Abstract
Most regulatory pathways are governed by the reversible phosphorylation of proteins. Recent developments in mass spectrometry-based technology allow the large-scale analysis of protein phosphorylation. Here, we show the application of immobilized metal affinity chromatography to purify phosphopeptides from Arabidopsis extracts. Phosphopeptide sequences were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS/MS). A total of 79 unique phosphorylation sites were determined in 22 phosphoproteins with a putative role in RNA metabolism, including splicing of mRNAs. Among these phosphoproteins, 12 Ser/Arg-rich (SR) splicing factors were identified. A conserved phosphorylation site was found in most of the phosphoproteins, including the SR proteins, suggesting that these proteins are targeted by the same or a highly related protein kinase. To test this hypothesis, Arabidopsis SR protein-specific kinase 4 (SRPK4) that was initially identified as an interactor of SR proteins was tested for its ability to phosphorylate the SR protein RSp31. In vitro kinase assays showed that all in vivo phosphorylation sites of RSp31 were targeted by SRPK4. These data suggest that the plant mRNA splicing machinery is a major target of phosphorylation and that a considerable number of proteins involved in RNA metabolism may be targeted by SRPKs.
INTRODUCTION
Reversible phosphorylation of proteins is an important posttranslational regulatory mechanism and can influence activity, subcellular localization, protein–protein interactions and turnover of the protein involved. In the past, however, the analysis of phosphorylation sites of proteins has been a great challenge. In the last few years there has been an explosive growth in the amount of studies describing the use of immobilized metal affinity chromatography (IMAC) (1) or other phosphopeptide purification methods such as metal oxide affinity chromatography (2) or TiO2 (3) coupled to mass spectrometric analysis. IMAC has been used most successfully for the large-scale identification of phosphorylation sites, although TiO2 has been shown recently to be highly selective (4) and might promise to be an alternative to IMAC in the future. Many of these studies focused on tyrosine phosphorylation in mammalian cells, since this represents a small and therefore easier analyzable component of the complex phosphoproteome.
In contrast to the wealth of phosphoproteomic investigations performed on yeast and animal systems, only a few studies have focused on plants. IMAC coupled to mass spectrometry has been used for small-scale analysis of Arabidopsis thylakoid membrane phosphoproteins, identifying around 10 sites per study (5,6). In a different report, this technique was used to isolate 253 phosphopeptides from the moss Physcomitrella patens in a complex mixture but failed to identify the sequences of most of the peptides (7). The first successful large-scale study described >300 phosphorylation sites of Arabidopsis plasma membrane proteins (8). This revealed that receptor-like kinases (RLKs) and transport systems are major targets of phosphorylation. The RLKs are targeted by kinases at unexpected regions, such as the juxtamembrane domain. Comparative analysis allowed the authors to group phosphorylation sites into conserved motifs, which may be targeted by identical or similar kinases. Thus, large-scale phosphoproteomics can increase our knowledge of posttranslational regulation of plant proteins because of its unbiased, global approach.
We have set up the IMAC-based mass spectrometric technology and used it for large-scale identification of in vivo phosphorylation sites of Arabidopsis proteins from nuclear and cytosolic extracts. This allowed the identification of both known and novel phosphorylation sites in two sucrose-phosphate synthase isoforms (9). Here, we focus specifically on proteins involved in RNA metabolism and identify 22 phosphoproteins. Most of the identified proteins are predicted to be involved in pre-mRNA splicing, including so-called Ser/Arg-rich (SR) proteins. SR proteins promote both constitutive and alternative splicing and have overlapping but distinct functions. They interact with specific RNA sequences mainly through RNA recognition motif (RRM) domains, sometimes with contributions of the RS domains. Protein interactions occur mainly through the RS domain and are regulated by phosphorylation. Phosphorylated SR proteins are recruited from sites of nuclear storage in speckles and are thought to stimulate splicing by interacting with proteins at the 5′- and 3′-splice site. Some SR proteins were shown to shuttle between the cytoplasm and nucleus playing a role in export of processed mRNA and in translation. All these events are regulated by phosphorylation/dephosphorylation of the RS domain (10).
Analysis of the Arabidopsis genome has revealed at least 19 SR proteins, which is almost twice as many as in animals (11). In plant cells, SR proteins mainly localize to nuclear speckles, and several can shuttle between the nucleus and the cytosol (12–15). As in animal cells, the subcellular localization of these proteins is affected by their phosphorylation status in plants (16). In addition, protein–protein interactions in RS containing plant proteins have been shown to be phosphorylation dependent (17) corroborating the importance of the phosphorylation status of SR proteins for their respective activity.
Our data now indicate that the plant splicing machinery is a major target of regulatory phosphorylation. We show that plant SR proteins, like their animal counterparts, are extensively phosphorylated at Ser residues in their RS domains. Analysis of the phosphorylation sites in the splicing factors suggests that related kinases target a set of conserved motifs. In conclusion, the protocol described here can serve for the global analysis of in vivo phosphorylation in plants and can be extended for quantitative analysis of signaling pathways in the future.
MATERIALS AND METHODS
Preparation of nuclear and cytosolic extracts
Dark-grown Arabidopsis root cell culture were shaken for 5 days in the dark at 150 r.p.m. and harvested by filtration and frozen in liquid nitrogen. Cells were ground with a mortar and a pestle. Crude nuclear extracts were prepared by slowly adding the cell material to 34 ml of ice-cold nuclear isolation buffer [20 mM Tris, pH 7.8, 70% glycerol, 50 mM sucrose, 5 mM MgCl2, 5 mM KCl, 1 mM DTT, 0.5 mM phenylmethlysulfonyl fluoride (PMSF), 2 µg/ml leupeptin, 1 µg/ml pepstatin and 1.8 µg/ml aprotinin]. The suspension was then stirred for 30 min at 4°C, filtered through a 20 µm nylon mesh without pressure, and centrifuged for 1 h at 4°C at 1790 g. The pellet was resuspended in 300 µl extraction buffer (20 mM Tris, pH 7.8, 5 mM MgCl2, 0.5 M NaCl, 1 mM DTT, 0.2 mM PMSF, 1 mM NaF, 0.5 mM Na3VO4 and 15 mM β-glycerophosphate). The pellet was resuspended by gentle shaking for 30 min at 4°C and centrifuged for 30 min 4°C at 20 120 g. The supernatant was taken and dialyzed to 50 mM Tris, pH 8.0. For rough cytosolic protein isolation, 400 µl buffer (25 mM Tris, pH 7.5, 80 mM NaCl, 2 mM DTT, 4 mM EDTA, 1 mM NaF, 0.5 mM Na3VO4, 15 mM β-glycerophosphate, 15 mM 4-nitrophenylphosphate, 0.5 mM PMSF, 5 µg/ml leupeptin and 10 µg/ml aprotinin) was added per ml of ground cell material. The sample was then centrifuged at 20 000 g for 40 min at 4°C. The supernatant fraction was used directly for the isolation of phosphopeptides.
Isolation of phosphopeptides from nuclear extracts by IMAC
Approximately 40 µg of nuclear extract were reduced with 2 µg of DTT, alkylated with 10 µg of iodoacetamide, and digested overnight at 37°C with 4 µg of trypsin (Roche). The digest was terminated by the addition of formic acid to a final concentration of 3%. To desalt the sample, it was loaded onto Oligo R3 (ABI), which was filled into constricted gel loader tips. The bound peptides were washed with 0.1 M acetic acid (HAc) and eluted with 0.1 M HAc containing 50% acetonitrile (Chromasolv®; Sigma–Aldrich, Seelze, Germany). The eluate was diluted to 30% acetonitrile and incubated in batch with ∼5 µl of POROS MC material (ABI), which was charged with FeCl3. After 30 min of incubation at room temperature the suspension was filled into a constricted gel loader tip. The bound peptides were washed with 0.1 M HAc containing 30% acetonitrile and eluted with 1 M NH4OH containing 30% acetonitrile.
