nt) with [13C5]-labeled riboses at specific positions and their application for the resonance assignments and for the structure determination of two alternative-splicing factors, polypyrimidine tract binding protein (PTB) and feminizing locus on X (Fox-1), in complex with RNA. For this purpose, we evaluated and optimized the various reported synthetic methods (9–12) for the preparation of [13C5]-ribonucleosides from [13C6]-D-glucose. These intermediates were then converted into the corresponding 2′-O-[(triisopropylsilyl)oxy]methyl (= 2′-O-TOM) protected ribonucleoside phosphoramidites, which allows an efficient preparation of RNA sequences under DNA coupling conditions (13,14). Chemical synthesis allows efficient 13C-labeling of very short RNAs and the possibility to introduce isotope-labeling at any position in the sequence, providing a fundamental advantage over isotope-labeling by in vitro transcription (1,2). Using chemically synthesized 13C-labeled RNA, RNA resonance assignment was greatly facilitated as several ambiguities could be resolved. Also a very high number of intermolecular and intramolecular sugar–sugar NOE cross-peaks could be assigned permitting to obtain structures of very high precision (15,16).
MATERIALS AND METHODS
General
Reagents and solvents (highest purity) from various suppliers, used without further purification, unless otherwise stated. [1,2,3,4,5,6–13C6] glucose (13C > 99%) from Spectra Stable Isotopes (Columbia, MD). Work up implies distribution of the reaction mixture between CH2Cl2 and saturated aqueous NaHCO3 solution, drying of the organic layer (MgSO4), and evaporation. TLC: pre-coated silica gel plates from Merck, stained by dipping into a solution of anisaldehyde (10 ml), H2SO4 (10 ml) and AcOH (2 ml) in EtOH (180 ml) and subsequent heating with a heat-gun. Column chromatography (CC): silica gel 60 (230–400 mesh) from Fluka. NMR (Bruker): chemical shift δ in p.p.m., relative to external standards (1H- and 13C: Me4Si, 31P: 85% aqueous H3PO4); coupling constants J in Hz; multiplicities (13C) according to DEPT-spectra. ESI-MS (positive mode): SSQ 710 (Finnegan), measurements in MeCN/H2O/AcOH 50:50:1; MALDI-MS (positive mode): Axima CFR Plus (Kratos/Shimadzu), matrix: 2,4,6-trihydroxyacetophenone (50 mg/ml), di-ammonium hydrogen citrate (15 mg/ml) in H2O/MeCN (1:1); relative intensity in % as indicated in brackets.
[1,2,3,4,5–13C5]-1,2-Di-O-acetyl-3,5-di-O-benzoyl-α,β-D-ribofuranose (1)
Prepared essentially according to Saito et al. (17), for experimental details see Supplementary Data.
[1′,2′,3′,4′,5′–13C5]-6-Allyloxy-2-amino-N2-isobutyryl-5′-O-(4,4′-dimethoxytrityl)purine (3)
A suspension of 1 (3.20 g, 7.15 mmol) and 2-amino-6-chloro-N2-isobutyrylpurine (2.06 g, 8.61 mmol) in (CH2Cl)2 (28 ml) was treated with N,O-bis(trimethylsilyl)acetamide (2.63 ml, 10.77 mmol). After 15 min at 65°C, TMS-OTf (4.53 ml, 25.13 mmol) was added and the mixture was stirred 30 min at 65°C. After usual work up, the intermediate 2 was dissolved in allyl alcohol (23 ml), treated with DABCO (1.21 g, 10.77 mmol) and DBU (1.61 ml, 10.77 mmol). After 1 h at 20°, the reaction mixture was diluted with CH2Cl2 (150 ml), extracted with 10% citric acid (200 ml), washed with saturated NaHCO3 (150 ml) and dried over MgSO4. After evaporation, the residue was dissolved in THF/MeOH 5:4 (180 ml), cooled to 0° and treated with 2N NaOH (18 ml). After 10 min at 0°, ion exchange resin IR 120 (H+ form, 30 ml) was added until the pH 7. The resin was filtered off and washed with THF/MeOH 5:4. The solvents were evaporated; the residue was co-evaporated with pyridine and dried under vacuum (0.01 mbar). The residue was dissolved in pyridine (30 ml) and treated with (MeO)2TrCl (2.9 g, 8.62 mmol). After 1 h at 20°, the reaction mixture was subjected to usual work up. CC [75 g SiO2, CH2Cl2 (+2% NEt3) → CH2Cl2/MeOH 97:3 (+2% NEt3)] gave 3 (3.26 g, 65%) as yellow foam. TLC (CH2Cl2/MeOH 9:1) Rf 0.58. 1H-NMR (13C decoupled, 400 MHz, CDCl3): 1.32, 1.34 (2d, J = 6.6, Me2CHCO); 2.62 (heptett, J = 6.8, Me2CHCO); 3.20 [d, J = 9.7, H–C(5′)]; 3.37 [d, J = 10.2, H′–C(5′)]; 3.45 (br. s, HO); 3.77 (s, 2 MeO); 3.79 (br. s, HO); 4.42 [m, H–C(4′)]; 4.50 [m, H–C(3′)]; 4.95 [s, H–C(2′)]; 5.09 (d, J = 5.4, OCH2CH=CH2);5.33 (dd, J = 10.4, 0.8, 1H, OCH2CH=CH2); 5.48 (dd, J = 17.3, 0.8 1H OCH2CH=CH2); 5.94 [d, J = 5.2, H–C(1′)]; 6.15, (m, OCH2CH=CH2); 6.68–6.75 (m, 4 arom. H); 7.09–7.40 (m, 9 arom. H); 7.46 [br. s, HN-C(2)]; 8.13 [s, H–C(8)]. 13C-NMR (100 MHz, CDCl3): 19.4 (Me2CHCO); 37.2 (Me2CHCO); 55.2 (MeO); 64.2 [d, J = 42, C(5′)]; 68.6 (OCH2CH=CH2); 72.2 [dd, J = 38, C(4′)]; 77.1 [dd, J = 38, C(2′)]; 87.4 [dd, J = 41, C(3′)]; 88.8 (arom. C); 92.6 [d, J = 39, C(1′)]; 113.5 (arom. CH); 119.2 (OCH2CH=CH2); 127.2, 128.2, 130.3, 130.4, 132.4 (5 arom. CH); 135.8, 135.9 (2 arom. C), 140.4 [C(8)]; 144.7 (arom. C); 151.5 [C(6)]; 152.0 [C(2)]; 158.88 (arom. C), 161.2 (arom. C); 175.7 (Me2CHCO). ESI-MS: 701.31 (100, [M + H]+).
