Abstract
Human cancer genome and epigenome projects aim to identify new cancer genes and targets for therapy that have been overlooked by conventional approaches. Here we integrated large-scale genomics and epigenomics of 31 human infiltrative gliomas and identified low-frequency deletion and highly recurrent epigenetic silencing of WNK2, encoding a putative serine/threonine kinase. Prior cancer genome sequencing projects also identified point mutations in WNK1–4, suggesting that WNK family genes may have a role in cancers. We observed consistent gene silencing in tumors with dense aberrant methylation across 1.3 kb of the CpG island but more variable expression when the 5′-most region remained unmethylated. This primary tumor data fit well with WNK2 promoter analysis, which showed strong promoter activity in the 5′-most region, equivalent to the simian virus 40 promoter, but no activity in the 3′ region. WT WNK2 exhibited autophosphorylation and protein kinase activity that was enhanced in cells exposed to hypertonic conditions, similar to WNK1. WNK2 inhibited up to 78% of colony formation by glioma cells but in an unexpectedly kinase-independent manner. The WNK2 silencing by epigenetic mechanisms was significantly associated (P < 0.01) with a known genetic signature of chemosensitive oligodendroglial tumors, 1p and 19q deletion, in two small but independent tumor sets. Taken together, the epigenetic silencing, occasional deletion and point mutation, and functional assessment suggest that aberrations of WNK2 may contribute to unregulated tumor cell growth. Thus, our integrated genetic and epigenetic approach might be useful to identify genes that are widely relevant to cancer, even when genetic alterations of the locus are infrequent.
Keywords: brain tumor, epigenomics, genomics
Tumor suppressor genes are typically discovered through analyses of familial cancers and through mapping allelic loss of heterozygosity in sporadic human cancers (1). Regions exhibiting recurrent, nonrandom deletion are selected for further identification of the tumor suppressor gene by identifying a second hit involving point mutation or homozygous deletion (2). Until recently, surveys for point mutations have thus been confined to regions of recurrent deletion, gain, or amplification. Theoretically, any gene that is activated or inactivated primarily by mutation could remain undiscovered. Recent proposals for sequencing entire cancer genomes aim to identify genes that have escaped detection by lower resolution approaches, to provide new targets for therapy, and to further improve experimental modeling of cancer. Pilot projects have proven the utility of this approach (3–5).
We and others have been addressing a related hypothesis by taking an unbiased approach to mapping nonrandom and tumor-type-specific epigenetic alterations that result in gene silencing (6–9). We proposed that there may be tumor suppressor genes that have escaped detection because they are seldom deleted but often silenced by epigenetic mechanisms (10). By using restriction landmark genome scanning (RLGS) (11), which allows quantitative analysis of the methylation status of >1,000 CpG islands at a time, we previously estimated that across the entire genome, hundreds of CpG islands may be aberrantly methylated in any given tumor, although the range across individual tumors varies significantly (9). A small subset of these methylation events are sufficiently recurrent to qualify as nonrandom events, potentially arising through selection of cells harboring a methylation-mediated silencing event that confers a growth advantage. Integrated genomic and epigenomic tumor profiles showed that the majority of loci affected by aberrant methylation are independent of recurrent deletions (10, 12), suggesting that these approaches are complementary for cancer gene discovery.
Here we used a combined genomic and epigenomic analysis of human tumors to identify candidate tumor suppressor genes and found highly recurrent aberrant methylation at the 5′ end of the gene WNK2 (with no lysine 2). WNK2 resides on chromosome 9q22.31 and belongs to a serine/threonine kinase subfamily having the catalytic lysine in a noncanonical position (13, 14). Of the four known WNK genes, deletion of WNK1 or mutations of WNK4 are found in hypertensive patients with pseudohypoaldosteronism type II (15, 16). Point mutations of WNK1 in breast and colon cancer and WNK2 in lung cancer were discovered by large-scale cancer genome sequencing (4, 5). Point mutations of WNK1, WNK2, WNK3, and WNK4 were also discovered in a spectrum of solid tumor types (17).