IMAC protocol used for isolation of phosphopeptides from cytosolic extracts
Proteins were precipitated from 300 µg of cytosolic extracts as described previously (18). The protein precipitate was dissolved in 8 M urea in 0.5 M NH4HCO3 and reduced and alkylated as described above. The solution was diluted to 0.8 M urea and proteins were digested with 5 µg of trypsin as described above. To desalt the sample, 40 µl of settled Oligo R3 was filled into constricted 200 µl tips. The sample was loaded and bound peptides were washed with 0.1% HAc and eluted with 70% acetonitrile in 0.1% HAc. Thereafter, the sample was dried in a speed vac. To reduce nonspecific binding of acidic peptides to the IMAC column, peptide carboxyl groups were esterified in methanolic HCl as described previously (19). Methylesterified peptides were dried in a speed vac and redissolved in a solution containing equal parts of methanol, acetonitrile and 0.02% HAc as described previously (20). The peptide solution was incubated with 5 µl of IMAC material (Phos-Select; Sigma). After 1 h of incubation the suspension was filled into a constricted gel loader tip, washed with 0.02% HAc and bound peptides were eluted with 125 mM of Na2HPO4 (pH 6.0).
Analysis of phosphopeptides by nano-LC-MS/MS and MS/MS/MS
IMAC eluates were separated on a nano-reversed phase high-performance liquid chromatography (HPLC) (Ultimate, Switchos, Famos, LC Packings, Amsterdam, The Netherlands). Samples were applied to a trapping column (PepMap C18, 300 µm × 5 mm) using 0.1% TFA (Pierce Biotechnology Inc., Rockford, IL, USA) at a flow rate of 20 µl/min. Bound peptides were eluted to a 75 µm × 150 mm analytical column of the same material at a flow rate of 250 nl/min. Elution was performed by applying a linear gradient of 2.5–40% ACN in 0.1% formic acid in 3 h. The HPLC was coupled online to an LTQ (ThermoElectron, San Jose, CA, USA) linear ion trap mass spectrometer, which was equipped with a nanoelectrospray ion source (Proxeon, Odense, Denmark). Distal coated silica nanospray capillaries of New Objective (Woburn, MA, USA) were used and the electrospray voltage was set to 1500 V. The mass spectrometer was operated in the data-dependent mode: 1 full scan (m/z 450–1600) was followed by MS/MS (MS2) scans of the four most abundant ions. These ions were excluded from further selection for 30 s. For each MS2 spectrum the neutral loss algorithm in the Xcalibur 1.4 software was enabled. In this mode, MS/MS/MS (MS3) experiments are automatically triggered, if a neutral loss of phosphoric acid (98, 49, 32.7 Da for singly, doubly or triply charged precursor ions, respectively) is detected among the eight most intense fragment ions in the preceding MS2 scan. The general settings of the LTQ were as follows: ion transfer tube temperature, 200°C; collision gas pressure, 3.6 bar; normalized collision energy, 35%; activation value q, 0.25; activation time, 30 ms; isolation with ±2 Da. These values were applied for both MS2 and MS3 scans.
Verification of phosphopeptide sequences
The obtained MS2 and MS3 spectra were searched against the non-redundant Arabidopsis thaliana protein database from NCBI using the Bioworks Browser 3.1 SR1 (Thermo Electron). The following modifications were entered in the search. Static: methylation of Asp/Glu and peptide C-termini (+14 Da); carbamidomethylation of Cys (+57 Da). Variable: oxidation of Met (+16 Da); phosphorylation of Ser/Thr/Tyr (+80 Da); loss of water from Ser/Thr (−18 Da; to interpret the MS3 spectra). Searches were done with tryptic specificity allowing two missed cleavages. The obtained result was filtered using Xcorr (+1, 2, 3) = 1.5, 2.0, 2.5 and all phosphopeptide spectra were subjected to stringent manual validation by at least two persons and mass spectra not shown in this article are available upon request. When only MS2 or MS3 spectra were given as hits in the initial database search, the corresponding MS3 (when available) and MS2 spectra, respectively, were checked for confirmation.
Expression and purification of glutathione S-transferase (GST) fusion proteins
RSp31 (At3g61860) and SRPK4 (At3g53030) were cloned into the EcoRI–NotI sites of pGex4T-1 as described elsewhere (T. Hufnagl, S. Lopato, C. Forstner, M. Kalyna and A. Barta, manuscript in preparation). Expression was performed in the Escherichia coli strain BL21. Purification of the GST-tagged proteins was performed as described in the Fast Flow column purification protocol from Amersham Biosciences.
Kinase assays
The in vitro phosphorylation of RSp31 by SRPK4 was performed in kinase buffer (20 mM HEPES, pH 7.5, 15 mM MgCl2, 5 mM EGTA and 1 mM DTT). GST–RSp31 fusion protein (5 µg) and GST–SRPK4 fusion proteins (200 ng) were added to a total volume of 20 µl kinase reaction mixture. The reaction was started with 2 µCi [γ-32]ATP and incubated at 22°C for 30 min. The reaction was stopped by the addition of 5 µl of 5× SDS loading buffer. Phosphorylation of RSp31 as a substrate of SRPK4 was monitored by autoradiography after SDS–PAGE.
RESULTS
Phosphoproteomic analysis of Arabidopsis nuclear and cytosolic extracts
A phosphoproteomic analysis was performed on Arabidopsis intracellular proteins (9). In short, protein extracts from Arabidopsis cell suspensions were subjected to trypsin digestion. From these complex mixtures, phosphopeptides were purified by IMAC by two different protocols (Materials and Methods). Esterification of peptides before IMAC has been shown to greatly improve the abundance of phosphopeptides in the isolated sample (19). The protocol that included the esterification step indeed gave the highest purity of phosphopeptides. Because the two protocols differed considerably, we analyzed the importance of the esterification step in more detail. Therefore, we divided a cytosolic extract into two aliquots that were either esterified or not before phosphopeptide purification by IMAC. The peptide mixtures were analyzed by liquid chromatography/electrospray ionization tandem mass spectrometry. The use of a Finnigan LTQ mass spectrometer allowed us to obtain a third stage of MS (MS/MS/MS or MS3). These fragmentations were performed only when a loss of phosphate was observed during MS/MS (MS2). Strikingly, the purity increased from ∼30% to ∼90% in the esterified sample. A bias towards multiply phosphorylated peptides has been ascribed to the higher affinity of the IMAC material for these peptides (19,21). However, of the manually verified esterified peptides >70% of the esterified peptides were singly phosphorylated. These results are comparable with those described for IMAC-purified esterified human phosphopeptides (22).
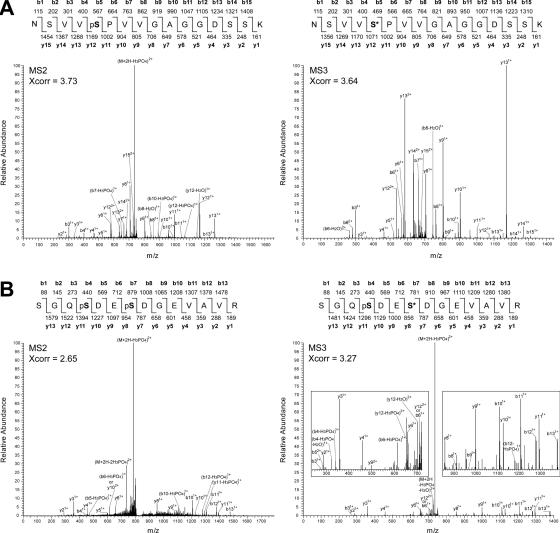
To analyze the phosphoproteome of Arabidopsis, phosphopeptides of nuclear and cytosolic samples were isolated by IMAC and analyzed by LC-ESI-MS/MS/MS. Peptide sequences were searched against the non-redundant Arabidopsis database at the National Center for Biotechnology Information. Subsequently, the sequences of the detected phosphopeptides were verified by careful examination of the MS2 and corresponding MS3 (when available) fragmentation spectra. Examples of MS2 and the corresponding MS3 fragmentation spectra of the peptide NSVVpSPVVGAGGDSSK of RSZ33 are shown in Figure 1A. It is clear that the MS3 spectrum greatly aids in confirmation of the peptide sequence. Because of low detection levels of some peptides, corresponding MS3 spectra are not always available. In spite of this limitation, many MS2 spectra could be confirmed by the corresponding MS3 spectra (Table 1). As a second example, the sequence of the doubly phosphorylated peptide SGQpSDEpSDGEVAVR of At1g32490 (see below) was validated by corresponding MS2 and MS3 spectra (Figure 1B). The sequence information provided by the MS3 spectrum is not optimal due to the presence of one phosphate group. Moreover, the loss of the phosphate group(s) from multiple fragments makes it more difficult to interpret the MS2 spectrum compared to the MS3 fragmentation pattern. In addition, the MS3 spectrum has much less background fragments than the MS2 spectrum and confirms the sequence. Thus, manual inspection of mass spectra of both singly and doubly phosphorylated peptides can be greatly facilitated by the MS3 spectra.