[1′,2′,3′,4′,5′–13C5]-5′-O-(4,4′-Dimethoxytrityl)-N2-(isobutyryl)guanosine (4)
A solution of 3 (3.44 g, 4.91 mmol) in CH2Cl2 (20 ml) was treated with Et2NH (1.28 ml, 12.3 mmol), PPh3 (296 mg, 1.13 mmol) and Pd(PPh3)4 (30 mg, 0.03 mmol). After 30 min at 20°, SiO2 (9 g) was added and the solvent was evaporated. The residue was purified by CC [10 g SiO2, CH2Cl2 (+2% Et3N) → CH2Cl2/MeOH 95:5 (+2% Et3N)]: 4 (2.90 g, 89%). TLC (CH2Cl2/MeOH 9:1) Rf 0.47. 1H-NMR (13C decoupled, 400 MHz, CDCl3): 0.81, 1.00 (2d, J ≈ 6, Me2CHCO); 2.83 (br. q, J ≈ 7, Me2CHCO); 3.19 [m, H–C(5′)]; 3.40 (br. d, J = 6.7, HO); 3.47 [m, H′–C(5′)]; 3.75, 3.76 (2s, 2 MeO); 4.31 [m, H–C(4′)]; 4.55 [m, H–C(3′)]; 5.14 [s, H–C(2′)]; 5.87 [d, J = 5.2, H–C(1′)]; 6.75–6.85 (m, 4 arom. H); 7.10–7.50 (m, 9 arom. H); 7.78 [s, H–C(8)]; 9.46 [br. s, HN-C(2)]; 12.16 [br. s, H-N(1)]. 13C-NMR (100 MHz, CDCl3): 18.5, 18.7 (Me2CHCO); 35.8 (Me2CHCO); 55.2 (MeO); 63.7 [d, J = 43, C(5′)]; [71.5 (t, J = 38), 73.9 (t, J = 39), 85.1 (t, J = 41): C(2′), C(3′), C(4′)]; 90.0 [d, J = 41, C(1′)]; 113.2 (arom. CH); 121.0 [C(5′)]; 126.9, 127.9, 128.1, 129.9, 130.0 (5 arom. CH); 135.7, 136.1 (2 arom. C); 138.9 [C(8)]; 144.7 (arom. C); 147.4 [C(4)]; 148.2 [C(2)]; 155.8 [C(6)]; 158.6 (arom. C); 179.7 (Me2CHCO). ESI-MS: 661.36 (100, [M + H]+).
[1′,2′,3′,4′,5′–13C5]-N6-Benzoyl-5′-O-(4,4′-dimethoxytrityl)adenosine (5)
A suspension of 1 (3.20 g, 7.15 mmol) and N6-benzoyladenine (2.05 g, 8.59 mmol) in (CH2Cl)2 (28 ml) was treated with N,O-bis(trimethylsilyl)acetamide (2.62 ml, 10.74 mmol). The mixture was stirred at 65°C for 15 min, treated with SnCl4 (2.9 ml, 25.06 mmol) and stirred 30 min at 65°C. After usual work up, the residue was dissolved in THF–MeOH 5:4 (250 ml), cooled to 0° and treated with 2N NaOH (25 ml). After 40 min at 0°, ion exchange resin IR 120 (H+ form, 30 ml) was added until the pH 7. The resin was filtered off and washed with THF–MeOH 5:4. The solvents were evaporated; the residue was co-evaporated with pyridine and dried under vacuum (0.01 mbar). The residue was dissolved in pyridine (30 ml) and treated with (MeO)2TrCl (3.20 g, 9.5 mmol). After 1 h at 20°, the reaction mixture was subjected to usual work up. CC [75 g SiO2, CH2Cl2 (+2% NEt3) → CH2Cl2/MeOH 24:1 (+2% NEt3)] gave 5 (3.79 g, 78%) as yellow foam. TLC (CH2Cl2/MeOH 9:1) Rf 0.44. 1H-NMR (13C decoupled, 400 MHz, CDCl3): 2.03 (br. s, HO); 3.35 [d, J = 9.7, H–C(5′)]; 3.48 [d, J = 9.7, H′–C(5′)]; 3.77 (s, 2 MeO); 4.42 [m, H–C(4′)]; 4.50 [m, H–C(3′)]; 4.93 [m, H–C(2′)]; 5.83 (br. s, HO); 6.10 [d, J = 4.2, H–C(1′)]; 6.75–6.79 (m, 4 arom. H); 7.16–7.38 (m, 9 arom. H); 7.49–7.69 (m, 3 arom. H); 8.02 (d, J = 7.3, 2 arom. H); 8.25 [s, H–C(8)]; 8.67 [s, H–C(2)]; 9.19 [br. s, HN-C(6)]. 13C-NMR (100 MHz, CDCl3): 55.2 (MeO); 63.5 [d, J = 43, C(5′)]; [72.2 (t, J = 38), 75.4 (dd, J = 37, 40), 85.6 (dd, J = 39, 43): C(2′), C(3′), C(4′)]; 86.5 (arom. C); 90.0 [d, J = 41, C(1′)]; 113.1 (arom. CH); 122.9 [C(5)]; 126.9, 127.7, 127.8, 127.9, 128.0, 128.3, 128.9, 129.1, 129.9 (9 arom. CH); 132.9, 135.4, 135.5 (3 arom. C); 141.6 [C(8)]; 144.4 (arom. C); 149.4 [C(6)]; 151.0 [C(4)]; 152.2 [C(2)]; 158.5 (arom. C); 164.7 (PhCO). ESI-MS: 679.39 (100, [M + H]+).
