Abstract
DNA polymerase lambda (Pol λ) is one of several DNA polymerases suggested to participate in base excision repair (BER), in repair of broken DNA ends and in translesion synthesis. It has been proposed that the nature of the DNA intermediates partly determines which polymerase is used for a particular repair reaction. To test this hypothesis, here we examine the ability of human Pol λ to extend mismatched primer-termini, either on ‘open’ template-primer substrates, or on its preferred substrate, a 1 nt gapped-DNA molecule having a 5′-phosphate. Interestingly, Pol λ extended mismatches with an average efficiency of ≈10−2 relative to matched base pairs. The match and mismatch extension catalytic efficiencies obtained on gapped molecules were ≈260-fold higher than on template-primer molecules. A crystal structure of Pol λ in complex with a single-nucleotide gap containing a dG·dGMP mismatch at the primer-terminus (2.40 Å) suggests that, at least for certain mispairs, Pol λ is unable to differentiate between matched and mismatched termini during the DNA binding step, thus accounting for the relatively high efficiency of mismatch extension. This property of Pol λ suggests a potential role as a ‘mismatch extender’ during non-homologous end joining (NHEJ), and possibly during translesion synthesis.
INTRODUCTION
The human genome encodes 16 DNA polymerases (1), classified into four discrete families (A, B, X and Y) based on differences in the primary structure of their catalytic subunits (2). Family X DNA polymerases are small enzymes present in different organisms ranging from viruses to higher eukaryotes (3–6). Members of this family vary considerably in their biochemical features and associated functions.
DNA polymerase λ (Pol λ) is a family X member that is widespread among higher eukaryotes, both in animals and plants. Human Pol λ is similar in sequence to human Pol β (32% amino acid identity), and both enzymes share many of their biochemical properties, including a dRP lyase activity, required to complement the DNA synthesis step associated with base excision repair (BER), as shown both in vitro and in vivo (7,8). Compared to Pol β, Pol λ has a higher affinity for dNTPs, leading to the suggestion that it would be the DNA repair enzyme of choice when intracellular dNTP concentrations are low (9). Moreover, structural analysis of Pol λ has revealed a high overall structural similarity with Pol β, but also different conformational changes associated with catalysis (10,11).
In eukaryotes and prokaryotes, double-strand breaks (DSBs) are repaired by non-homologous end joining (NHEJ) and homologous recombination (HR) (12–16). NHEJ is the predominant pathway in multicellular eukaryotes. In this process, DNA ends are rejoined using sequence microhomology (17,18). Generation and resolution of NHEJ intermediates frequently requires nucleolytic processing and DNA synthesis in addition to the ligation step. Multiple family X enzymes have been implicated in DSB repair processes, including yeast Pol IV (19), TdT for a specialized form of NHEJ, V(D)J recombination (20) and Pol μ for NHEJ during immunoglobulin light chain recombination (21,22). Although the exact biological functions of Pol λ are not yet certain, the observation that Pol λ has an extraordinary ability to generate frameshift errors (23) suggests an ability to use NHEJ intermediates. In fact, filling short gaps during XRCC4-LigaseIV-dependent rejoining of DSBs requires the presence of Pol λ in HeLa cell extracts, suggesting that Pol λ contributes to NHEJ in human cells (24). More recently, it has been shown that Pol λ is capable of performing junctional additions during NHEJ in vitro, based on a BRCT-mediated efficient interaction with the NHEJ factors Ku, XRCC4 and Ligase IV (18,25,26). Thus, at least three different human family X polymerases are implicated in NHEJ reactions. The choice of which polymerase participates in a given repair reaction has been suggested to partly depend on differences in the nature of the DNA ends that must be joined (18). One goal of the present study is to further test this hypothesis.
All the family X DNA polymerases implicated in NHEJ lack intrinsic exonucleolytic proofreading activity, which is undoubtedly relevant to their ability to use misaligned and/or mismatched template-primer regions with only short regions of homology. Indeed, it has recently been shown that Pol λ can accommodate and efficiently extend misaligned substrates containing as little as one correct terminal base pair with an unpaired nucleotide upstream of the polymerase active site (27). Gap-filling during NHEJ may require the ability to extend not only misaligned substrates but also those containing base–base mismatches. Also, Pol λ has been implicated in translesion DNA synthesis (TLS) (28,29), a process that may also benefit from an ability to extend substrates with aberrant geometry, including mismatched substrates. For example, Pol ζ, Pol η, Pol ι and Pol κ are all implicated in TLS, and all four are relatively promiscuous in extending mismatched primer-termini (30–33). For these reasons, and because the ability of Pol λ to extend base–base mismatches has not been reported, here we determine the ability of Pol λ to extend mispairs both on ‘open’ template-primer and 1 nt gapped molecules. These data are considered in comparison to other polymerases implicated in BER, NHEJ and TLS, and in light of a new crystal structure of Pol λ complexed with a 1 nt gapped-DNA containing a dG·dGMP mismatched primer-terminus.