Figure 1.
Mass spectra of singly and doubly phosphorylated peptides. (A) Corresponding MS2 and MS3 spectra of the NSVVpSPVVGAGGDSSK peptide of RSZ33. (B) Corresponding MS2 and MS3 spectra of the peptide of SGQpSDEpSDGEVAVR of DEAH box RNA helicase encoded by At1g32490. pS indicates a phospho-Ser residue; S* indicates a phospho-Ser residue that has lost its phosphate group.
Table 1.
Phosphopeptides of Arabidopsis RNA metabolism proteins
| Gene IDa | Protein name | Peptide sequence | Sites | Xcorrb | Confirmed by MS3 | Process involved |
|---|---|---|---|---|---|---|
| SR proteins | ||||||
| At3g49430 | SRp34a | pSRSPpSKpSPPK | 3 | 2.47 | Pre-mRNA splicing | |
| At3g61860 | RSp31 | ARpSPEYDRc | 1 | 2.31 | Yes | Pre-mRNA splicing |
| RpSLpSPVYRc | 2 | 2.50 | ||||
| At4g25500 | RSp40/SRp35 | SRpSPRpSPPADEd | 2 | 2.32 | Pre-mRNA splicing | |
| At5g52040 | RSp41 | ARLpSPDYK | 1 | 2.61 | Pre-mRNA splicing | |
| GEpSRpSPPPYEK | 2 | 2.72 | ||||
| GEpSRpSPPPYEKR | 2 | 2.84 | ||||
| pSKSSPENGQVEpSPGQIMEVEAGR | 2 | 3.25 | ||||
| At5g52040.1 | RSp41 splice variant 1 | GYDGADpSPIREpSPSRpSPPAEE3 | 3 | 3.83 | Yes | |
| At4g31580 and/or At2g24590 | SRZ22/RSZ22 and/or RSZ22a | ARpSPPPPR | 1 | 2.27 | Pre-mRNA splicing | |
| At3g53500 | RSZ32 | KVIDApSPK | 1 | 2.23 | Yes | Pre-mRNA splicing |
| DQSLpSPDRK | 1 | 2.12 | ||||
| At2g37340 | RSZ33 | NSVVpSPVVGAGGDSSK | 1 | 3.73 | Yes | Pre-mRNA splicing |
| MDDpSLpSPR | 2 | 2.56 | ||||
| IIDGpSPPPSPK | 2 | 2.80 | Yes | |||
| DRpSPVLDDEGpSPK | 2 | 4.04 | Yes | |||
| SPVLDDEGpSPK | 1 | 2.55 | Yes | |||
| At5g64200 | SC35 | KVDAASRpSQpSPYAAEd | 2 | 2.63 | Yes | Pre-mRNA splicing |
| SNERpSPpSPGpSPAPLR | 3 | 3.11 | Yes | |||
| At1g55310 | SR33/SCL33 | SRpSGDYYpSPPPR | 2 | 2.44 | Yes | Pre-mRNA splicing |
| At3g55460 | SCL30 | GRpSPPPPPPR | 1 | 2.56 | Yes | Pre-mRNA splicing |
| SRpSYpSPAPR | 2 | 2.04 | Yes | |||
| SRpSVEVpSPR | 2 | 2.84 | Yes | |||
| RpSYpSPGYEGAAAAAPDR | 2 | 3.64 | Yes | |||
| pSYpSPGYEGAAAAAPDR | 2 | 2.26 | ||||
| RYpSPPYYpSPPR | 2 | 2.62 | Yes | |||
| YpSPPYYpSPPR | 2 | 2.59 | ||||
| At3g13570 | SCL30a | HQpSRpSVpSPQDR | 3 | 2.72 | Pre-mRNA splicing | |
| GYNpSPPAKR | 1 | 2.16 | ||||
| At1g16610 | SR45 | GRpSPPPPPSK | 1 | 3.05 | Yes | Pre-mRNA splicing |
| GRpSPpSSPPPR | 2 | 2.35 | Yes | |||
| VpSSPPKPVSAAPK | 1 | 3.17 | ||||
| RS domain | ||||||
| At1g07350.1 | Tra-2-like protein splice variant 1 | DRpSYpSPYYR | 2 | 2.50 | Pre-mRNA splicing | |
| DRpSPYYMR | 1 | 2.39 | ||||
| DRpSYpSPHYQGR | 2 | 2.09 | ||||
| At3g63400 | CypRS64 | NFpSPGDVSDR | 1 | 2.03 | Pre-mRNA splicing | |
| At3g63400.1 | CypRS64 splice variant 1 | SFRpSPpSPSGVPK | 2 | 2.48 | Yes | |
| At2g29210 | HsSRm160-like | NRpSPpSPLYR | 2 | 2.08 | Pre-mRNA splicing | |
| RpSPpSPLYR | 2 | 2.17 | ||||
| HTpSPSHIKQDGpSMpSPVR | 3 | 2.91 | ||||
| LPpSPPVAQRLPpSPPPR | 2 | 3.11 | ||||
| LPpSPSIEQR | 1 | 2.33 | ||||
| RNA helicases | ||||||
| At1g32490 | DEAH box RNA helicase | SGQpSDEpSDGEVAVR | 2 | 3.27 | Yes | Pre-mRNA splicing |
| At5g08610 | DEAD box RNA helicase (RH26) | GKFTpSDEDNADPEVVR | 1 | 4.10 | Unknown | |
| GKFpTpSDEDNADPEVVR | 2 | 3.52 | Yes | |||
| FTpSDEDNADPEVVR | 1 | 3.56 | ||||
| Other | ||||||
| At1g04080 | Similar to yeast PRP39 splicing factor | SQVDGSTEQpSPK | 1 | 3.18 | Yes | Pre-mRNA splicing |
| At4g17720 | RRM-containing protein | VHLSEpSPK | 1 | 2.17 | Yes | Unknown |
| At5g04430 | NOVA-like RNA-binding protein | RpSPEPHDSSEADSAEKPTHIR | 1 | 5.20 | Pre-mRNA splicing | |
| pSPEPHDpSSEADSAEKPTHIR | 2 | 3.05 | ||||
| At5g48650 | NTF2-containing RNA-binding protein | DKFGVPAVSLPpSPK | 1 | 2.49 | Yes | Nucleocytoplasmic transport |
| At5g51300 | Splicing factor 1 (SF1)-like | MLQSGMPLDDRPEGQR pSPpSPEPVYDNMGIR | 2 | 4.39 | Pre-mRNA splicing | |
aTAIR database entry. Phosphorylation sites are indicated by a lower case ‘p’ preceeding the phosphoamino acid. Unambiguous sites are indicated in boldface, and ambiguous pSer sites are indicated in italics.
bOf the assigned peptide.
cFound to be phosphorylated in vitro by SRPK4 (Figure 6).
dC-terminus of the protein.
We have manually assigned sequences of 57 phosphopeptides defining 87 unique sites from the analysis of two nuclear samples using a protocol without esterification. In addition, using the protocol that contains an esterification step, 129 phosphopeptides defining 155 unique sites were determined from a single cytosolic sample. To classify the phosphoproteins into different functional groups, the domain composition of the corresponding phosphoproteins was analyzed using the SMART database (23). Until now we have identified 151 phosphoproteins from both the nuclear and cytosolic samples that were assigned to 8 groups (9). The largest functional group consisted of 22 phosphoproteins that are predicted to function in RNA metabolism, and here we will focus on this group.