[1′,2′,3′,4′,5′–13C5]-5′-O-(4,4′-Dimethoxytrityl)uridine (6)
A suspension of 1 (1.70 g, 3.80 mmol) and uracil (0.52 g, 4.64 mmol) in MeCN (19 ml) was treated with N,O-bis(trimethylsilyl)acetamide (2.80 ml, 11.1 mmol) and stirred at 60°C for 30 min. Then, TMS-OTf (2.44 ml, 13.3 mmol) was added and the mixture was stirred 30 min at 60°C. After usual work up, the resulting intermediate was dissolved in 8 M MeNH2 in EtOH (19 ml) and stirred overnight at 20°. The solvent was evaporated; the residue was co-evaporated with pyridine and dried overnight under vacuum (0.01 mbar). The solid was dissolved in pyridine (19 ml) and treated with (MeO)2TrCl (1.80 g, 5.32 mmol). After 1 h at 20°, the reaction mixture was subjected to usual work up. CC (40 g SiO2, CH2Cl2 (+2% Et3N) → CH2Cl2/MeOH 19:1 (+2% Et3N)) gave 7 (1.49 g, 71%) as pink foam. TLC (CH2Cl2/MeOH 9:1) Rf 0.50. 1H-NMR (13C decoupled, 400 MHz, CDCl3): 3.48 [d, J = 10.4, H–C(5′)]; 3.55 [d, J = 10.5, H–C(5′)]; 3.79 (s, 2 MeO); 4.19 [d, J ≈ 4.1, H–C(4′)]; 4.34 [m, H–C(2′)]; 4.43 [m, H–C(3′)]; 5.38 [d, J = 8.1, H–C(5)]; 5.92 [br. s, H–C(1′)]; 6.83–6.88 (m, 4 arom. H); 7.22–7.41 (m, 9 arom. H); 8.00 [d, J = 8.1, H–C(6)]. 13C-NMR (100 MHz, CDCl3): 55.2 (MeO); 61.8 [d, J = 43, C(5′)]; [69.6 (t, J = 69), 75.4 (dd, J = 37, 41), 83.7 (dd, J = 43, 39): C(2′), C(3′), C(4′)]; 86.9 (arom. C); 90.4 (d, J = 42), [C(1′)]; 102.2 [C(5)]; 113.2 (arom. CH); 127.1, 128.0, 128.1, 130.0, 130.1 (5 arom. CH); 135.1, 135.3 (2 arom. C); 140.4 [C(6)]; 144.3 (arom. C); 151.2 [C(2)]; 158.6 (arom. C); 163.9 [C(4)]. MALDI-MS: 574.5 (100, [M + Na]+).
[1′,2′,3′,4′,5′–13C5]-5′-O-(4,4′-Dimethoxytrityl)-N2-isobutyryl-2′-O-{[(triisopropylsilyl)oxy]methyl}guanosine (7), [1′,2′,3′,4′,5′–13C5]-N6-benzoyl-5′-O-(4,4′-dimethoxytrityl)-2′-O-{[(triisopropylsilyl)oxy]methyl}adenosine (8) and [1′,2′,3′,4′,5′–13C5]-5′-O-(4,4′-dimethoxytrityl)-2′-O-{[(triisopropylsilyl)oxy]methyl}uridine (9)
Prepared essentially according to Pitsch et al. (14), for experimental details see Supplementary Data.
[1′,2′,3′,4′,5′–13C5]-N4-Acetyl-5′-O-(4,4′-dimethoxytrityl)-2′-O-{[(triisopropylsilyl)oxy]methyl}cytidine (10)
Prepared according to Wenter et al. (18), for experimental details see Supplementary Data.
[1′,2′,3′,4′,5′–13C5]-N2-isobutyryl-5′-O-(4,4′-dimethoxytrityl)-2′-O-{[(triisopropylsilyl) oxy]methyl}guanosine 3′-(2-cyanoethyl diisopropylphosphoramidite) (11), [1′,2′,3′,4′,5′–13C5]-N6-Benzoyl-5′-O-(4,4′-dimethoxytrityl)-2′-O-{[(triisopropylsilyl)oxy]methyl} adenosine 3′-(2-cyanoethyl diisopropylphosphoramidite) (12), [1′,2′,3′,4′,5′–13C5]-5′-O-(4,4′-Dimethoxytrityl)-2′-O-{[(triisopropylsilyl)oxy]methyl}uridine 3′-(2-cyanoethyl diisopropylphosphoramidite) (13) and [1′,2′,3′,4′,5′–13C5]-N4-cetyl-5′-O-(4,4′-dimethoxytrityl)-2′-O {[(triisopropylsilyl)oxy]methyl}cytidine 3′-(2-cyanoethyl diisopropylphosphoramidite) (14)
Prepared according to Pitsch et al. (14), for experimental details see Supplementary Data.
Solid supports
Prepared according to Pitsch et al. (14), for structures and experimental details see Supplementary Data.
Assembly and deprotection of RNA sequences S1–S6
The RNA sequences S1–S6 were assembled from the 13C5-labeled phosphoramidites 11–14 and the corrresponding 2′-O-TOM protected, unlabeled phosphoramidites on a Gene Assembler Plus (Pharmacia) in a 1 μmol scale according to (14). After the assembly, the solid supports were treated with a 1:1 mixture of 12 M MeNH2 in H2O/8 M MeNH2 in EtOH (1 ml) for 3 h at 35°. By centrifugation, the supernatant solution was separated from the solid support and evaporated, and the residue was dissolved in 1 M Bu4NF·3 H2O solution in THF (1 ml). After 14 h at 20°, 1 M Tris–HCl buffer (pH 7.4, 1 ml) was added and the solution concentrated to half its volume.