MATERIALS AND METHODS
Materials
Synthetic oligonucleotides purified by PAGE were obtained from Invitrogen or Eurogentec. Template-primer molecules were generated by annealing four P1 primers (5′-GATCACAGTGAGTAN, where N is A, C, G or T) to four T6 templates (5′-TCTATCNTACTCACTGTGATC, where N is A, C, G or T). One nucleotide gapped molecules with a 5′ phosphate group were obtained by annealing the four P1 primers to four T13 templates (5′-AGAAGTGTATCTCNTACTCACTGTGATC where N is A, C, G or T) and to downstream oligonucleotide Dg1P (5′-AGATACACTTCT, with a 5′ phosphate group). All primers were labeled at its 5′ end with [γ-32P]ATP (3000 Ci/mmol, Amersham) and T4 polynucleotide kinase (New England Biolabs). The primers were then hybridized to one (template) or two (template and downstream) oligonucleotides to generate all different molecules in the presence of 50 mM Tris–HCl (pH 7.5) and 0.3 M NaCl and heating to 80°C for 10 min before slowly cooling to room temperature over night.
The full-length versions of human Pol μ, Pol λ and the human Pol λ core fragment (39 kDa) were expressed and purified as described previously (9,10,34). Human Pol β was a generous gift of Dr S.H. Wilson (NIEHS, Research Triangle Park, NC).
Primer extension assays
The incubation mixture (20 μl) contained 50 mM Tris–HCl (pH 7.5), 2.5 mM MgCl2, 1 mM DTT, 4% glycerol, 0.1 mg/ml BSA, 5 nM labeled primer strand and 360 nM of each DNA polymerase (human Pol β, human Pol μ and human Pol λ full-length). Extension of the labeled primer strand in the presence of each nucleotide (5 μM) was analyzed by 8 M urea and 20% PAGE and autoradiography.
Steady-state kinetics assays
The incubation mixture (20 μl) contained 50 mM Tris–HCl (pH 7.5), 2.5 mM MgCl2, 1 mM DTT, 4% glycerol, 0.1 mg/ml BSA, 0.25 μM DNA and 2.5 or 5 nM of human Pol λ core fragment (39 kDa) for matched ‘open’ template-primers, and 75 or 100 nM for mismatched ones. dGTP concentrations ranged from 0.05 to 100 μM for the matched ‘open’ template-primers, and from 1 to 1000 μM for the mismatched ones. On 1 nt 5′-phosphate gapped molecules, 5 nM of Pol λ core fragment was added for matched and mismatched template-primers, with the exception of the dC·dCMP pair that required 20 nM Pol λ. On gap molecules, dGTP concentrations ranged from 0.0005 to 0.2 μM for the matched template-primers, and from 0.005 to 1000 μM for the mismatched ones. Reactions were started by adding the indicated concentration of dGTP and incubated at 37°C for 20 min. After incubation, reactions were stopped by adding loading buffer (10 mM EDTA, 95% formamide, 0.03% bromophenol blue and 0.3% cyanol blue). Samples were then run on 20% polyacrylamide sequencing gels containing 8 M urea, to separate the unextended and extended DNA primers. Gel band intensities were quantified using a BAS reader 1500 (Fujifilm). The observed rate of nucleotide incorporation (extended primer) was then plotted as a function of nucleotide concentration, and the apparent Km and Vmax parameters were obtained from the best fit to the Michaelis–Menten equation {V = Vmax · [dNTP] / (Km + [dNTP])} using nonlinear regression (Kaleidagraph, Synergy Software, www.synergy.com). In all cases the quality of the fit of the data were acceptable. The intrinsic efficiency of mismatch extension, , a constant that represents the efficiency of extending mismatched versus matched termini, was calculated using the following equation: = (kcat/Km)mismatch/(kcat/Km)match, where kcat = Vmax / [enzyme].
Protein crystallization
Crystals were formed using the hanging drop method, mixing 2 μl of the protein solution containing DNA with 2 μl of the reservoir solution containing 0.1 M sodium cacodylate (pH 5.5), 40 mM MgCl2 and 7% (w/v) MPD. The crystal was transferred to a solution containing 0.1 M sodium cacodylate (pH 5.5), 50 mM MgCl2, 100 mM NaCl, 8% (w/v) MPD and then transferred into cryo-solution containing 0.1 M sodium cacodylate (pH 5.5), 50 mM MgCl2, 100 mM NaCl and 35% (w/v) MPD for flash cooling. The crystal was transferred into the cryo-solution in four steps of increasing cryoprotectant concentration, frozen in liquid nitrogen and then mounted on a goniometer in a cold stream of nitrogen at −178°C for data collection.
Data collection and processing
Data was collected on a RAXISIV area detector system mounted on a RU3H rotating anode generator equipped with MSC confocal blue mirrors, and processed using the HKL2000 data processing software (35).
Refinement
The PDB 1XSL coordinates were used as a starting model for refinement. This model was then refined with CNS (36) using the torsion angle dynamics and individual B-factor refinement protocols. Model building was performed using iterative cycles of manual model building using the program O (37) and refinement with CNS. The electron density maps were of sufficient quality to build most side chains, all backbone atoms from 249–575 of Pol λ and all nucleotides of the primer, template and downstream primer. Superimposition of the four molecules present in the asymmetric unit revealed no noticeable difference between them. The molecule with the lowest mean B value (chain M) was used to generate the figures.
The quality of the model was assessed using Molprobity (38) and it was found to have good stereochemistry (see Table 1).
Table 1.