We report the discovery of genetic and epigenetic silencing of WNK2 in human adult glial tumors and investigate its potential role as a tumor suppressor. By using extensive methylation data and WNK2 expression from human tumors, and analysis of WNK2 function, we provide evidence that WNK2 is involved in glial tumorigenesis and is one part of a pathway taken by tumors that are more chemosensitive. Taken together with the recent discovery of point mutations in WNK genes in common tumor types, our data highlight the unique discovery potential afforded by integrating genomics and epigenomics for cancer gene identification and for identifying molecular markers of clinical response.
Results
The CpG Island Encompassing the 5′ End of WNK2 Is Aberrantly Methylated in Human Gliomas.
To examine the relative contribution of genetic and epigenetic alterations to the genesis of adult infiltrative gliomas, aberrant CpG island methylation was screened with RLGS, and copy number change was assessed by using array comparative genomic hybridization (CGH). We have previously shown that the majority of aberrantly methylated genes in human tumors are independent of the loci affected by recurrent deletion (10). To investigate the significance of this finding, we studied in depth one of the most recurrent sites of aberrant methylation in human infiltrative gliomas, corresponding to RLGS fragment 1D05. The NotI site of this DNA fragment corresponds to a CpG island locus at chr9q22.31, which encompasses the putative transcription start site of the WNK2 gene encoding a putative serine/threonine kinase (Fig. 1A). RLGS profiles show that the intensity of the NotI fragment is reduced in 29 of 31 infiltrative gliomas [supporting information (SI) Fig. 6A and SI Table 1], which suggests that WNK2 is frequently affected by either aberrant CpG island methylation or deletion, or both.
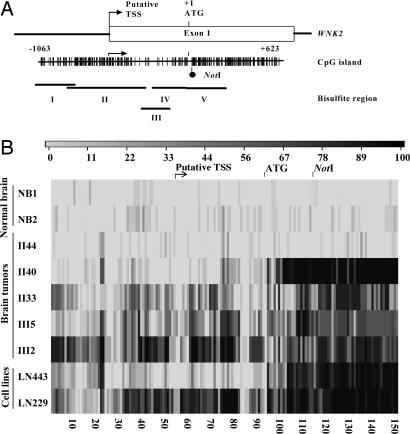
Fig. 1.
Human adult gliomas exhibit aberrant methylation of WNK2. (A) Diagram of the 5′ end of the WNK2 gene showing the position of the putative transcriptional start site (TSS) and first exon and the translation start site (ATG). The vertical bars represent the CpG dinucleotides within the CpG island, and the RLGS fragment NotI site is indicated. The open rectangle represents exon 1 and includes the putative transcription start site based on the prediction from the FirstEF algorithm (32) and our cDNA mapping (data not shown). The translational start site is designated position +1. The locations of the five contiguous regions for bisulfite sequencing are depicted by the horizontal lines. The CpG island sequence has a CpG ratio of 0.91 and a GC content of 69.3%. (B) Unique patterns of aberrant methylation of the WNK2 CpG island in normal brain, human gliomas (grade II: 40, 44, and 33; grade III: 5 and 2), and glioma cell lines (LN443 and LN229). The methylation status of the CpG island was analyzed by sequencing subclones of PCR products from bisulfite-treated DNA. The grayscale of each bar represents the percentage of methylation of one CpG assessed from at least eight clones, as indicated in the key.
By using array CGH, hemizygous deletion of the 9q22 locus was observed, but only in two of 31 cases (SI Fig. 6B and SI Table 1). One case involved the entire chromosome, whereas the other was ≈37 megabases, excluding the p16INK4A locus. Deletion of the p16INK4A locus at 9p21.3 was found in three of 31 tumors. These data suggest that deletion of WNK2 occurs but that it is infrequent in this set of predominantly low-grade gliomas.