Desalting and purification of RNA sequences r([13C′5]Cp[13C′5]Up[13C′5]Cp[13C′5]U) (S1), r(Cp[13C′5]UpCp[13C′5]U) (S2), r([13C′5]Up[13C′5]Cp[13C′5]Up[13C′5]Cp[13C′5]U) (S3) and r([13C′5]UpCp[13C′5]UpCp[13C′5]U) (S4)
The remaining solution from above (1 ml) was applied on a NAP-10 cartridge (Pharmacia) and eluted with H2O. Two fractions were collected: fraction 1 (1.5 ml) and fraction 2 (1 ml). Fraction 2 was again applied to a NAP-10 cartridge and eluted with H2O. The first 1.5 ml solution was combined with fraction 1 and purified by anion exchange–high-performance liquid chromatography (HPLC): Pharmacia Source 15Q column (4.5 × 100 mm), flow 2 ml/min, eluent A: 50 mM NEt3·H2CO3, eluent B: 1 M NEt3·H2CO3, detection at 260 nm, elution at 20°, gradient 0–45% B in 40 min, 1 injection. The fractions containing pure product were pooled and subjected to lyophilization. In order to completely remove the remaining NEt3·H2CO3, 0.5 ml H2O were added to the residue, followed by lyophilization (this procedure was carried out twice). The residue was dissolved in H2O (0.5 ml), treated with NaHCO3 (5 mg) and lyophilized twice. Final desalting on a NAP-10 cartridge was performed as described above, resulting in pure RNA sequences S1–S4 as Na+ salts. Their amount was determined spectrophotometrically. S1 (2 syntheses, 0.68 mg, 0.58 μmol, 29% yield), MALDI-MS (negative mode): 1182 a.m.u. [M+H]+; S2 (2 syntheses, 0.66 mg, 0.56 μmol, 28% yield), MALDI-MS (negative mode): 1172 a.m.u. [M+H]+; S3 (2 syntheses, 1.27 mg, 0.85 μmol, 43% yield), MALDI-MS (negative mode): 1493 a.m.u. [M+H]+; S4 (2 syntheses, 1.26 mg, 0.85 μmol, 43% yield), MALDI-MS (negative mode): 1483 a.m.u. [M+H]+.
Desalting and purification of RNA sequences r([13C′5]UpGp[13C′5]CpAp[13C′5]UpGpU) (S5) and r(Up[13C′5]GpCp[13C′5]ApUp[13C′5]Gp[13C′5]U) (S6)
The remaining solution from above (1 ml) was applied on a NAP-10 cartridge (Pharmacia) and eluted with H2O. The first 1.5 ml solution was purified by anion exchange–HPLC: Pharmacia Source 15Q column (4.5 × 100 mm), flow 2 ml/min, eluent A: 12.5 mM Tris–HCl (pH 7.5); eluent B: 12.5 mM Tris–HCl (pH 7.5), 1 M NaCl; detection at 260 nm, eluent at r.t., gradient 0–50% B in 45 min, two injections. Fractions containing pure product were treated with 1 M aqueous Et3N·H2CO3 to a final 0.1 M concentration and applied to a Sepak cartridge [Waters, conditioned by washing with MeCN (10 ml) and 0.1 M aqueous Et3N·H2CO3 (10 ml)]. The cartridge was washed with 20 mM Et3N·H2CO3 (10 ml) and the RNA sequences were eluted with MeCN/H2O 1:1 (4 ml). In order to completely remove the remaining NEt3·H2CO3, 0.5 ml H2O were added to the residue, followed by lyophilization (this procedure was carried out twice). The residue was dissolved in H2O (0.5 ml), treated with NaHCO3 (10 mg) and lyophilized twice. Final desalting on a NAP-10 cartridge was performed as described above, resulting in pure RNA sequences S5 and S6 as Na+ salts. Their amount was determined spectrophotometrically. S5 (4 syntheses, 4.50 mg, 2.05 μmol, 51% yield), MALDI-MS (negative mode): 2197 a.m.u. [M+H]+; S6 (4 syntheses, 3.43 mg, 1.56 μmol, 39% yield), MALDI-MS (negative mode): 2201 a.m.u. [M+H]+.
Preparation of protein, NMR-measurements, RNA resonance assignments and structure calculation
The preparation of PTB and Fox-1 proteins and the related NMR-measurements are reported in Oberstrass et al. (16) and Auweter et al. (15), respectively. The structure calculation of the Fox-1 complex with the reduced set of NOE distance constraints from the unlabeled RNA sequence was performed in a restrained simulated annealing run using AMBER 7.0, following the same protocol that was employed for the structure calculation of the Fox-1 complex with the entire set of constraints; see (15).