Crystallographic data statistics
| Data collection statistics | |
| Unit cell dimensions (a × b × c) | 192.374 × 98.268 × 104.960 α = β = γ = 90° |
| Space group | P21212 |
| Number of observations | 351 477 |
| Unique reflections | 146 433 |
| Rsym (%) (last shell)a | 12.4 (47.0) |
| I/σI (last shell) | 10.1 (2.4) |
| Completeness (%) (last shell) | 98.7 (94.5) |
| Refinement statistics | |
| Resolution (Å) | 2.4 |
| Rcryst (%)b | 20.9 |
| Rfree (%)c | 25.2 |
| Number of complexes in asymmetric unit | 4 |
| Number of waters | 799 |
| Mean B value (Å) | 45.39 |
| Rms deviation from ideal values | |
| Bond length (Å) | 0.006 |
| Bond angle (°) | 1.135 |
| Dihedral angle (°) | 21.533 |
| Improper angle (°) | 0.885 |
| Ramachandran statistics | |
| Residues in: | |
| Favored regions | 96.99 |
| Allowed regions | 100 |
| Disallowed regions | 0 |
aRsym = ∑ (|Ii−<I>|)/∑(Ii), where Ii is the intensity of the ith observation and is the mean intensity of the reflection.
bRcryst = ∑ ‖Fo|−|Fc‖/∑|Fo|, calculated from working dataset.
cRfree is calculated from 5% of data randomly chosen not to be included in refinement.
RESULTS
Mismatch extension ability of the members of the family X of DNA polymerases
In humans, all three template-dependent DNA polymerases from family X (Pol β, Pol λ and Pol μ) lack a proofreading 3′→5′ exonuclease, but they differ in their interaction with DNA and, consequently, in their biological function during DNA repair (1).
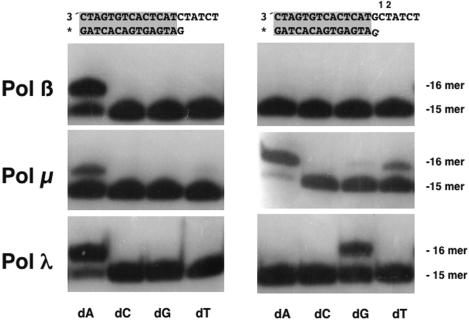
Each of the three enzymes preferentially incorporates the correct nucleotide (dAMP) into a matched primer-terminus (Figure 1, left panels). However, when the primer-terminus is not correctly paired to a template base (as an example, a dG·dGMP mispair at the primer-terminus), the behavior of the three enzymes is significantly different (Figure 1, right panels). While Pol β is not able to extend the mispaired primer-terminus with any of the 4 nt, Pol μ and Pol λ use different solutions for extension. In agreement with previous findings (39), Pol μ prefers to stabilize/induce a relocation of the primer to form a dC·dGMP correct pair (dC at position 1), that allows incorporation of a complementary dA residue opposite the next templating base (dT at position 2). In agreement with that mechanism, when the template base at position 2 was changed, the inserted nucleotide changed accordingly (data not shown). With lower efficiency, Pol μ is also able to insert an untemplated dT residue, perhaps due to its intrinsic terminal transferase activity (34,40). The least efficient solution is the insertion of dG, suggesting that Pol μ is poorly able to directly extend the mismatch.
Figure 1.
Human Pol λ directly extends a mismatched end. Two different template-primer structures were used, differing in the 3′ end nucleotide of the primer. Extension of the 5′ end labeled primer (*) was examined by PAGE. Reactions were carried out as described in Materials and Methods.
In clear contrast, when human Pol λ encounters the dG·dGMP mispair, it directly extends the mismatch by incorporating a dG residue, complementary to the first available templating base (dC at position 1).
Therefore, both Pol μ and Pol λ are able to extend mismatched DNA primers but with very different consequences. The action of Pol μ on the mismatch would mainly produce a −1 deletion associated with extension of the mismatch, whereas Pol λ would predominantly extend the mismatch without producing additional errors.
Steady-state kinetic analysis of mismatch extension efficiency of human Pol λ
A truncated form of Pol λ corresponding to the Pol β-like core has been recently crystallized in the presence of DNA and dNTP (11). This 39 kDa form of human Pol λ, lacking the first 241 amino acids encoding the BRCT and Ser/Pro-rich domains, but having wild-type like activity (10), was used to examine the steady-state kinetics of primer extension for the four correctly paired and 12 mispaired termini in ‘open’ template-primer structures. Direct mismatch extension by Pol λ on these substrates was assessed in the presence of dGTP. The rate of nucleotide incorporation, obtained as described in Materials and Methods, was plotted as a function of nucleotide concentration and Vmax and Km apparent values for extension of each primer-terminus were then determined. From these values, the frequency of mismatch extension (fext°), which is the ratio of the apparent kcat/Km of extension from the mispair to the apparent kcat/Km of extension from a correct base pair, was then calculated.
As shown in Table 2, human Pol λ is capable of extending most mismatches, although the efficiency of extension strongly varies depending on the nature of the mismatch. Thus, while Pol λ has low efficiency for extension of mismatches having a pyrimidine as the template base (dC·dAMP; dC·dCMP; dC·dTMP; dT·dCMP; dT·dTMP), it is relatively efficient at extending mismatches when the template base is a purine. The exception to this trend is the mismatch dT·dGMP, extended with a catalytic efficiency that is similar to those obtained with mismatches employing purines as template bases. Interestingly, this is one of the most efficiently inserted and extended mismatches by DNA polymerases (41). As it has been reported for Bacillus stearothermophilus DNA polymerase (42), extension of dT·dGMP by Pol λ could be favored by a special geometry in which the mismatched primer base (dG) adopts a conformation similar to that of a cognate base at that position.