In vivo phosphorylation of proteins involved in RNA metabolism
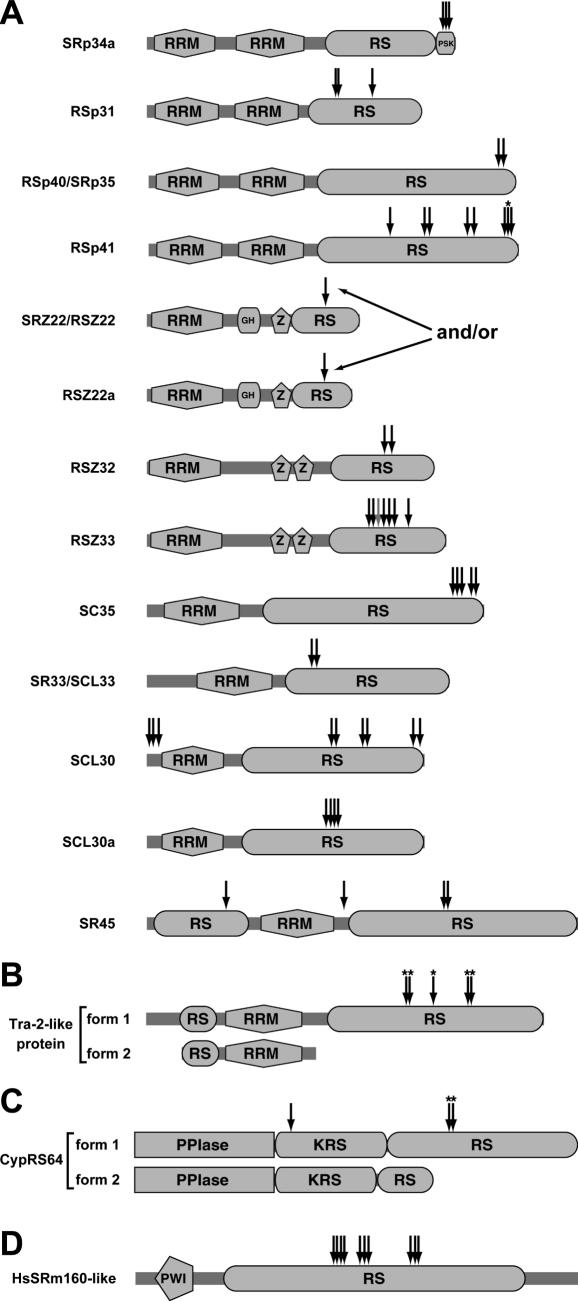
In total, 79 phosphorylation sites of RNA metabolism-related proteins were determined (Table 1), representing roughly one-third of the sites that were identified from nuclear and cytosolic samples. Supplementary Table 1 shows an overview of phosphopeptides that were identified in nuclear and cytosolic extracts. As described above, these numbers indicate that optimization of the IMAC protocol reduced the bias towards multiply phosphorylated phosphopeptides. The majority (19 proteins) of the phosphoproteins are predicted to function in pre-mRNA splicing (Table 1). For instance, so-called SR proteins, a homolog of the animal splicing factor 1 (SF1), a cyclophilin (CypRS64) and an SR ribonucleoprotein related to animal transformer-2 (Tra-2) were found. SR proteins typically contain one or two RRMs and a C-terminal Arg/Ser-rich (RS) domain (11). Of the 19 Arabidopsis SR proteins, 12 were found to be phosphorylated (Figure 2A). These include plant-specific splicing factors, such as the highly conserved SR45 protein. SR proteins have been shown to be phosphoproteins in both animal (24) and plant cells (17,25,26). Animal SR proteins are highly phosphorylated on Ser residues in their RS domains (24). In contrast, no phosphorylation sites of plant SR proteins have been described. In addition to the SR proteins, we found three other proteins that contain RS domains. The cyclophilin CypRS64 and the Tra-2-like protein display splicing form-specific phosphorylation (see below). Of the third protein, the PWI domain-containing protein encoded by At2g29210, 10 phosphorylation sites were determined (Table 1 and Figure 2). PWI domains are commonly found in splicing factors, and Arabidopsis has two proteins with PWI domains. The sequence of At2g29210 is related to human SRm160 (SR nuclear matrix protein of 160 kDa), which is highly phosphorylated in human cells (27). Of the two Arabidopsis PWI domain-containing proteins, only At2g29210 is structurally similar to HsSRm160. All sites of the HsSRm160-like protein are located in the central RS domain (Figure 2D).
Figure 2.
Position of phosphorylation sites within proteins containing Arg/Ser-rich (RS) domains. (A) In vivo phosphorylation sites of SR proteins. Structures are modified from Ref. (15). (B) Splice form-specific phosphorylation sites of the transformer-2-like protein (Tra-2-like). Scaling of the protein isoforms is the same as in A. (C) Splice form-specific and -nonspecific phosphorylation sites of CypRS64. (D) Phosphorylation of a PWI domain-containing splicing factor that is homologous to human SRm160. Phosphorylation sites are indicated by arrows. Asterisks denote splice variant-specific phosphorylation sites. The gray arrow (at RSZ33) represents a pSer also identified in non-phosphorylated form. GH, glycine hinge; KRS, Lys/Arg/Ser-rich region; PPIase, peptidyl-prolyl cis/trans isomerase domain; PSK, Pro/Ser/Lys-rich region; PWI, PWI motif-containing domain in splicing factors; RRM, RNA recognition motif; RS, Arg/Ser-rich domain; Z, Zinc knuckle.
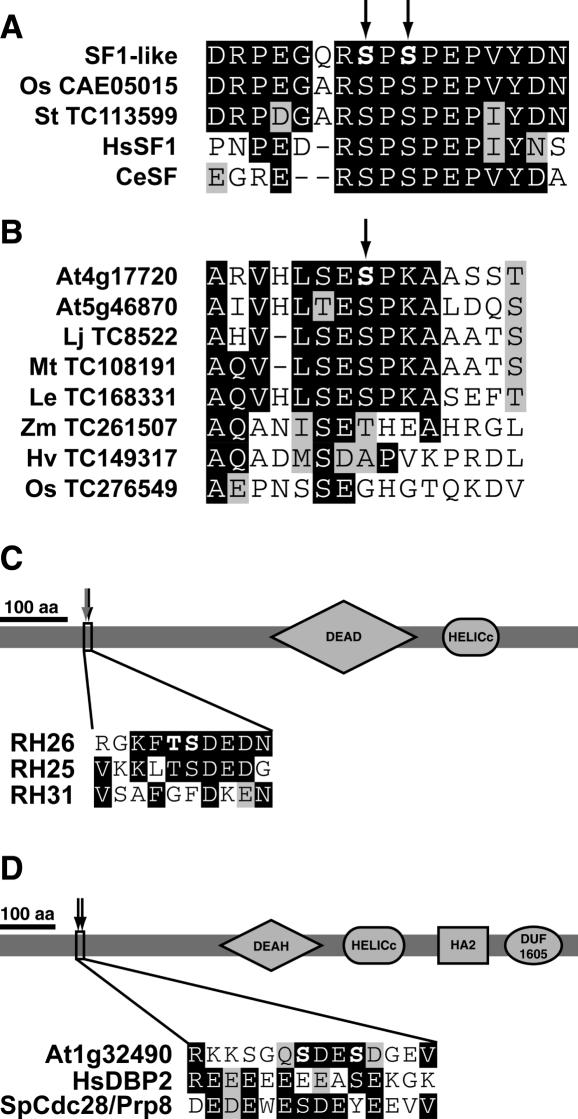
Among the other phosphoproteins is the SF1-like protein (Table 1). This protein is the closest HsSF1 homolog in Arabidopsis (first blast hit value of 4E−54). The two determined phosphorylation sites of the Arabidopsis SF1-like protein are conserved in both animal and other plant SF1-like sequences (Figure 3A). Strikingly, exactly these two sites were found recently to be phosphorylated in phosphoproteomic analyses of human and mouse cells as well (27,28). The Tra-2-like protein encoded by At1g07350 is the closest homolog of Drosophila melanogaster Tra-2 (BLAST hit 2E−11). Tra-2 mediates alternative splicing by recruiting SR proteins to regulatory elements in D.melanogaster (29). At1g04080 is the second most related Arabidopsis protein to Saccharomyces cerevisiae Prp39 (BLAST hit 3E−19), a U1 small nuclear ribonucleoprotein involved in mRNA splicing (30). At5g04430 is related to human NOVA-1 (BLAST hit 2E − 23), which is involved in splicing in human brain cells (31). The peptide of the NOVA-like protein was isolated as singly and doubly phosphorylated forms (Table 1).
Figure 3.