RESULTS AND DISCUSSION
Preparation of the 13C5-ribose-labeled ribonucleoside phosphoramidites
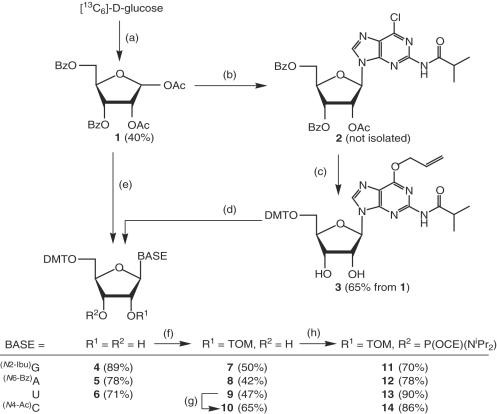
For the preparation of the four canonical, 13C5-ribose-labeled 2′-O-TOM protected ribonucleoside phosphoramidites and solid supports, we adapted and optimized existing reaction conditions with the purpose of minimizing the number of time-consuming purification steps (Scheme 1). The four-step synthesis of the protected 13C5-ribose derivative 1 from 13C6-labeled d-glucose (40% yield) was carried out essentially according to Saito et al. (17) and is described in the Supplementary Data. The protected adenosine and uridine derivatives 5 and 6 were obtained by nucleosidation of 1 with N6-benzoyladenosine (→ 5) and uracil (→ 6), respectively, under optimized Vorbrüggen conditions, followed by partial deacylation and dimethoxytritylation. The guanosine derivative 6 was prepared according to a modified method originally described for the synthesis of the corresponding ribopyranosylguanosine phosphoramidite by Pitsch et al. (19). In the presence of Me3SiOTf and in 1,2-dichloroethane, 1 reacted smoothly and completely regioselectively with in situ trimethylsilylated N2-isobutyryl-6-chlor-2-aminopurine to the purine derivative 2. Without purification, it was treated with 1,4-diazabicyclo[2,2,2]octane (DABCO) and 1,8-diazabicyclo[5,4,0]undec-7-ene (DBU) in allyl alcohol, resulting in the corresponding O6-allyl derivative which was partially deacylated with NaOH in THF/MeOH/H2O and then dimethoxytritylated with (MeO)2TrCl in pyridine (→ 3, 65% based on 1). Cleavage of the O6-allyl group was achieved by treating the intermediate 3 with Et2NH in the presence of [Pd(PPh3)4], resulting in formation of the guanosine derivative 4 (89% yield).
Scheme 1.
Preparation of the [13C5]-ribose-labeled 2′-O-TOM protected ribonucleoside phosphoramidites 11–14. Abbreviations: Ac = acetyl, Bz = benzoyl, Ibu = isobutyryl, CE = cyanoethyl, DMT = (4,4′-dimethoxy)trityl, TOM = (triisopropylsilyl)oxymethyl. Reagents and conditions: (a) Adapted from Saito et al. (17), detailed procedure in Supplementary Data: 1. FeCl3, MgSO4, acetone, 20°; 2. pyridinium dichromate, Ac2O, CH2Cl2, reflux; 3. H5IO6, THF, 20°; 4. NaBH4, THF/EtOH 1:1, 20°; 5. BzCl, pyridine, 20°; 6. Ac2O, AcOH, H2SO4, 20°. (b) 6-Chloro-N2-isobutyrylpurine-2-amine, N,O-bis(trimethylsilyl)acetamide (BSA), Me3SiOTf, 1,2-dichloroethane, 65°. (c) 1. Allyl alcohol, DABCO, DBU, 20°; 2. NaOH, THF/MeOH/H2O, 0°; 3. DMT-Cl, pyridine, 20°. (d) Pd(Ph3P)4, HNEt2, PPh3, CH2Cl2, 20°. (e) Synthesis of 5: 1. N6-benzoyladenine, BSA, SnCl4, 1,2-dichloroethane, 65°; 2. NaOH, THF/MeOH/H2O, 0°; 3. DMT-Cl, pyridine, 20°; synthesis of 6: 1. uracil, BSA, Me3SiOTf, MeCN, 60°; 2. MeNH2, EtOH, 20°; 3. DMT-Cl, pyridine, 20°. (f) Bu2SnCl2, iPr2NEt, TOM-Cl, 1,2-dichloroethane, 80° according to (14). (g) 1. Ac2O, DMAP, pyridine 25°; 2. 4-chlorophenyl phosphorodichloridate, 1H-1,2,4-triazole, iPr2NEt, MeCN, 4° → 20°; 3. aqueous NH3, dioxane/MeCN, 20°; 4. NaOH, THF/MeOH/H2O, 4° 5. Ac2O, DMF, 20°, according to (18). (h) 2-Cyanoethyldiisopropylphosphoramidochloridite, iPr2NEt, CH2Cl2, 20°, according to (14).
The introduction of the 2′-O-TOM protecting group into nucleosides 4, 5 and 6 was carried out according to our standard procedure (14), with Bu2SnCl2, iPr2NEt and TOM-Cl in 1,2-dichloroethane at 80° (detailed procedure in Supplementary Data). The product mixture was separated into the 2′-O-TOM protected products 7–9, the unreacted starting materials, and the 3′-O-TOM protected regioisomers. The latter were deprotected with Et4NF in MeCN, the products were combined with the unreacted starting materials and subjected again to alkylation with TOM-Cl. By this procedure, the 2′-O-TOM protected nucleosides 7–9 were obtained in fair yields of 42–50% (Scheme 1). From the 2′-O-TOM protected uridine 9, the corresponding cytidine derivative 10 was obtained according to our published method (18), in a yield of 65%. Under standard conditions, the four 2′-O-TOM protected nucleosides 7–10 were converted with 2-cyanoethyl diisopropylphosphoramidochloridite/iPr2NEt in CH2Cl2 into their corresponding phosphoramidite building blocks (Scheme 1). The solids supports were prepared according to (14) by first synthesizing the 2′-O-(nitrophenylheptane)dioates from the 3′-O-tom nucleosides of adenosine, uridine and cytosine and the 3′-O-(nitrophenylheptane)dioate from the 2′-O-TOM guanosine derivative. Then these activated esters were immobilized on 500 Å (aminoalkyl)-functionalized controlled pore glass (CPG) with loadings from 25 to 35 μmol/g (a detailed description for the preparation of phosphoramidites 7–14 and the corresponding solid supports is given in the Supplementary Data).
Chemical synthesis of the short 13C-labeled RNA oligonucleotides
The short and partially labeled RNA sequences S1–S6 (Table 1) were assembled in 1 μM batches from conventional 2′-O-TOM protected ribonucleoside phosphoramidites and solid supports, and the corresponding 13C-labeled building blocks 11–14, respectively, on an oligonucleotide synthesizer according to our standard protocols (14). The isolation and purification protocols of the shortest 4 and 5mer oligonucleotides S1–S4 had to be modified, because of the small size and small number of negative charges present (for a detailed procedure see Materials and Methods). Finally, all RNA sequences were converted into their Na+ salts and characterized by MALDI-MS (Table 1).