Table 2.
Frequencies of extension from matched and mismatched termini by human Pol λ on ‘open’ template-primer molecules
| T.P | Vmax μM min−1 | Km μM | kcat min−1 | kcat min−1/Km μM | foext |
|---|---|---|---|---|---|
| A·A | (1.44 ± 0.45) × 10−4 | 15.51 ± 1.21 | 1.92 10−3 | 1.24 10−4 | 1.04 10−2 |
| A·C | (5.71 ± 0.86) × 10−4 | 9.87 ± 1.22 | 7.61 10−3 | 7.71 10−4 | 6.48 10−2 |
| A·G | (3.25 ± 0.65) × 10−4 | 7.75 ± 0.68 | 3.91 10−3 | 5.09 10−4 | 4.28 10−2 |
| A·T | (2.86 ± 0.46) × 10−3 | 7.84 ± 0.83 | 9.18 10−2 | 1.19 10−2 | |
| C·A | n.d. | n.d. | n.d. | n.d. | n.d. |
| C·C | n.d. | n.d. | n.d. | n.d. | n.d. |
| C·G | (7.49 ± 0.82) × 10−4 | 9.61 ± 1.22 | 2.44 10−1 | 2.54 10−2 | |
| C·T | n.d. | n.d. | n.d. | n.d. | n.d. |
| G·A | (1.98 ± 0.26) × 10−4 | 15.28 ± 2.35 | 1.98 10−3 | 1.30 10−4 | 7.83 10−3 |
| G·C | (6.49 ± 0.54) × 10−4 | 7.83 ± 0.52 | 1.30 10−1 | 1.66 10−2 | |
| G·G | (1.78 ± 0.13) × 10−4 | 9.37 ± 1.85 | 2.37 10−3 | 2.53 10−4 | 1.52 10−2 |
| G·T | (4.47 ± 0.48) × 10−4 | 7.97 ± 0.48 | 4.99 10−3 | 6.25 10−4 | 3.77 10−2 |
| T·A | (1.81 ± 0.29) × 10−4 | 1.48 ± 0.27 | 3.62 10−2 | 2.45 10−2 | |
| T·C | n.d. | n.d. | n.d. | n.d. | n.d. |
| T·G | (2.29 ± 0.45) × 10−4 | 8.05 ± 1.61 | 2.76 10−3 | 3.54 10−4 | 1.45 10−2 |
| T·T | n.d. | n.d. | n.d. | n.d. | n.d. |
Primer extension was measured in the presence of dGTP, the next correct nucleotide. Data are means (± standard error) from at least three independent experiments. n.d. not detected.
Even when a purine is the template base of the mispair, the catalytic efficiencies are comparatively lower than those observed when extending matched primer ends (varying over a 100-fold range). Consequently, human Pol λ extends from mispairs on ‘open’ template-primer molecules with an average frequency of 2.76 × 10−2. Therefore, the kinetic difference for the efficiency of extending a matched terminus versus a mismatched terminus is only about 36-fold on average, when considering the seven mismatches that were extended.
The presence of a downstream oligonucleotide with a 5′ phosphate group allows Pol λ to perform mismatch extension on 12 possible mispairs
DNA molecules having small gaps are the preferred DNA substrate for Pol λ. In these substrates, the 8 kDa domain of the enzyme, which contains its intrinsic dRP lyase activity (7), establishes interactions with the downstream end of the gap (10) that modulate the activity of the enzyme. Thus, the presence of a 5′ phosphate group flanking the gap produces a significant increase in the formation of the enzyme–DNA binary complex (A. J. Picher and L. Blanco, unpublished data), and also increases the processivity of the enzyme (9). Both in vivo roles attributed to Pol λ [i.e. gap-filling synthesis during BER and repair of double-strand breaks through NHEJ (18,24–26)] involve synthesis on a gapped substrate. Therefore, it was important to measure the capacity of Pol λ as a mismatch extender when acting on its preferred substrate, a single-nucleotide gap having a flanking 5′ phosphate.
As shown in Table 3, the catalytic efficiency for extending the four complementary primer-termini (dA·dTMP; dC·dGMP; dG·dCMP; dT·dAMP) on a 1 nt 5′-phosphate gap molecule was 320-, 130-, 270- and 340-fold higher respectively, than that for a ‘open’ template-primer molecule having identical primer DNA sequence. Interestingly, extension of all 12 different mismatches was detected on the gapped-DNA (Table 3, see also Figure 3), in contrast with what was observed on the ‘open’ template-primer substrates (Table 2). This significant increase in the catalytic efficiency was similar to the average increase obtained for the matched primer-termini (260-fold). As a mean value, human Pol λ extends from mispairs on gapped-DNA molecules with an average frequency of 1.53 × 10−2. Thus, the kinetic differences for extending a matched versus mismatched terminus on gapped-DNA is about 65-fold.
Table 3.