Conservation of phosphorylation sites in homologous proteins from other species and phosphorylation of DExD/H box RNA helicases. (A) Alignment of the region surrounding the Arabidopsis SF1-like protein phosphorylation site and related sequences from rice (Os, Oryza sativa), potato (St, Solanum tuberosum), human (Hs, Homo sapiens) and Caenorhabditis elegans (Ce) homologs. The two corresponding Ser residues of HsSF1 are phosphorylated in vivo as well (27,28). (B) Alignment of the phosphosite in At4g17720 and corresponding regions of homologs from the dicot species Arabidopsis (At, A.thaliana), Lotus japonicus (Lj), Medicago truncatula (Mt), tomato (Le, Lycopersicon esculentum), and the monocots maize (Zm, Zea mays), barley (Hv, Hordeum vulgare), and rice (Os, O.sativa). (C) Positioning of the phosphorylation sites in the DEAD box RNA helicase RH26. The phosphorylation sites are both conserved in AtRH25, but not in AtRH31. The gray arrow (at RSZ33) represents a pSer also identified in non-phosphorylated form. (D) Phosphorylation of the DEAH box RNA helicase encoded by At1g32490. One of the two pSer residues are positionally conserved in the human homolog DBP2 and the fission yeast homolog Cdc28/Prp8. DEAD/H, DEAD/H-like helicase domain; HELICc, helicase superfamily c-terminal domain; HA2, Helicase associated domain (PFAM accession no. PF04408), DUF1605, domain of unknown function (PFAM accession no. PF07717). DUF1605 is always found in association with HA2 in helicases.
Two phosphoproteins are members of the superfamily of DexD/H box RNA helicases (32). Based on conserved motifs of the different subfamilies of DexD/H box RNA helicases (33), At5g08610 encodes a DEAD box helicase and At1g32490 codes for a DEAH box helicase. At5g08610 has been classified as RH26 (32), and its phosphorylation sites are conserved in the most closely related member RH25 but not in the more distantly related RH31 (Figure 3C). At1g32490 shares high sequence homology with S.cerevisiae Prp22, Schizosaccharomyces pombe Cdc28/Prp8 and human DBP2, which are all DEAH box RNA helicases involved in pre-mRNA splicing (34–36). This suggests that the Arabidopsis homolog may function in splicing as well. Although highly similar in their C-terminal regions, the N-termini of At1g32490, HsDBP2 and SpCdc28/Prp8 are not very well conserved. Nevertheless, both HsDBP2 and SpCdc28/Prp8 contain one Ser residue at the corresponding position, which are followed by an acidic residue (Asp or Glu) as well (Figure 3D). Three phosphorylation sites of the two Arabidopsis RNA helicases are followed by an Asp (Table 1) and all sites are located in the N-terminus of the proteins (Figure 3C and D).
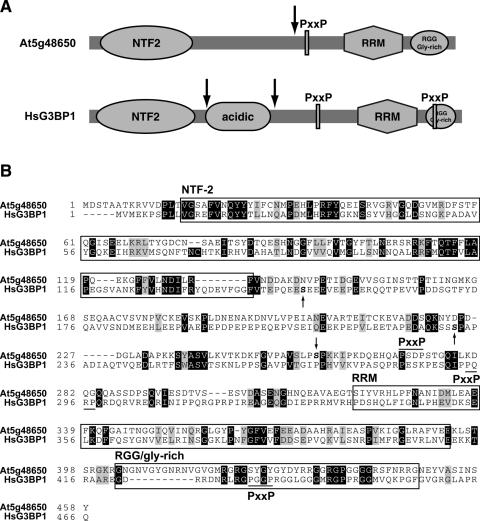
Besides the DEAD box helicase, only 2 of the 22 phosphoproteins predicted to function in RNA metabolism could not be directly related to RNA splicing. The first is encoded by At5g48650, and this protein contains a nuclear transport factor 2 (NTF2)-like domain and an RRM domain (Figure 4A). Its structure suggests that the protein may be involved in the nucleocytoplasmic transport of RNA or RNA-containing complexes. Arabidopsis At5g48650 has a similar domain organization as S.cerevisiae Bre5 that acts as a cofactor for the Ubp3 de-ubiquitinating enzyme (37). Structurally similar mammalian proteins are Ras-GTPase-activating protein SH3 domain-binding proteins (G3BPs). A closer inspection of the polypeptide sequence showed that the Arabidopsis protein, like HsG3BP1, also has a potential SH3 domain-binding site and a C-terminal RGG/glycine-rich domain, but lacks the acidic domain (Figure 4). Human G3BPs are implicated in signal transduction downstream of Ras and may act as endonucleases (38). Phosphorylation of HsG3BP1 at Ser-149 (Figure 4B) plays a role in regulation of its endonuclease activity and subcellular localization (39). HsG3BP1 is phosphorylated at a second site, Ser-232 (Ser-231 in mouse G3BP1), which is followed by a Pro (27,28,39). Interestingly, the determined site of the Arabidopsis homolog is followed by a Pro as well, and is located at a similar position (Figure 4A). Therefore it is tempting to speculate that similar kinases may target these homologs in both plants and mammals. The nature of the kinase targeting Ser-232 in HsG3BP1 and the functional effect of this phosphorylation event is unknown. Several proteins with the same domain organization are encoded by the Arabidopsis genome, but the region containing the phosphorylation site that we determined is not conserved in these homologs (data not shown).
Figure 4.
Phosphorylation of an NTF2-RRM protein homologous to human G3BP1. (A) Schematic representation of the NTF2-RRM domain containing protein (At5g48650) and its phosphorylation sites. As a comparison, the human G3BP1 protein is shown with its two known phosphorylation sites. Phosphorylation sites are indicated by arrows. Domain names are explained below. (B) Boxes surround domains are chosen for the Arabidopsis protein, but are essentially the same for HsG3BP1. pSer residues are indicated in bold lettering, either above the protein sequence (for the Arabidopsis protein) or below (for HsG3BP1). NTF2, nuclear transport factor 2-like; PxxP, potential SH3 domain-binding site, as indicated by a black line above (for Arabidopsis) or below (for HsG3BP1) the sequence; RRM, RNA recognition motif.
The second protein (encoded by At4g17720) contains an RRM domain and its C-terminal phosphorylation site is conserved in homologs of other dicot species, but absent in monocot homologs (Figure 3B and Supplementary Table 2). Arabidopsis contains a closely related protein (encoded by At5g46870) that is 78% identical to At4g17720. The phosphorylation site is conserved in this homolog (Figure 3B).
In conclusion, our analysis shows that many Arabidopsis SR proteins are phosphoproteins and that they are almost exclusively phosphorylated in their RS domains. In total, 50 of the 79 phosphorylation sites were on SR proteins, 18 sites on other RS domain-containing proteins and the remaining 11 sites on proteins lacking RS domains.
Phosphorylation of alternative splice isoforms
Different isoforms of many plant SR proteins and other RS domain-containing proteins are generated by alternative splicing (40). Interestingly, we found that several of the alternative isoforms lack one or more of the observed phosphorylation site(s). Alternatively spliced mRNAs of the Tra-2-like protein, CypRS64 and RSp41, encode proteins that differ in their RS domains, and several or all of the observed phosphorylation sites are specific for single isoforms, all encoded by their full-length splice variant 1 (Figure 2A–C and Table 1). In the case of Tra-2-like protein, the RS domain is completely absent in splice variant 2 (Figure 2B). The splice variant 2 of CypRS64 contains a distinct and shorter RS domain compared to splice variant 1 (Figure 2C), whereas the two RSp41 isoforms differ by only a single amino acid (TAIR database). Strikingly, the splice variant 2 of RSp41 contains an insertion of a Ser residue directly N-terminal (position −1) of the phosphorylated Ser residue in splice variant 1.