Table 1.
Preparation of selectively 13C-labeled RNA sequences
| Synthesisa | Purificationb and MALDI-MSc | |||||
|---|---|---|---|---|---|---|
| Sequence | Scale | Yield | Condition | [M+H+]calc | [M+H+]found | |
| (μmol) | (μmol) | (%) | (a.m.u.) | (a.m.u.) | ||
| S1: 5′-CUCU-3′ | 2 | 0.58 | 29 | A | 1182 | 1182 |
| S2: 5′-CUCU-3′ | 2 | 0.56 | 28 | A | 1172 | 1172 |
| S3: 5′-UCUCU-3′ | 2 | 0.85 | 43 | A | 1493 | 1493 |
| S4: 5′-UCUCU-3′ | 4 | 0.85 | 43 | A | 1483 | 1483 |
| S6: 5′- UGCAUGU-3′ | 4 | 2.05 | 51 | B | 2196 | 2197 |
| S7: 5′- UGCAUGU-3′ | 4 | 1.56 | 39 | B | 2202 | 2201 |
aPreparation and deprotection of RNA-sequences according to Pitsch et al.(14); underlined letters in sequences indicate 13C5-ribose-labeled nucleosides.
bPurification and isolation: condition A: 1. Desalting on size-exclusion cartridges according to a modified protocol, 2. anion exchange–HPLC with aq. Et3N·H2CO3 gradients, 3. Removal of buffer by repeated lyophilization, 4. Transformation into Na+salt form by addition of NaHCO3, repeated lyophilization and desalting on size-exclusion cartridges; condition B: 1. Desalting on size-exclusion cartridges according to the standard protocol, 2. Anion exchange–HPLC with NaCl gradients, 3. Removal of eluent on C18-RP cartridges according to a modified protocol, 4. Transformation into Na+-salt form by addition of NaHCO3, repeated lyophilization and desalting on size-exclusion cartridges.
cMatrix: 2,4,6-trihydroxyacetophenone, di-ammonium citrate.
NMR study of the 13C5-sugar labeled UCUCU and CUCU in complex with PTB
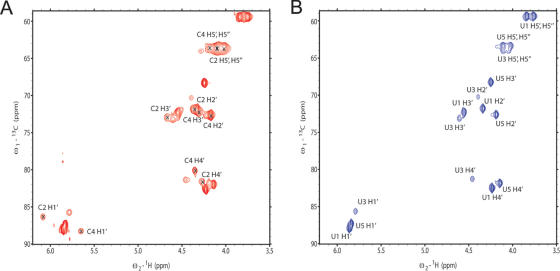
The oligo(CU) sequences S1–S4 (Supplementary Table) containing 13C5-sugar labeled nucleotides at various positions were used for structure determination of PTB in complex with its target RNA sequence. We had to use these very short tetramers and pentamers, because PTB RBD1, RBD2 and RBD4 recognize only 3 nt and RBD3 only 5 nt (16). As illustrated in the 13C HSQC spectrum of S3 bound to PTB RBD34 H134A, complete or partial 13C-labeling of the sugar moieties of these short RNAs allows resolving the strong proton resonance overlap in the sugar region of the RNA in complex with PTB (Figure 1A). The RNAs containing only labeled uracils (S2 and S4) were essential to confirm the resonance assignments of the bound RNAs (Figure 1B) and to unambiguously derive intermolecular NOEs between the sugar moiety and the protein side chains. Isotope-labeling of the RNAs was essential to this structure determination as 50 and 67% of the intermolecular NOEs involved RNA sugar resonances in the complex with RBD3 and RBD4 of PTB, respectively (16).
Figure 1.
(A) 13C HSQC spectra of UCUCU (S3) bound to PTB34 H134A (red). (B) 13C HSQC spectra of UCUCU (S4) bound to PTB34 H134A. In S4, only the uridines are 13C-labeled. The assignment is indicated and in spectrum (A), a cross indicates the position of the cross-peaks from cytidine resonances.
NMR study of the 13C5-sugar labeled UGCAUGU in complex with Fox-1
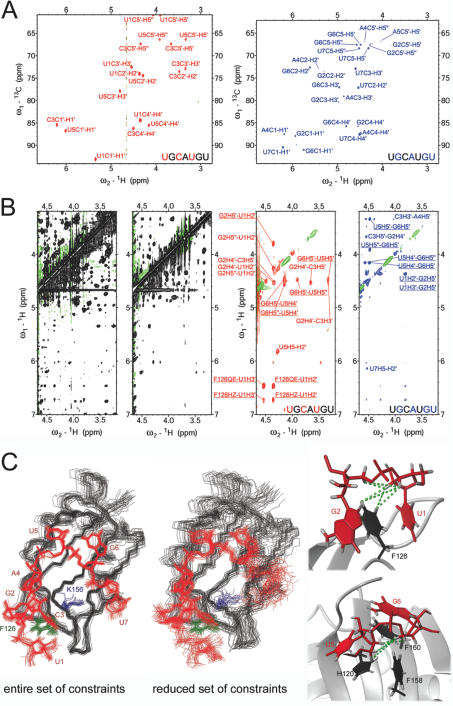
The partially 13C-labeled RNA oligonucleotides S5 and S6 were used to determine the structure with Fox-1 (16). The two RNAs were essential to unambiguously assign the RNA resonances and to collect a high number of intra- and intermolecular NOE-derived constraints for UGCAUGU in complex with Fox-1. Without isotopically labeled RNA, the resonance assignment of the 7 nt RNA in complex was difficult and not unambiguous. While the proton resonances within each individual nucleoside could be linked using 2D NOESY and 2D TOCSY spectra, it was not possible to unambiguously place each individually assigned nucleotide spin-systems in the RNA sequence. In particular, we were doubtful about which spin-systems were corresponding to U5 and U7. With the 2D 13C HSQC spectra of the partially 13C-labeled RNA S6 and S7 in complex with Fox-1, these ambiguities could be solved, as S6 contains a 13C5-labeled U5 and S7 a 13C5-labeled U7 (Figure 2A). Importantly, with the particular labeling scheme present in S6 and S7, in which 13C5-labeled and unlabeled nucleotides are present in alternation, unusual internucleotide sugar–sugar NOEs could be assigned using X-filter NOESY spectra (20) (Figure 2B). In total, 39 internucleotide sugar–sugar NOEs were assigned, which corresponds to 68% of the internucleotide constraints and one-third (31%) of the intra-RNA constraints. Only five of these internucleotide sugar–sugar NOEs could be assigned using an unlabeled RNA, the remaining 34 could only be resolved using labeled S6 and S7 due to ambiguity or resonance overlap. Similarly, of the 149 intermolecular distance constraints, which were used to calculate the Fox-1 complex structure, 83 originate from the sugar protons of the RNA and 57 of these could not have been assigned without the help of the labeled oligonucleotides (15). A structure calculation without the distance constraints from the 