Frequencies of extension from matched and mismatched termini by human Pol λ on 1 nt 5′-phosphate gapped molecules
| T.P | Vmax μM min−1 | Km μM | kcat min−1 | kcat min−1/Km μM | foext |
|---|---|---|---|---|---|
| A·A | (1.12 ± 0.16) × 10−3 | 7.48 ± 0.54 | 2.24 10−1 | 3.00 10−2 | 8.00 10−3 |
| A·C | (2.27 ± 0.23) × 10−3 | 1.85 ± 0.25 | 4.54 10−1 | 2.45 10−1 | 6.53 10−2 |
| A·G | (1.46 ± 0.17) × 10−3 | 7.99 ± 1.65 | 2.92 10−1 | 3.66 10−2 | 9.76 10−3 |
| A·T | (2.25 ± 0.34) × 10−3 | 0.12 ± 0.01 | 4.50 10−1 | 3.75 | |
| C·A | (2.01 ± 0.41) × 10−4 | 42.82 ± 6.49 | 4.02 10−2 | 9.39 10−4 | 2.94 10−4 |
| C·C | (4.36 ± 0.76) × 10−4 | 6.84 ± 0.17 | 2.18 10−2 | 3.19 10−3 | 1.00 10−3 |
| C·G | (5.59 ± 0.96) × 10−3 | 0.35 ± 0.04 | 1.12 | 3.19 | |
| C·T | (4.21 ± 0.49) × 10−4 | 8.27 ± 0.78 | 8.42 10−2 | 1.02 10−2 | 3.19 10−3 |
| G·A | (3.91 ± 0.78) × 10−4 | 4.48 ± 0.21 | 7.82 10−2 | 1.75 10−2 | 3.93 10−3 |
| G·C | (3.78 ± 0.73) × 10−3 | 0.17 ± 0.04 | 7.56 10−1 | 4.45 | |
| G·G | (2.49 ± 0.45) × 10−3 | 5.73 ± 1.29 | 4.98 10−1 | 8.69 10−2 | 1.95 10−2 |
| G·T | (2.44 ± 0.30) × 10−3 | 1.94 ± 0.09 | 4.88 10−1 | 2.52 10−1 | 5.66 10−2 |
| T·A | (3.73 ± 0.61) × 10−4 | 0.09 ± 0.02 | 7.46 10−1 | 8.29 | |
| T·C | (3.55 ± 0.61) × 10−4 | 5.51 ± 0.71 | 7.10 10−2 | 1.29 10−2 | 1.56 10−3 |
| T·G | (6.41 ± 0.66) × 10−4 | 1.24 ± 0.28 | 1.28 10−1 | 1.03 10−1 | 1.24 10−2 |
| T·T | (1.84 ± 0.36) × 10−4 | 8.17 ± 0.54 | 3.68 10−2 | 2.00 10−2 | 2.41 10−3 |
Primer extension was measured in the presence of dGTP, the next correct nucleotide. Data are means (± standard error) from at least three independent experiments.
Structural analysis of mismatch extension by Pol λ
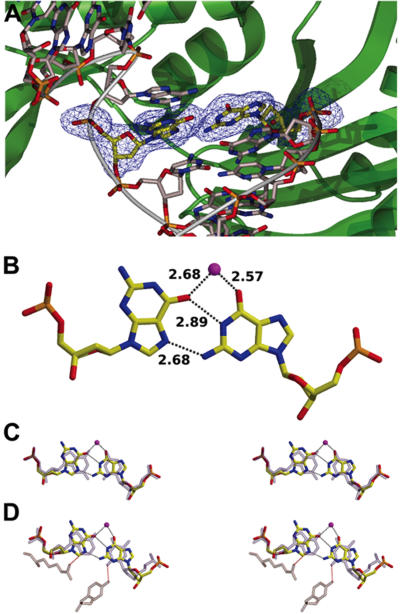
Mismatch extension kinetics for other polymerases indicated that, in general, purine–purine mispairs are extended with lower efficiencies than pyrimidine–pyrimidine or purine–pyrimidine mispairs (41,43,44). This is consistent with the importance of the size and shape of the base pair for DNA polymerase fidelity (45). In contrast, Pol λ appears to be capable of extending purine–purine mismatches with a similar efficiency (even higher in some cases) than pyrimidine–pyrimidine or purine–pyriminidine mismatches. To understand this observation we decided to crystallize Pol λ with a DNA duplex containing a single-nucleotide gap with a dG·dGMP mispair in the primer-terminus. This mispair is known to be extended with very low efficiency by some DNA polymerases (43).
Crystals diffracting at 2.4 Å were obtained, and the structure was refined to a R-factor of 21.04% and a R-free of 25.45% (Table 1; see Materials and Methods). In this complex, Pol λ is in a closed conformation, similar to that observed in a complex with a correctly matched substrate [r.m.s.d. of 0.208 Å for 327 C-α atoms; (10)]. In the new structure, a dG·dGMP mispair is observed at the primer-terminus, where the templating dG has rotated about the N-glycosidic bond and is in a syn conformation (Figure 2). Interestingly, no significant amino acid side chain movements are required to accommodate the mispair in the primer-terminus, and no significant perturbation in the DNA backbone is observed, indicating that Pol λ is capable of easily accommodating even a bulky purine–purine mispair, consistent with the high relative efficiency of mismatch extension reported here. However, extension from a dG·dGMP mismatch is about 50-fold less efficient that extension from a correct dG·dCMP pair, and part of this effect might be related to a slight difference in the conformation of the ribose (and the 3′ OH nucleophile) of the primer-terminal nucleotide. Moreover, upon dNTP binding, Pol λ undergoes a conformational change that results in a shift in the position of the template strand through the establishment of protein interactions of the Arg517 and Tyr505 side chains with the minor groove of this base pair (11). These interactions are believed to be critical for correct positioning of the substrates, and thus for catalysis. Optimal geometry, as determined by those interactions, will thus strongly depend on the geometry of the terminal base pair. Modeling of these Pol λ catalytic conformational changes on the Pol λ / dG·dGMP crystal suggests that this mispair will not properly establish those interactions (Figure 2), thus likely having a negative effect on the rate of the reaction.