We next tested for aberrant methylation at 149 contiguous CpGs of the WNK2 CpG island, including the NotI site, predicted promoter region and exon 1 (Fig. 1A). Normal brain samples showed very little methylation in the entire 1.3-kb region. In contrast, the tumor data confirmed and extended the aberrant methylation at the NotI site. For tumors II33, III2, III5, we observed dense aberrant methylation across the 1.3-kb region. For tumor II40, however, the methylation pattern consisted of a 3′ portion showing extensive methylation followed by an abruptly demarcated 5′ zone lacking methylation (Fig. 1B, SI Fig. 7, and SI Table 2).
WNK2 Expression Is Down-Regulated in Human Gliomas.
To explore the consequence of the different patterns and extent of aberrant methylation on WNK2 expression in human gliomas, quantitative RT-PCR analysis of WNK2 expression was performed. WNK2 was expressed at significant levels in normal brain relative to the control gene, GusB. In contrast, the majority of gliomas showed a dramatic reduction in the WNK2 RT-PCR product, especially those tumors exhibiting aberrant methylation over the entire CpG island region, suggesting that WNK2 expression is significantly repressed in these tumors (Fig. 2A, SI Fig. 8, and SI Table 3). In tumor II44, which had little or no aberrant methylation, the decrease in WNK2 expression was small (2- to 3-fold) relative to that in tumors with extensive aberrant methylation (20- to 100-fold). Interestingly, in tumor II40, which exhibits aberrant methylation in the 3′ but not the 5′ zone, the level of WNK2 expression was similar to normal brain. Taken together, the methylation and expression data suggest that WNK2 silencing is most strongly associated with aberrant methylation of the 5′ zone. It is within this 5′ region that the putative core promoter of WNK2 is predicted to reside (Fig. 1A).
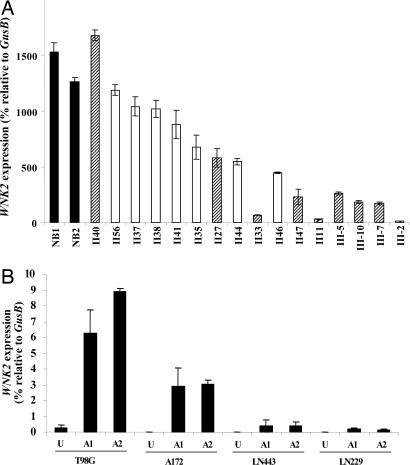
Fig. 2.
Expression of WNK2 was down-regulated in human gliomas and reactivated in glioma cell lines by a methylation inhibitor. (A) Expression of WNK2 in human gliomas was determined by real-time quantitative RT-PCR and is expressed relative to a control gene, GusB. Data are presented as the average of triplicates ± SD. Three independent experiments yielded similar results. A filled bar indicates normal brain (NB), an open bar indicates astrocytoma, and a hatched bar indicates oligodendroglioma. (B) WNK2 is reactivated by treatment with 5-aza-2′-deoxycytidine in four glioma cell lines. U, untreated cells. A1 and A2 are independent cell cultures treated with 5 μM 5-aza-2′-deoxycytidine for 3 days.
WNK2 Reactivation in Human Glioma Cells Treated with a Demethylating Agent.
To investigate whether methylation maintains WNK2 silencing, we treated glioma cells with 5-aza-2′-deoxycytidine, a chemical inhibitor of DNA methyltransferase 1, and tested for reactivation of WNK2 expression. In four untreated glioma cell lines, there was little or no WNK2 mRNA. After 3 days of drug treatment, however, WNK2 expression was modestly reactivated to various degrees in all four cell lines (Fig. 2B, SI Fig. 9, and SI Table 3). It should be noted that WNK2 expression level after 5-aza-2′-deoxycytidine is substantially lower than that of uncultured tissue, presumably because of long-term culturing, additional epigenetic silencing mechanisms, and the nature of this drug reactivation mechanism. In LN443 cells, a more robust reactivation was observed with the histone deacetylase inhibitor trichostatin A, either alone or in combination with 5-aza-2′-deoxycytidine (SI Fig. 10A). By using a chromatin immunoprecipitation assay, increased histone H3 acetylation was detected at the WNK2 promoter after trichostatin A alone or trichostatin A and 5-aza-2′-deoxycytidine (SI Fig. 10B and SI Table 4). These data suggest that for some glioma cells, demethylation, and/or histone acetylation causes WNK2 reactivation, through direct or indirect mechanisms.