13C-labels resulted in a set of structures in which the RNA is significantly less defined: the RMSD of the heavy atoms of the RNA is 1.02 Å as compared to 0.55 Å in the case when all NOE distance constraints were included (Figure 2C). While each nucleotide is affected, the effect is strongest at the 5′ and 3′ terminal ends of the RNA. In particular, nucleotide U7 does not converge to one particular conformation (Figure 2C). Furthermore, the relative orientation of protein and RNA is less precise (Figure 2C). The complex structure of Fox-1 using all constraints is extraordinarily well defined, allowing a detailed analysis of the individual hydrogen bond, van der Waals and electrostatic interactions at the protein–RNA interface (15). In the structures obtained with the reduced set of constraints, an equal level of precision cannot be reached. Especially the intermolecular contacts to the RNA backbone are more heterogeneous. For example, a salt bridge between lysine 156 and the phosphate between G6 and U7 is observed in 28 of 30 structures (Figure 2C, left). In contrast, this salt bridge is observed in only 11 of the 30 structures obtained with the smaller number of NOE distance constraints (Figure 2C, middle). Hence, these NOEs were critical to determine with high precision the very unusual structure that the RNA adopts when bound to the RBD of Fox-1.
Figure 2.
(A) 13C HSQC spectra of ∼1mM Fox-1 in complex with S6 (red) or S7 (blue). (B) Left: section of a 2D NOESY spectrum of ∼1 mM Fox-1 (unlabeled) in complex with UGCAUGU (unlabeled) showing the extreme overlap in the RNA sugar resonances. Middle left: the same section of a ω1, ω2-filtered 2D NOESY spectrum of ∼1 mM Fox-1 (uniformly 13C-labeled) in complex with UGCAUGU (unlabeled). In this spectrum, NOE cross-peaks arise only from intra-RNA correlations; overlap is reduced but assignments remain ambiguous. Middle right: the same section of a ω1-filtered, ω2-edited 2D NOESY spectrum of ∼1 mM Fox-1 (unlabeled) in complex with S6. NOE cross-peaks arise from correlations of protons attached to 13C (ω2) with protons attached to 12C (ω1). Right: the same section of a ω1-filtered, ω2-edited 2D NOESY spectrum of ∼1 mM Fox-1 (unlabeled) in complex with S7. NOE cross-peaks arise from correlations of protons attached to 13C (ω2) with protons attached to 12C (ω1). Underlined NOE assignments are displayed in the structure of the complex with green dashed lines in (C). Overlap and ambiguity are significantly reduced in the spectra with S6 and S7 and many internucleotide cross-peaks can be assigned due to the alternating labeling scheme. (C) Left: overlay of the 30 final structures of Fox-1 in complex with UGCAUGU calculated using the entire set of NOE distance constraints [PDB code: 2ERR] (15). Middle: overlay of the 30 final structures of Fox-1 in complex with UGCAUGU calculated using only those NOE distance constraints that could be assigned with unlabeled RNA oligonucleotides. The structures are superimposed on the heavy atoms of the RNA; the protein backbone is shown in black, the heavy atoms of the RNA are in red, and the side chains of F126 and K156 are in green and blue, respectively. Right: sections of one representative structure of the Fox-1-UGACUGU complex calculated using the entire set of NOEs. The protein backbone is depicted in ribbon representation (gray), aromatic protein side chains are shown in black, and the RNA is shown in red. Protons are displayed in gray.
CONCLUSION AND PERSPECTIVES
In conclusion, we present a short and efficient synthetic access to 2′-O-TOM protected ribonucleoside phosphoramidites containing 13C-labeled sugar moieties. The site-specific introduction of these 13C-labeled nucleotides into RNA sequences permitted a straightforward and unambiguous resonance assignment of their complexes with PTB and Fox-1. The analysis of sets of RNA sequences with alternating labeling patterns allowed the assignments of numerous unusual sugar–sugar and intermolecular NOEs, which substantially improved the precision and accuracy of the resulting structures. Use of chemically synthesized 13C-labeled RNA sequences is likely to facilitate the study of single-stranded RNA binding proteins in complex with their target RNAs (21), particularly RNA binding proteins targeting low complexity or repeat sequences that are numerous in posttranscriptional gene regulation (22–24).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by the EPFL, the NCCR ‘Structural Biology’ and the Swiss National Science Foundation (Grant No. 2000-06890) to S.P. and by the grants from the Swiss National Science Foundation and the ETH Zurich through the NCCR Structural Biology and by the Roche Research Fund for Biology at the ETH Zurich to F.H.T.A. F.H.T.A. is an EMBO Young Investigator. Funding to pay the Open Access publication charges for this article was provided by the EPFL.
Conflict of interest statement. None declared.