Figure 2.
3D-structure of the human Pol λ core complexed with a 1 nt gapped-DNA having a dG·dGMP mismatched primer-terminus. (A) View of the Pol λ active site showing the dG·dGMP mismatch (yellow). The template and primer strands are shown in gray and the protein is shown in green. A simulated annealing Fobs-Fcalc omit electron density map where the terminal base pair was omitted, contoured at 2.5 σ, is shown (blue). (B) Bonding distances between the template base (dG) and the 3′ primer nucleotide (dG) forming a dG·dGMP mismatch corresponding to the structure shown in (A). Hydrogen bonds are shown as dotted lines. Water molecule is represented as a purple ball. (C) Stereo-view showing the superimposition of the dG·dGMP terminal mismatch (colored) and a correct dG·dCMP base pair (gray) derived from a binary complex of Pol λ (PDB entry 1XSL). (D) Stereo-view showing the superimposition of the dG·dGMP terminal mismatch (colored) and a correct dA·dTMP base pair (gray) derived from the ternary complex of Pol λ (PDB entry 1XSN).
DISCUSSION
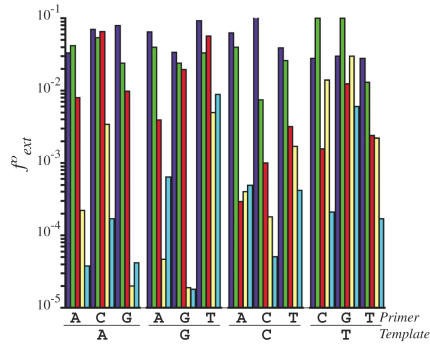
Family X DNA polymerases include those implicated in several different DNA repair processes, such as BER, NHEJ and V(D)J recombination. Crucial differences in their biochemical properties enable each member of this family to take part in different synthetic reactions. Here, we show that Pol λ is relatively efficient in extending single base–base mispairs, supporting a role for Pol λ in DNA synthesis processes that may require the extension of mismatches. Mismatch extension is a key event for mutagenesis, and it is also believed to be important for processes, such as TLS. Indeed, efficient mismatch extension is a characteristic of several polymerases involved in TLS, such as Pol ζ, Pol η, Pol ι and Pol κ (30–33,46,47). Pol λ has been proposed to play a role in TLS for incorporation opposite a lesion (28,29). Interestingly, concerning its ability to extend from mismatches, Pol λ resembles these DNA polymerases, suggesting its capacity to participate in the TLS extension step. On ‘open’ template-primer DNA molecules that may be relevant to TLS, for certain mismatches Pol λ shows extension frequencies (fext°) similar to those of the most promiscuous mismatch extenders known, Pol κ and Pol ζ (30,31). Moreover, on 1 nt 5′-phosphate gapped-DNA molecules that may be particularly relevant to DNA repair, Pol λ showed an efficient mismatch extension ability for all possible mispairs (Table 3). When a purine is the template base of the mismatch, Pol λ shows values close to the ones of Pol κ and Pol ζ, whereas, when a pyrimidine is the template base, the mismatch extension efficiencies decrease and are similar to those obtained by Pol β [(48); see Figure 3). Pol α from Drosophila melanogaster displays a much more modest ability to extend mispairs [(43); see Figure 3).
Figure 3.
Mismatch extension efficiencies of different DNA polymerases. Mismatch extension efficiencies (fext°) of different DNA polymerases: human Pol κ [dark blue; (40)], yeast Pol ζ [green; (41)], human Pol λ (red; obtained from Table 3), human Pol β [yellow; (48)] and Drosophila melanogaster Pol α [light blue; (37)].
The in vivo relevance of the mismatch extension capacity of Pol λ could be related to its role in gap-filling during the NHEJ repair process. During NHEJ, a DNA polymerase is needed for any event that requires filling of gaps or extension of the 3′ end at 5′ overhangs. Recent in vitro studies have shown that both Pol λ and Pol μ are able to bind the NHEJ proteins and perform gap-filling synthesis during rejoining (18,21,25,26). Moreover, both Pol λ and Pol μ are able to catalyze NHEJ of ends with very limited complementarity. The ability of Pol λ to directly extend mismatches distinguishes it from Pol μ, which appears not to be an efficient direct extender of mispaired primer-termini. Family X DNA polymerases thus seem to have become highly specialized, able to perform different tasks even in the same general process of DNA repair. While there seems to be certain redundancy of function within the gap-filling step of NHEJ, the specific DNA polymerase that operates on a particular joining reaction appears to be determined by the sequence of the ends. Thus, while Pol μ would be particularly suited for joining reactions where the two strands display no homology (18), Pol λ would be more suited for substrates with microhomology, particularly when implying misalignments (27) or, as shown here, implying a mismatched primer-terminus.