Analysis of phosphorylation sites
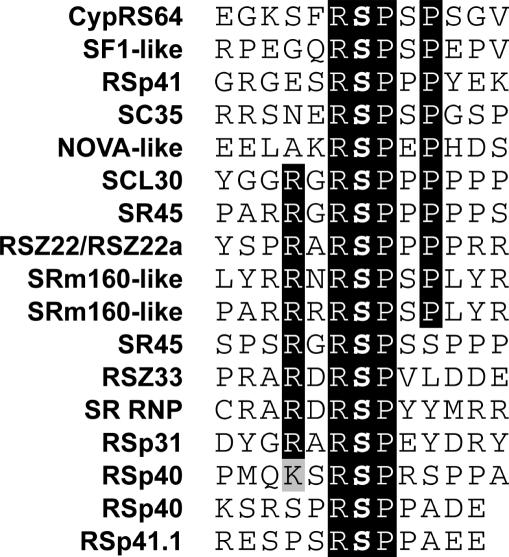
All the phosphorylation sites of the phosphoproteins other than the RS domain-containing proteins mapped outside their functional domains as predicted by SMART (Supplementary Table 2). This has also been observed for Arabidopsis plasma membrane phosphoproteins (8). Strikingly, all of the phosphorylation sites were on Ser residues. This can be explained by the fact that the RS domains of the detected phosphoproteins hardly contain Thr residues, and that phosphorylation on Ser is much more abundant (12-fold in the phosphoproteome analyzed by us) than on Thr. Of the 75 unambiguously determined phosphorylation sites, 57 could be assigned as pSP sites (76%), which are potential mitogen-activated protein kinases or cyclin-dependent kinases phosphorylation sites. The overrepresentation of pSP sites is very likely not due to a technical bias, since the other seven groups of phosphoproteins (see above) did not show such a high percentage of pSP sites. Among the pSP sites, we found 17 RpSP sites in 13 of the phosphoproteins (Figure 5), suggesting that similar or the same kinase(s) target(s) these sites. In most cases, the RpSP motif was flanked by an Arg at the −3 and/or a Pro at the +3 positions (Figure 5). In seven other cases, the phosphorylated Ser was preceded by a Tyr at position −1 (Table 1).
Figure 5.
Identification of a conserved phosphorylation motif within RNA metabolic phosphoproteins. The phosphorylated Ser residue is indicated in boldface. Identity and homology of amino acids are marked by black and gray shades, respectively.
Phosphorylation of the SR protein RSp31 by an SR protein-specific kinase
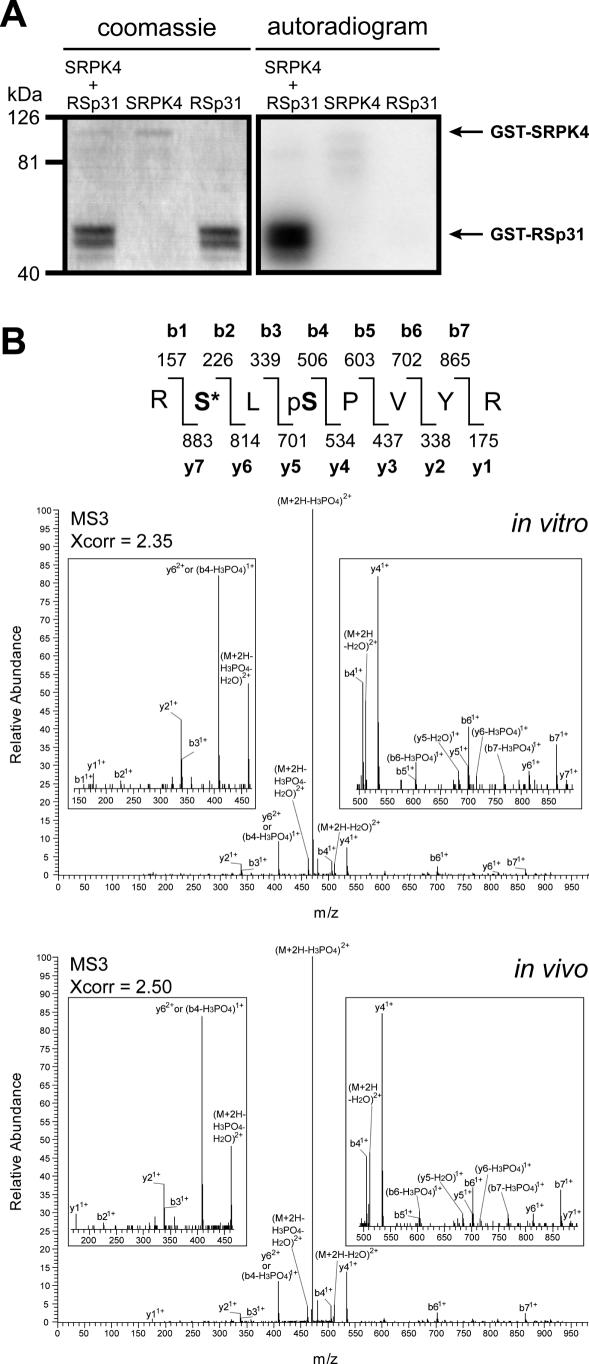
In a yeast two-hybrid screen using SR33/SCL33 as bait, the SR protein-specific kinase SRPK4 was isolated (C. Forstner and A. Barta, unpublished data). It was tested whether one of the SR proteins in our list of in vivo phosphorylated proteins, RSp31, could be phosphorylated in vitro by SRPK4. For this purpose, both proteins were purified from E.coli as a GST fusion protein (Figure 6A, left panel). In a radioactive in vitro kinase assay SRPK4 showed low autophosphorylation activity, but strongly phosphorylated RSp31 (Figure 6A, right panel).
Figure 6.
SR protein-specific kinase 4 phosphorylates splicing factor RSp31 in vitro. (A) Purification of SR protein-specific kinase 4 (SRPK4) and RSp31 from E.coli (left panel). Autoradiogram of SRPK4 and RSp31 incubated together in a kinase buffer-containing radioactive ATP (right panel). As controls, SRPK4 and RSp31 were separately incubated in the same buffer. The faint signal in the SRPK4 lane represents low autophosphorylation activity of the kinase. GST alone was not phosphorylated at all by SRPK4 (data not shown). (B) Mass spectra of the RpSLpSPVYR peptide. The upper panel shows an MS3 spectrum of the peptide of the in vitro phosphorylated protein. The lower panel shows an MS3 spectrum of the peptide found in the in vivo analysis (Table 1). pS indicates a phospho-Ser residue; S* indicates a phospho-Ser residue that has lost its phosphate group.
These data suggested that RSp31 could be targeted by SRPK4 in vivo. Therefore we determined the actual sites of RSp31 that were targeted by SRPK4, by using RSp31 as a substrate in a non-radioactive in vitro kinase assay. After phosphorylation, the GST–RSp31 protein was digested with trypsin and subjected to LC-MS3 analysis. Among the phosphopeptides that were found, both peptides that were identified in the in vivo phosphoproteomic analysis (ARpSPEYDR and RpSLpSPVYR) were also present (Table 1). The mass spectra of the peptides observed in the in vitro and in vivo analyses were very similar (see the example of RpSLpSPVYR MS3 spectra in Figure 6B), showing that the peptide sequences are identical. One of these sites is an RpSP site that is conserved among other members of the RNA metabolism proteins (Figure 5), and thus SRPK4 could be one of the kinases that target these sites. These results independently validate the usefulness of mapping in vivo sites by large-scale IMAC-based mass spectrometry.
DISCUSSION
Analysis of the Arabidopsis intracellular phosphoproteome
The use of mass spectrometry to determine phosphopeptide sequences has great advantages for the identification of phosphorylation sites, but brings about practical difficulties as well (41). For instance, phosphopeptides represent only a very small fraction within a mixture of peptides obtained from a complex extract. Additionally, the sequencing of phosphopeptides is often difficult because the most widely used phosphopeptide fragmentation techniques in mass spectrometry induce dissociation primarily of the phosphate bond. This leads to the observation of a prominent loss of phosphate from the peptide but low fragmentation between amino acids, yielding low sequence information. In spite of these difficulties, the recent development of mass spectrometry-based phosphoproteomics technology that uses IMAC purification of phosphopeptides from complex mixtures has allowed rapid and large-scale identification of numerous in vivo phosphorylation sites (8,19,42). IMAC is based on the affinity of positively charged trivalent metal ions for negatively charged phosphate groups. The conversion of carboxylic acid groups to methyl esters prior to IMAC has greatly increased the specificity of phosphopeptide isolation (19). This modification reduces the binding of the otherwise acidic carboxyl groups to the metal ions (19,43).
Several protocols have been used successfully for studying the phosphoproteome of Arabidopsis (2,8,21). Our data show that a protocol that was initially developed for the analysis of phosphorylation sites of yeast proteins (19), is very suitable for plant work. We have used the combination of IMAC and LC-MS/MS/MS to determine novel phosphorylation sites of intracellular Arabidopsis proteins, allowing the large-scale identification of phosphopeptides in a single LC-MS/MS/MS run. As a proof of concept, we focused here on a group of highly phosphorylated proteins that are related to RNA metabolism. The data show that proteins containing RS domains are common targets of phosphorylation. Since these domains are present in many proteins in plants, these data may be generally applicable for other RS proteins. The conservation of the determined phosphorylation sites suggests that some of our results might even be extrapolated to other eukaryotes.