REFERENCES
- 1.Allen M., Varani L., Varani G. Nuclear magnetic resonance methods to study structure and dynamics of RNA–protein complexes. Meth. Enzymol. 2001;339:357–376. doi: 10.1016/s0076-6879(01)39322-9. [DOI] [PubMed] [Google Scholar]
- 2.Wu H., Finger L.D., Feigon J. Structure determination of protein/RNA complexes by NMR. Meth. Enzymol. 2005;394:525–545. doi: 10.1016/S0076-6879(05)94022-6. [DOI] [PubMed] [Google Scholar]
- 3.Varani G., Aboulela F., Allain F.H.T. NMR Investigation of RNA Structure. Prog. Nucl. Magn. Reson. Spectrosc. 1996;29:51–127. [Google Scholar]
- 4.Batey R.T., Inada M., Kujawinski E., Puglisi J.D., Williamson J.R. Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res. 1992;20:4515–4523. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikonowicz E.P., Sirr A., Legault P., Jucker F.M., Baer L.M., Pardi A. Preparation of 13C and 15N labelled RNAs for heteronuclear multi-dimensional NMR studies. Nucleic Acids Res. 1992;20:4507–4513. doi: 10.1093/nar/20.17.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hudson B.P., Martinez-Yamout M.A., Dyson H.J., Wright P.E. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nature Struct. Mol. Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z., Luyten I., Bottomley M.J., Messias A.C., Houngninou-Molango S., Sprangers R., Zanier K., Kramer A., Sattler M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- 8.Lingel A., Simon B., Izaurralde E., Sattler M. Nucleic acid 3′ end recognition by the Argonaute2 PAZ domain. Nature Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 9.Cromsigt J., van Buuren B., Schleucher J., Wijmenga S. Resonance assignment and structure determination for RNA. In: James T., Dotsch V., Schmitz U., editors. Nuclear Magnetic Resonance of Biologica Macromolecules. Vol. 338. London: Academic Press; 2001. pp. 371–399. Part A. [DOI] [PubMed] [Google Scholar]
- 10.Milecki J., Foldesi A., Fischer A., Adamiak R.W., Chattopadhyaya J. Synthesis of multiply labelled ribonucleosides for sequence-specific labelling of oligo-RNA. J. Labelled Comp. Radiopharm. 2001;44:763–783. [Google Scholar]
- 11.Tolbert T.J., Williamson J.R. Preparation of specifically deuterated and C-13-labeled RNA for NMR studies using enzymatic synthesis. J. Am. Chem. Soc. 1997;119:12100–12108. [Google Scholar]
- 12.Quant S., Wechselberger R.W., Wolter M.A., Worner K.H., Schell P., Engels J.W., Griesinger C., Schwalbe H. Chemical synthesis of C-13-labeled monomers for the solid-phase and template controlled enzymatic-synthesis of DNA and RNA oligomers. Tetra. Lett. 1994;35:6649–6652. [Google Scholar]
- 13.Wu X., Pitsch S. Synthesis and pairing properties of oligonucleotide analogues containing a metal-binding site attached to β-d-allofuranosyl cytosine. Nucleic Acids Res. 1998;26:4315–4323. doi: 10.1093/nar/26.19.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pitsch S., Weiss P.A., Jenny L., Stutz A., Wu X.L. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2′-O-[(triisopropylsilyl)oxy]methyl(2′-O-tom)-protected phosphoramidites. Helvetica Chimica Acta. 2001;84:3773–3795. [Google Scholar]
- 15.Auweter S.D., Fasan R., Reymond L., Underwood J.G., Black D.L., Pitsch S., Allain F.H.-T. Molecular basis of RNA recognition by human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oberstrass F., Auweter S.D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Pitsch S., Black D.L., Allain F.H.-T. RNA binding by PTB: specific recignition and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 17.Saito Y., Zevaco T.A., Agrofoglio L.A. Chemical synthesis of C-13 labeled anti-HIV nucleosides as mass-internal standards. Tetrahedron. 2002;58:9593–9603. [Google Scholar]
- 18.Wenter P., Pitsch S. Synthesis of selectively N-15-labeled 2′-O-{[(triisopropylsilyl)oxy]methyl}(=tom)-protected ribonucleoside phosphoramidites and their incorporation into a bistable 32mer RNA sequence. Helvetica Chimica Acta. 2003;86:3955–3974. [Google Scholar]
- 19.Pitsch S., Wendeborn S., Krishnamurthy R., Holzner A., Minton M., Bolli M., Miculca C., Windhab N., Micura R., Stanek M., et al. Pentopyranosyl oligonucleotide systems—9th communication—the beta-D-ribopyranosyl-(4′→2′)-oligonucleotide system (‘pyranosyl-RNA’): synthesis and resume of base-pairing properties. Helvetica Chimica Acta. 2003;86:4270–4363. [Google Scholar]
- 20.Peterson R.D., Theimer C.A., Wu H., Feigon J. New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA-protein complexes. J. Biomol. NMR. 2004;28:59–67. doi: 10.1023/B:JNMR.0000012861.95939.05. [DOI] [PubMed] [Google Scholar]
- 21.Messias A.C., Sattler M. Structural basis of single-stranded RNA recognition. Acc. Chem. Res. 2004;37:279–287. doi: 10.1021/ar030034m. [DOI] [PubMed] [Google Scholar]
- 22.Hui J., Bindereif A. Alternative pre-mRNA splicing in the human system: unexpected role of repetitive sequences as regulatory elements. Biol. Chem. 2005;386:1265–1271. doi: 10.1515/BC.2005.143. [DOI] [PubMed] [Google Scholar]
- 23.Grabowski P.J. A molecular code for splicing silencing: configurations of guanosine-rich motifs. Biochem. Soc. Trans. 2004;32:924–927. doi: 10.1042/BST0320924. [DOI] [PubMed] [Google Scholar]
- 24.Singh R., Valcarcel J. Building specificity with nonspecific RNA-binding proteins. Nature Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.