The ability of Pol λ to extend mispaired primer-termini prompted us to investigate the structural aspects of mismatch extension for this polymerase. Previous work (42,49) has shown that mismatches at the primer-terminus can disrupt significantly the polymerase active site, impairing catalysis. This is true even when the mismatch is located a few base pairs upstream of the primer-terminus (42). This suggests that DNA polymerases discriminate against terminal mismatches both at the binding step and the insertion step. For replicative polymerases, this is believed to be a mechanism by which they can favor proofreading of the misincorporated nucleotide: these enzymes monitor correct base pairing through minor groove interactions with several base pairs upstream of the active site (50), and these interactions can be disrupted by a mismatch (42). Pol λ establishes similar interactions with the DNA duplex, but only with the terminal base pair and in the catalytic conformation. The fewer number of interactions has been suggested to be one of the structural reasons why Pol λ can tolerate significant distortion upstream of the template-primer. In addition, the present structure suggests that, at least before those minor groove interactions are established, Pol λ is capable of binding mismatched template-primers. Thus, at least for dG·dGMP mispairs, Pol λ seems to lack a mechanism to discriminate against the terminal mispair during DNA binding. However, the minor groove interactions that are established during the incoming nucleotide binding are unlikely to form, thus accounting for the lower catalytic efficiency on mismatched substrates. This structure thus confirms the importance of the Pol λ conformational change to polymerase selectivity, and further supports the unusual tolerance of Pol λ for potentially mutagenic substrates.
Acknowledgments
The authors thank Dr Samuel Wilson for providing human Pol β. This work was supported by Ministerio de Ciencia y Tecnología Grant BMC 2003-00186 to L.B., by the Division of Intramural Research, NIEHS, NIH, DHHS to T.A.K. and by an institutional grant to Centro de Biología Molecular ‘Severo Ochoa’ from Fundación Ramón Areces. A.J.P. was recipient of a fellowship from the Ministerio de Educación y Ciencia. Funding to pay the Open Access publication charges for this article was provided by Ministerio de Ciencia y Tecnología.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bebenek K., Kunkel T.A. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 2.Burgers P.M., Koonin E.V., Bruford E., Blanco L., Burtis K.C., Christman M.F., Copeland W.C., Friedberg E.C., Hanaoka F., Hinkle D.C., et al. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 2001;276:43487–43490. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 3.Ito J., Braithwaite D.K. Compilation and alignment of DNA polymerases sequences. Nucleic Acids Res. 1991;19:4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliveros M., Yanez R.J., Salas M.L., Salas J., Vinuela E., Blanco L. Characterization of an African swine fever virus 20 kDa DNA polymerase envolved in DNA repair. J. Biol. Chem. 1997;272:30899–30910. doi: 10.1074/jbc.272.49.30899. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Diaz M., Dominguez O., Lopez-Fernandez L.A., de Lera L.T., Saniger M.L., Ruiz J.F., Parraga M., Garcia-Ortiz M.J., Kirchhoff T., del Mazo J., et al. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potencial role in meiosis. J. Mol. Biol. 2000;301:851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- 6.Nick McElhinny S.A., Ramsden D.A. Sibling rivalry: competition between Pol X family members in V(D)J recombination and general double strand break repair. Immunol. Rev. 2004;200:156–164. doi: 10.1111/j.0105-2896.2004.00160.x. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Diaz M., Bebenek K., Kunkel T.A., Blanco L. Identification of an intrinsic 5′-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: a possible role in base excision repair. J. Biol. Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 8.Braithwaite E.K., Kedar P.S., Lan L., Polosina Y.Y., Asagoshi K., Poltoratsky V.P., Horton J.K., Miller H., Teebor G.W., Yasui A., et al. DNA polymerase lambda protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J. Biol. Chem. 2005;280:31641–31647. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Diaz M., Bebenek K., Sabariegos R., Dominguez O., Rodriguez J., Kirchhoff T., Garcia-Palomero E., Picher A.J., Juarez R., Ruiz J.F., et al. DNA polymerase lambda, a novel DNA repair enzyme in human cells. J. Biol. Chem. 2002;277:13184–13191. doi: 10.1074/jbc.M111601200. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Diaz M., Bebenek K., Krahn J.M., Blanco L., Kunkel T.A., Pedersen L.C. A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology. Mol. Cell. 2004;13:561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Diaz M., Bebenek K., Kranh J.M., Kunkel T.A., Pedersen L.C. A closed conformation for the Pol lambda catalytic cycle. Nature Struct. Mol. Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 12.Valerie K., Povirk L.F. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 13.Puchta H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 2005;56:1–14. doi: 10.1093/jxb/eri025. [DOI] [PubMed] [Google Scholar]
- 14.Daley J.M., Palmbos P.L., Wu D., Wilson T.E. Nonhomologous end joining in yeast. Annu. Rev. Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 15.Aylon Y., Kupiec M. DSB repair: the yeast paradigm. DNA Repair. 2004;3:797–815. doi: 10.1016/j.dnarep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Wilson T.E., Topper L.M., Palmbos P.L. Non-homologous end-joining: bacteria join the chromosome breakdance. Trends Biochem. Sci. 2003;28:62–66. doi: 10.1016/S0968-0004(03)00005-7. [DOI] [PubMed] [Google Scholar]