The 5′-Most Region, but Not the 3′ Region, of the WNK2-Associated CpG Island Functions as a Promoter.
The unique patterns of aberrant methylation in the WNK2 CpG island, and their association with expression levels in glial tumors, might give clues to which region(s) is important for normal promoter activity. Promoter–reporter constructs were, therefore, designed to include 5′ regions of the CpG island, 3′ regions, or both and then tested for promoter activity in human cell lines (Fig. 3A). WNK2 promoter constructs 1, 3 and 5, which contain only the 5′ region, exhibited remarkably high induction of luciferase activity, comparable with that in our positive control, a simian virus 40 viral promoter construct (Fig. 3B). By contrast, WNK2 promoter construct 4, which contains primarily the 3′ region, did not exhibit significant reporter gene induction in these four cell lines. Inclusion of the 5′ region and 3′ region resulted in little or no induction of luciferase activity, suggesting inhibition of the promoter activity in the 5′ region by sequences in the 3′ region. In LN443 and T98G cells, all constructs exhibited significantly lower promoter activities, although the relative activities among the 5 constructs are quite similar in the four cell lines tested. Taken together, these results suggest that the 5′ zone is critical for WNK2 promoter function in vitro and in vivo.
Fig. 3.
The 5′-most region of the WNK2-associated CpG island exhibits strong promoter activity. (A) Diagram of the 5′ end of WNK2 and the associated CpG island showing the five different DNA fragments that were cloned into pGL3 basic luciferase (Luc) vector. The nucleotide positions are given relative to the translation start site. (B) Human embryonic kidney cells (HEK293) and three glioma cell lines were transiently transfected with each of the five constructs. The luciferase activities were measured and normalized with an internal control, Renilla luciferase activity. The data are presented as the averages ± SD of the fold induction of the putative promoter construct over that in cells transfected with the pGL3 basic vector. (Inset) The results of LN443 and T98G cells by using a smaller range on the y axis, because all constructs exhibit lower activity in these cells. Two additional independent transfections showed similar results.
WNK2 Exhibits Protein Kinase Activity and Restoration of WNK2 Expression Suppressed Colony Formation.
To assess the function of WNK2, in vitro kinase assays were performed in HEK293 cells transfected with WT WNK2, WNK2 mutants of the putative kinase domain, or vector alone. We created mutations of the putative kinase domain based on a similar analysis of WNK1 (13, 18), including mutation of the catalytic lysine WNK2–K207A (K), and double mutant S352A, S356A (S). A truncation mutant encoding amino acids 1–195 (195) that excluded the kinase domain also was created. WT WNK2 was capable of both autophosphorylation and phosphorylation of a generic protein substrate, myelin basic protein. Exposure of cells to 0.5 M NaCl for 15 min enhanced WNK2 kinase activity (Fig. 4A), as has been reported for WNK1 and WNK4 (19). In contrast, very little kinase activity was seen with the three mutants.
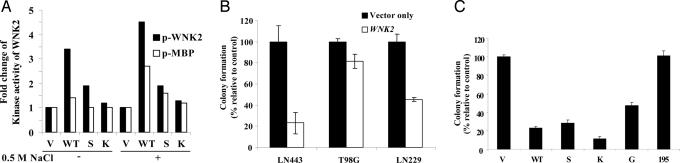
Fig. 4.