In vivo phosphorylation of SR proteins
By studying the nuclear and cytosolic phosphoproteomes of Arabidopsis globally, we determined 79 in vivo phosphorylation sites derived from 22 proteins involved in different aspects of RNA metabolism. SR proteins represented a prominent part of this group. Although several SR proteins have been shown to be phosphorylated in plants, none of the identified sites was described before. Our evidence shows that plant SR proteins, like their animal counterparts (24), are extensively phosphorylated at Ser residues in their RS domains. In extracts from a single cell type (Arabidopsis root cells), 12 of the 19 SR proteins were identified as phosphoproteins. One peptide could not be assigned to either SRZ22/RSZ22 or RSZ22a (Table 1), but since this peptide was detected six times it is possible that it is derived from both proteins. Although the other seven members may not be phosphoproteins, it is more likely that they are not expressed in the root cell culture used for our analysis or that they are of too low abundance to be detected. In total, we identified phosphorylation sites in the RS domains of 15 proteins. This indicates that RS domains of plant proteins are general targets of phosphorylation. The conservation of SR proteins suggests that our results can be extrapolated to other plant species. Since the RS domains of SR proteins are involved in protein–protein interactions, which can be phosphorylation-dependent (17), the highly repetitive RS/SP motifs can possibly generate multiple protein-binding sites depending on the phosphorylation status of each motif. It can thus be speculated that the highly phosphorylated RS domains provide different binding surfaces for different proteins, such as the RS domain containing cyclophilin CypRS64 (17), other SR proteins and additional components of the splicing machinery (15). The effect of the phosphorylation-dependent interaction between SR proteins with CypRS64 is the relocalization of CypRS64 from nuclear bodies to speckles (17). It will be valuable to determine the phosphorylation status of SR proteins and other spliceosome components during stresses that affect splicing, such as high temperature (44), or during different phases of spliceosome assembly. The latter might be reached by the isolation of early and late spliceosomes by the immunoprecipitation of specific components of these complexes (45).
Several plant SR proteins and other proteins that contain RS domains occur in distinct splice variants, which differ often in their C-terminal RS domains (40). Importantly, we found that alternative splice forms lack one or more phosphorylation sites and thus may be impaired in binding other components of the splicing machinery. Strikingly, the differential splice variant 2 of RSp41 contains an insertion of one amino acid adjacent to the phosphorylated residue in splice variant 1. It will be interesting to test whether splice form 2 is phosphorylated at the corresponding residue and how its protein interaction network is affected by this modification. A general question concerns the function of these alternative isoforms that contain a shorter RS domain or lack this domain at all. One could speculate that they act as dominant negative proteins that bind to the RNA/spliceosome and block the recruitment of additional splicing factors, which otherwise would bind to the full-length isoform. This may then lead to blockage of the splice site, causing the selection of a neighboring splice site or skipping of the site at all.
Identification of kinases that are responsible for the phosphorylation of SR proteins
In general, the RS/SP motifs within the RS domains of SR proteins are phosphorylated, affecting multiple functions of this class of proteins (10). The phosphorylation of SR proteins is mediated by both Clk (or LAMMER-type) kinases and SR protein-specific kinases (SRPKs). Recently it was shown that animal SRPKs and Clks have different specificities. Although SRPKs specifically phosphorylate Ser residues in a particular stretch of RS domain of an SR protein, Clk kinases can target all Ser residues in the RS domain (46). Several SR proteins are phosphorylated by members of these kinase groups and Clk kinases are involved in splicing of pre-mRNAs and development in Arabidopsis (47–49). In addition to Clk kinases and SRPKs, other proline-directed kinases may target SR proteins. Interestingly, the SR proteins SRZ21, SR1/SRp34 and the Tra-2-like protein are in vitro phosphorylated by the MAP kinases MPK6 and/or MPK3 (50). Whether other SR proteins are substrates and whether some of our determined in vivo pSP sites are targeted by MAP kinases remains to be tested.
Confirming our findings by an alternative method, we show that an SRPK can in vitro phosphorylate all three sites of RSp31 that were determined in this study. Moreover, one of these sites is an RpSP site, which we found to be phosphorylated in multiple SR and other proteins (Figure 5). This may suggest that these conserved phosphorylated sites (and likely other sites) found in the other phosphoproteins are targeted by SRPKs in vivo as well. In line with this hypothesis, CypRS64 is an SRPK4 substrate (17) and we identified an RpSP site in this protein (Table 1). Concluding, the combination of in vivo and in vitro analyses described here is suitable to establish kinase–substrate pairs.
RNA helicases are targets of phosphorylation
DexD/H box RNA helicases are involved in different facets of RNA metabolism, such as RNA transport, transcription, editing, ribosome biogenesis, translation and splicing (33). We revealed phosphorylation sites in DEAD box and DEAH box RNA helicases that belong to this protein superfamily. The conservation of the phosphorylation sites suggests that the same or similar protein kinase(s) may target both proteins. Other members of the DEAD box family are translation initiation factors, and we have found these to be phosphorylated as well (S. de la Fuente van Bentem, D. Anrather, E. Roitinger and H. Hirt, unpublished data). Human RNA helicases are phosphorylated at Ser residues and several of these sites are followed by an acidic residue at the +1 position as well (27,51), suggesting that similar kinases may target these proteins in both animals and plants. Except for translation initiation factor 4, there is no evidence for in vivo phosphorylation of RNA helicases in plants, and no phosphorylation site has been described.
In conclusion, the protocol described here can be used for studying the plant phosphoproteomes of different intracellular organelles. Furthermore, it can be used in combination with recently developed quantitative methods to measure changes in phosphoproteomes (52). Application of these methods to phosphoproteomic analysis identified novel components of even well-described signaling pathways (53–55). Although these technological improvements have been achieved, serious challenges have to be faced in the future to allow global quantitative analysis of signaling pathways by mass spectrometry (56). Encouraged by several promising studies, however, it seems evident that future studies applying these techniques to study the dynamics of the phosphoproteome during development or stress will significantly enhance our knowledge on the role of phosphorylation in signal transduction and regulatory processes in plants.