- 17.Lieber M.R., Ma Y., Pannicke U., Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nature Rev. Mol. Cell. Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 18.Nick McElhinny S.A., Havener J.M., Garcia-Diaz M., Juarez R., Bebenek K., Kee B.L., Blanco L., Kunkel T.A., Ramsden D.A. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Wilson T.E., Lieber M.R. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J. Biol. Chem. 1999;274:23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- 20.Benedict C.L., Gilfillan S., Thai T.H., Kearney J.F. Terminal deoxynucleotidyl transferase and repertoire development. Immunol. Rev. 2000;175:150–157. [PubMed] [Google Scholar]
- 21.Nick McElhinny S.A., Ramsden D.A. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertocci B., De Smet A., Berek C., Weill J.C., Reynaud C.A. Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity. 2003;19:203–211. doi: 10.1016/s1074-7613(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 23.Bebenek K., Garcia-Diaz M., Blanco L., Kunkel T.A. The frameshift infidelity of human DNA polymerase lambda. Implications for function. J. Biol. Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 24.Lee J.W., Blanco L., Zhou T., Garcia-Diaz M., Bebenek K., Kunkel T.A., Wang Z., Povirk L.F. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 25.Ma Y., Lu H., Tippin B., Goodman M.F., Shimazaki N., Koiwai O., Hsieh C.L., Schwarz K., Lieber M.R. A biochemically defined system for mammalian nonhomologous end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 26.Fan W., Wu X. DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interacy with XRCC4-ligase IV complex. Biochem. Biophys. Res. Commun. 2004;323:1328–1333. doi: 10.1016/j.bbrc.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Diaz M., Bebenek K., Krahn M.J., Pedersen L.C., Kunkel T.A. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 28.Maga G., Villani G., Ramadan K., Shevelev I., Tanguy Le Gac N., Blanco L., Spadari S., Hubscher U. Human DNA polymerase lambda functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J. Biol. Chem. 2002;277:48434–48440. doi: 10.1074/jbc.M206889200. [DOI] [PubMed] [Google Scholar]
- 29.Blanca G., Villani G., Shevelev I., Ramadan K., Spadari S., Hubscher U., Maga G. Human DNA polymerases lambda and beta show different efficiencies of translesion DNA synthesis past abasic sites and alternative mechanisms for frameshift generation. Biochemistry. 2004;43:11605–11615. doi: 10.1021/bi049050x. [DOI] [PubMed] [Google Scholar]
- 30.Washington M.T., Johnson R.E., Prakash L., Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc. Natl Acad. Sci. USA. 2002;99:1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson R.E., Washington M.T., Haracska L., Prakash S., Prakash L. Eukaryotic polymerases iota and zeta act sequentally to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 32.Vaisman A., Tissier A., Frank E.G., Goodman M.F., Woodgate R. Human DNA polymerase iota promiscuous mismatch extension. J. Biol. Chem. 2001;276:30615–30622. doi: 10.1074/jbc.M102694200. [DOI] [PubMed] [Google Scholar]
- 33.Washington M.T., Johnson R.E., Prakash S., Prakash L. Mismatch extension ability of yeast and human DNA polymerase eta. J. Biol. Chem. 2001;276:2263–2266. doi: 10.1074/jbc.M009049200. [DOI] [PubMed] [Google Scholar]
- 34.Dominguez O., Ruiz J.F., Lain de Lera T., Garcia-Diaz M., Gonzalez M.A., Kirchhoff T., Martinez-A C., Bernad A., Blanco L. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowsky Z., Minor V. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 38.Lovell S.C., Davis I.W., Arendall W.B., 3rd, de Bakker P.I., Word J.M., Prisant M.G., Richardson J.S., Richardson D.C. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y., Wu X., Yuan F., Xie Z., Wang Z. Highly frequent frameshift DNA synthesis by human DNA polymerase mu. Mol. Cell. Biol. 2001;21:7995–8006. doi: 10.1128/MCB.21.23.7995-8006.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Covo S., Blanco L., Livneh Z. Lesion bypass by human DNA polymerase mu reveals a template-dependent, sequence-independent nucleotidyl transferase activity. J. Biol. Chem. 2004;279:859–865. doi: 10.1074/jbc.M310447200. [DOI] [PubMed] [Google Scholar]
- 41.Joyce C.M., Sun X.C., Grindley N.D. Reactions at the polymerase active site that contribute to the fidelity of Escherichia coli DNA polymerase I (Klenow fragment) J. Biol. Chem. 1992;267:24485–24500. [PubMed] [Google Scholar]
- 42.Johnson S.J., Beese L.S. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 43.Mendelman L.V., Petruska J., Goodman M.F. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J. Biol. Chem. 1990;265:2338–2346. [PubMed] [Google Scholar]
- 44.Huang M.M., Arnheim N., Goodman M.F. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 1992;20:4567–4573. doi: 10.1093/nar/20.17.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kunkel T.A. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 46.Lawrence C.W. Cellular functions of DNA polymerase zeta and Rev1 protein. Adv. Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- 47.Prakash S., Johnson R.E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 48.Beard W.A., Shock D.D., Wilson S.H. Influence of DNA structure on DNA polymerase beta active site function: extension of mutagenic DNA intermediates. J. Biol. Chem. 2004;279:31921–31629. doi: 10.1074/jbc.M404016200. [DOI] [PubMed] [Google Scholar]
- 49.Krahn J.M., Beard W.A., Wilson S.H. Structural insights into DNA polymerase beta deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure. 2004;12:1823–1832. doi: 10.1016/j.str.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 50.Doublie S., Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr. Opin. Struct. Biol. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]