WNK2 has protein kinase activity and restoration of WNK2 expression inhibited colony formation in glioma cell lines. (A) WNK2 is a protein kinase with enhanced activity under hypertonic stress. In vitro kinase assays were performed in the presence of [γ-32P]dATP by using whole-cell protein extracts prepared from HEK293 cells transfected with WNK2 WT or kinase dead mutants, S or K. WNK2 was immunoprecipitated and in vitro kinase assays were performed to assess the autophosphorylation, and substrate [myelin basic protein (MBP)] phosphorylation catalytic activities of WNK2 from control cells or cells exposed to hypertonic stress (0.5 M NaCl for 15 min). Fold changes in activity are presented relative to empty vector. Two additional experiments showed similar results. (B) Restoration of WNK2 expression inhibited colony formation in glioma cell lines. Glioma cells were transfected in triplicate, selected, and stained, and colonies of >50 cells were counted and presented as a percentage relative to the control from the same cell line. Two additional independent experiments yielded similar results. (C) Similar experiment as in B in LN443 cells, including vector alone (V5), and vectors containing full-length WNK2 WT, or the mutant S, K, G, or 195.
To gain further insight into the potential consequence of WNK2 silencing in gliomas, we investigated whether restoration of WNK2 expression would impact the growth of WNK2-negative glioma cells. Glioma cells were transfected with control vector or WT WNK2 construct and tested for their ability to form colonies in vitro. WNK2-transfected LN443 and LN229 cells formed significantly fewer colonies: only 20% and 40% of controls, respectively. In contrast, expression of exogenous WNK2 had only a marginal effect on colony formation by T98G cells (Fig. 4B), suggesting some specificity of WNK2-mediated inhibition of growth for the cellular context in which it is expressed. Kinase dead mutants (S and K mutants) did not reverse the inhibitory effect of WNK2, indicating that the growth inhibition is kinase-independent. In contrast, a truncation mutant, WNK2-195 was unable to inhibit growth. One of the lung cancer mutants, WNK2-G1619E, exhibited approximately one-half the growth inhibition compared with WNK2 WT (Fig. 4C). The presence of the exogenous WNK2 protein was confirmed by immunocytochemical staining (SI Fig. 11A) and by Western blotting (SI Fig. 11B) with antibodies against the V5 epitope. These data suggest that WNK2 silencing is not a passenger epimutation but could confer a growth advantage.
Decreased WNK2 mRNA Is Significantly Associated with Chromosome 1p and 19q Deletion.
Histologically, infiltrating gliomas are classified into three major subtypes, including astrocytomas, oligodendrogliomas, and mixed oligoastrocytomas (20). Approximately 50% of oligodendroglial tumors exhibit combined deletion of the 1p and 19q chromosome arms, with an incidence of ≈60–70% in pure oligodendrogliomas and 40% in mixed oligoastrocytomas (21–24). Few astrocytomas exhibit these hallmark deletions. Combined deletion of the 1p and 19q chromosome arms is a powerful predictor of positive response to chemotherapy and of a significantly longer overall and progression-free survival among patients with oligodendroglial tumors (21, 25–27).
Many of our oligodendroglioma tumor samples had a greater reduction in WNK2 mRNA levels compared with the pure astrocytomas. In tumors for which both DNA and RNA were available, we therefore tested for a potential relationship between WNK2 mRNA levels and the presence or absence of combined 1p and 19q deletion, assessed by array CGH (SI Fig. 12). Combined 1p/19q deletion was detected in eight of nine pure oligodendrogliomas and in none of seven astrocytomas, which was consistent with previous results. These results suggest a potential relationship between WNK2 silencing and 1p/19q deletion (Fig. 5A).
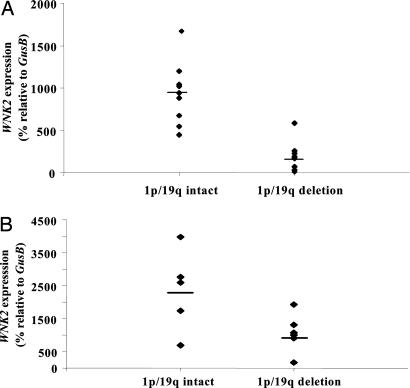
Fig. 5.
Reduction of WNK2 expression is significantly associated with the combined deletion of chromosome 1p and 19q. (A) WNK2 down-regulation is significantly associated with 1p/19 deletion. Array CGH was used to determine the chromosome 1p and 19q status (SI Fig. 12). The horizontal bar indicates the mean value for each group. WNK2 silencing is significantly associated with 1p/19q deletion (P < 0.01; Student's t test). (B) An independent set of oligodendroglial tumors, predominately mixed oligoastrocytomas, also shows a significant association between WNK2 silencing and 1p/19q deletion (P = 0.045).