Supplementary Material
Acknowledgments
We thank Andriy Belokurov for assistance with the Arabidopsis cell culture. This work was supported by the Austrian Science Foundation, the Vienna Science and Technology Fund and the European Union. Funding to pay the Open Access publication charges for this article was provided by the Vienna Science and Technology Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Andersson L., Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 1986;154:250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 2.Wolschin F., Wienkoop S., Weckwerth W. Enrichment of phosphorylated proteins and peptides from complex mixtures using metal oxide/hydroxide affinity chromatography (MOAC) Proteomics. 2005;5:4389–4397. doi: 10.1002/pmic.200402049. [DOI] [PubMed] [Google Scholar]
- 3.Pinkse M.W., Uitto P.M., Hilhorst M.J., Ooms B., Heck A.J. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 4.Larsen M.R., Thingholm T.E., Jensen O.N., Roepstorff P., Jorgensen T.J. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Hansson M., Vener A.V. Identification of three previously unknown in vivo protein phosphorylation sites in thylakoid membranes of Arabidopsis thaliana. Mol. Cell. Proteomics. 2003;2:550–559. doi: 10.1074/mcp.M300050-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Vener A.V., Harms A., Sussman M.R., Vierstra R.D. Mass spectrometric resolution of reversible protein phosphorylation in photosynthetic membranes of Arabidopsis thaliana. J. Biol. Chem. 2001;276:6959–6966. doi: 10.1074/jbc.M009394200. [DOI] [PubMed] [Google Scholar]
- 7.Heintz D., Wurtz V., High A.A., Van Dorsselaer A., Reski R., Sarnighausen E. An efficient protocol for the identification of protein phosphorylation in a seedless plant, sensitive enough to detect members of signalling cascades. Electrophoresis. 2004;25:1149–1159. doi: 10.1002/elps.200305795. [DOI] [PubMed] [Google Scholar]
- 8.Nühse T.S., Stensballe A., Jensen O.N., Peck S.C. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell. 2004;16:2394–2405. doi: 10.1105/tpc.104.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Fuente van Bentem S., Roitinger E., Anrather D., Csaszar E., Hirt H. Phosphoproteomics as a tool to unravel plant regulatory mechanisms. Physiol. Plant. 2006;126:110–119. [Google Scholar]
- 10.Huang Y., Steitz J.A. SRprises along a messenger's journey. Mol. Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 11.Lorkovic Z.J., Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002;30:623–635. doi: 10.1093/nar/30.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ali G.S., Golovkin M., Reddy A.S. Nuclear localization and in vivo dynamics of a plant-specific serine/arginine-rich protein. Plant J. 2003;36:883–893. doi: 10.1046/j.1365-313x.2003.01932.x. [DOI] [PubMed] [Google Scholar]
- 13.Fang Y., Hearn S., Spector D.L. Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Mol. Biol. Cell. 2004;15:2664–2673. doi: 10.1091/mbc.E04-02-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorkovic Z.J., Hilscher J., Barta A. Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol. Biol. Cell. 2004;15:3233–3243. doi: 10.1091/mbc.E04-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy A.S. Plant serine/arginine-rich proteins and their role in pre-mRNA splicing. Trends Plant Sci. 2004;9:541–547. doi: 10.1016/j.tplants.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Tillemans V., Dispa L., Remacle C., Collinge M., Motte P. Functional distribution and dynamics of Arabidopsis SR splicing factors in living plant cells. Plant J. 2005;41:567–582. doi: 10.1111/j.1365-313X.2004.02321.x. [DOI] [PubMed] [Google Scholar]
- 17.Lorkovic Z.J., Lopato S., Pexa M., Lehner R., Barta A. Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 small nuclear ribonucleoprotein-specific proteins suggest their involvement in pre-mRNA splicing. J. Biol. Chem. 2004;279:33890–33898. doi: 10.1074/jbc.M400270200. [DOI] [PubMed] [Google Scholar]
- 18.Wessel D., Flügge U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 19.Ficarro S.B., McCleland M.L., Stukenberg P.T., Burke D.J., Ross M.M., Shabanowitz J., Hunt D.F., White F.M. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 20.Garcia B.A., Busby S.A., Barber C.M., Shabanowitz J., Allis C.D., Hunt D.F. Characterization of phosphorylation sites on histone H1 isoforms by tandem mass spectrometry. J. Proteome Res. 2004;3:1219–1227. doi: 10.1021/pr0498887. [DOI] [PubMed] [Google Scholar]
- 21.Nühse T.S., Stensballe A., Jensen O.N., Peck S.C. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Moser K., White F.M. Phosphoproteomic analysis of rat liver by high capacity IMAC and LC-MS/MS. J. Proteome Res. 2006;5:98–104. doi: 10.1021/pr0503073. [DOI] [PubMed] [Google Scholar]
- 23.Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourgeois C.F., Lejeune F., Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- 25.Lopato S., Mayeda A., Krainer A.R., Barta A. Pre-mRNA splicing in plants: characterization of Ser/Arg splicing factors. Proc. Natl Acad. Sci. USA. 1996;93:3074–3079. doi: 10.1073/pnas.93.7.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopato S., Kalyna M., Dorner S., Kobayashi R., Krainer A.R., Barta A. atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 1999;13:987–1001. doi: 10.1101/gad.13.8.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beausoleil S.A., Jedrychowski M., Schwartz D., Elias J.E., Villen J., Li J., Cohn M.A., Cantley L.C., Gygi S.P. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shu H., Chen S., Bi Q., Mumby M., Brekken D.L. Identification of phosphoproteins and their phosphorylation sites in the WEHI-231 B lymphoma cell line. Mol. Cell. Proteomics. 2004;3:279–286. doi: 10.1074/mcp.D300003-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Tian M., Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 30.Lockhart S.R., Rymond B.C. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol. Cell. Biol. 1994;14:3623–3633. doi: 10.1128/mcb.14.6.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen K.B., Dredge B.K., Stefani G., Zhong R., Buckanovich R.J., Okano H.J., Yang Y.Y., Darnell R.B. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 32.Aubourg S., Kreis M., Lecharny A. The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res. 1999;27:628–636. doi: 10.1093/nar/27.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanner N.K., Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 34.Company M., Arenas J., Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 35.Imamura O., Saiki K., Tani T., Ohshima Y., Sugawara M., Furuichi Y. Cloning and characterization of a human DEAH-box RNA helicase, a functional homolog of fission yeast Cdc28/Prp8. Nucleic Acids Res. 1998;26:2063–2068. doi: 10.1093/nar/26.9.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundgren K., Allan S., Urushiyama S., Tani T., Ohshima Y., Frendewey D., Beach D. A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol. Biol. Cell. 1996;7:1083–1094. doi: 10.1091/mbc.7.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen M., Stutz F., Belgareh N., Haguenauer-Tsapis R., Dargemont C. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nature Cell Biol. 2003;5:661–667. doi: 10.1038/ncb1003. [DOI] [PubMed] [Google Scholar]
- 38.Irvine K., Stirling R., Hume D., Kennedy D. Rasputin, more promiscuous than ever: a review of G3BP. Int. J. Dev. Biol. 2004;48:1065–1077. doi: 10.1387/ijdb.041893ki. [DOI] [PubMed] [Google Scholar]
- 39.Tourriere H., Gallouzi I.E., Chebli K., Capony J.P., Mouaikel J., van der Geer P., Tazi J. RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol. Cell. Biol. 2001;21:7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalyna M., Barta A. A plethora of plant serine/arginine-rich proteins: redundancy or evolution of novel gene functions? Biochem. Soc. Trans. 2004;32:561–564. doi: 10.1042/BST0320561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mann M., Ong S.E., Gronborg M., Steen H., Jensen O.N., Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 42.Kim J.E., Tannenbaum S.R., White F.M. Global phosphoproteome of HT-29 human colon adenocarcinoma cells. J. Proteome Res. 2005;4:1339–1346. doi: 10.1021/pr050048h. [DOI] [PubMed] [Google Scholar]
- 43.Ficarro S.B., Salomon A.R., Brill L.M., Mason D.E., Stettler-Gill M., Brock A., Peters E.C. Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun. Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- 44.Shin C., Feng Y., Manley J.L. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- 45.Makarov E.M., Makarova O.V., Urlaub H., Gentzel M., Will C.L., Wilm M., Luhrmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 46.Velazquez-Dones A., Hagopian J.C., Ma C.T., Zhong X.Y., Zhou H., Ghosh G., Fu X.D., Adams J.A. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J. Biol. Chem. 2005;280:41761–41768. doi: 10.1074/jbc.M504156200. [DOI] [PubMed] [Google Scholar]
- 47.Golovkin M., Reddy A.S. An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1–70K protein. J. Biol. Chem. 1999;274:36428–36438. doi: 10.1074/jbc.274.51.36428. [DOI] [PubMed] [Google Scholar]
- 48.Savaldi-Goldstein S., Sessa G., Fluhr R. The ethylene-inducible PK12 kinase mediates the phosphorylation of SR splicing factors. Plant J. 2000;21:91–96. doi: 10.1046/j.1365-313x.2000.00657.x. [DOI] [PubMed] [Google Scholar]
- 49.Savaldi-Goldstein S., Aviv D., Davydov O., Fluhr R. Alternative splicing modulation by a LAMMER kinase impinges on developmental and transcriptome expression. Plant Cell. 2003;15:926–938. doi: 10.1105/tpc.011056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feilner T., Hultschig C., Lee J., Meyer S., Immink R.G., Koenig A., Possling A., Seitz H., Beveridge A., Scheel D., et al. High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics. 2005;4:1558–1568. doi: 10.1074/mcp.M500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Ballif B.A., Villen J., Beausoleil S.A., Schwartz D., Gygi S.P. Phosphoproteomic analysis of the developing mouse brain. Mol. Cell. Proteomics. 2004;3:1093–1101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Mumby M., Brekken D. Phosphoproteomics: new insights into cellular signaling. Genome Biol. 2005;6:230. doi: 10.1186/gb-2005-6-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blagoev B., Ong S.E., Kratchmarova I., Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 54.Gruhler A., Olsen J.V., Mohammed S., Mortensen P., Faergeman N.J., Mann M., Jensen O.N. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y., Wolf-Yadlin A., Ross P.L., Pappin D.J., Rush J., Lauffenburger D.A., White F.M. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the EGF receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 56.Chen W.G., White F.M. Proteomic analysis of cellular signaling. Expert Rev. Proteomics. 2004;1:343–354. doi: 10.1586/14789450.1.3.343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.