Given the relative paucity of pure oligodendrogliomas without 1p/19q deletion in this tumor set, we further assessed whether WNK2 silencing is more strongly associated with 1p/19q deletion than with the oligodendroglial phenotype itself. Toward this objective, we assessed an independent set of tumors composed of 11 predominately mixed oligoastrocytoma tumor specimens for which both DNA and RNA were available. Six of 11 tumors demonstrated combined 1p/19q deletion, consistent with previous reports. Fig. 5B shows that WNK2 silencing is significantly associated with 1p/19q deletion in this independent set of oligodendroglial tumors, including both grade II and grade III tumors. We conclude that the extent of WNK2 epigenetic silencing is significantly associated with the genetic signature of 1p/19q deletion in these tumors.
Discussion
Our integrated genomic and epigenomic analyses identified an association between a common epigenetic event and a recurrent genetic alteration in adult infiltrative tumors. Epigenetic inactivation of WNK2 expression correlates significantly with combined deletion of the 1p and 19q chromosome arms, a powerful marker of response to chemotherapy and longer survival in patients with oligodendroglial tumors (21, 25–27). Combined 1p/19q deletion may result in improved response and survival because of the specific, as-yet-unidentified genes that are altered. Alternatively, 1p/19q deletion, recently attributed to translocation (28), may be a marker of a biological pathway or specific type of transformed cell resulting in a more clinically favorable tumor. Within the group of tumors exhibiting 1p/19q deletions, however, there are individuals that do not experience the predicted tumor chemosensitivity or survival benefit, suggesting that discovery of further markers of response could be useful (21, 25). Our data propose a distinction in the biology of these chemosensitive tumors in that the tumorigenic pathway may involve both 1p/19q deletion/translocation and WNK2 down-regulation, potentially releasing the target cells from growth inhibition. It will be of significant interest to determine how well WNK2 down-regulation and 1p/19q alteration are associated in a larger, uniform set of tumors and whether WNK2 in combination with 1p/19q is useful in predicting the clinical behavior and guiding the therapeutic management of these infiltrative gliomas. Thus, our integrated approach to tumor genomes is a powerful way to identify unique and unexpected associations between genetic and epigenetic mechanisms of tumorigenesis.
Two observations suggest that WNK2 silencing is more strongly associated with 1p/19q deletion than with the pure oligodendroglioma phenotype itself. First, the single oligodendroglioma without 1p/19q deletion also did not show evidence of WNK2 silencing. Second, we tested an independent set of tumors that was composed primarily of oligoastrocytomas, and again we observed a significant association between WNK2 and 1p/19q deletion. Although distinguishing oligodendrogliomas, oligoastrocytomas, and astrocytomas based on histology alone is difficult (29), here we included only the most classic cases of oligodendroglioma in our first tumor set, which could explain why most of these tumors exhibit 1p/19q deletion.
We propose that WNK2 had escaped detection previously because of the very low frequency of deletion of the 9q22.31 locus in gliomas (30) and because of the strong bias introduced by focusing on highly recurrent deletions. Consistent with this proposal, just two of 31 infiltrative gliomas exhibited 9q hemizygous deletion in our tumor series. In one of the WNK2-deleted cases, the entire chromosome 9 was lost, whereas the other case encompassed ≈37 megabases, including the WNK2 locus but excluding p16/INK4A. In the context of genetic analyses alone, these deletions would be overlooked because of their infrequency. In light of our discovery of highly recurrent epigenetic alteration of WNK2, however, the deletions might represent the rare but important occasion when genetic and epigenetic alterations combine to inactivate WNK2 and potentially other genes in this locus.
We assessed methylation at 149 contiguous CpGs over 1.3 kb in the WNK2 promoter region. Most methylation analyses in tumors to date assess a few CpGs with high sensitivity, or assess each CpG within a bisulfite PCR amplicon of up to ≈300 bp in length. Our more extensive analyses of WNK2 emphasize the importance of the exact location for methylation analysis and for determining the true frequency of the aberrant methylation that has a functional consequence on gene expression. These data also have implications for the proposed human cancer epigenome project (31), particularly if aberrant promoter methylation is “sampled” rather than assessed in its entirety.
The informative patterns of aberrant methylation in WNK2, taken together with WNK2 expression levels, immediately suggested to us that much of the promoter activity could reside in a discrete 5′ zone of the CpG island. This conclusion is supported by the promoter–reporter assays and by the exact overlap of this 5′ zone with the WNK2 core promoter prediction by FirstEF (32). However, in the established glioma cell line LN443, WNK2 was not expressed despite retaining the unmethylated state of the 5′ zone. Alternatively, our data suggest that the histone acetylation state may play a more dominant role in WNK2 silencing in LN443 cells. This methylation difference between the primary tumors and LN443 cells may be related to cell culture, the fact that these and nearly all glioma cell lines are derived from glioblastomas and not low grade gliomas like our primary tumor samples, or perhaps may reflect a transitional step in the WNK2 silencing. We propose that, in general, patterns of aberrant methylation in tumors might aid in identification of core promoter elements in normal cells.
The fact that the WNK2-negative glioma cell lines still support WNK2 promoter activity suggests that they contain the factors necessary for WNK2 transcription, and that aberrant methylation and perhaps chromatin mechanisms actively prevent transcription. It is also likely that in vivo, this proposed core promoter requires additional cis- and trans-acting factors.
Our data support a functional role for WNK2 in glioma genesis, although further mechanisms of WNK2-mediated growth suppression will require more in-depth investigation. For example, WNK1 shares 91% homology with the amino acid sequence of WNK2 within the catalytic domain (14) and functions as a tetramer. Although there is no direct evidence of a stable interaction between WNK1 and WNK2, WNK1 phosphorylates WNK2 and WNK4 in vitro (19). Thus, it may be necessary to determine whether the level of other WNK proteins influences the response of cells to reexpression of WNK2. It will also be important to identify upstream and downstream targets of WNK2 and determine how they might influence WNK2 function. It is noteworthy that WNK1, independent of its kinase domain, is a negative regulator of insulin-stimulated cell proliferation, possibly through activation of its downstream target, SGK1. WNK1 activation of SGK1 is mediated by AKT, in part through phosphorylation of threonine 58 in the N-terminal amino acids 1–220, whereas the kinase domain resides in amino acids 221–479 (33). The point mutations of WNK2 in lung cancer also are well outside the kinase domain (4). Our limited sequence analysis in infiltrating gliomas did not reveal mutations of the WNK2 kinase domain or the local region encompassing the lung cancer mutation sites (SI Table 5). Further sequencing of the 29 exons of WNK2, as well as exons of WNK1, WNK3, and WNK4, in a larger set of tumors will be required to determine whether point mutation contributes to WNK family dysfunction in gliomas.
Experimental Procedures
Human Tissues and Glioma Cell Lines.
Human tumor samples were obtained from the Neurosurgery Tissue Bank at the University of California, San Francisco, including seven World Health Organization, grade II astrocytomas, 11 grade II oligodendrogliomas, and 13 grade III oligodendrogliomas. Nontumor brain tissues include three samples from different regions of one autopsy brain (autopsy 1, white matter; autopsy 4, gray matter; autopsy 13, white matter and gray matter) and two surgical samples from individuals with epilepsy (NB4 and NB8). A second set of 11 tumors, predominately mixed oligoastrocytomas, were used for the association of 1p/19q deletion and WNK2 expression. All samples were obtained with informed consent, and their usage was approved by the Committee on Human Research at the University of California.
Expression Constructs.
A partial cDNA for human WNK2 was generously donated by Osamu Ohara (Kazusa DNA Research Institute, Chiba, Japan) (34). The full-length sequence was constructed by adding the missing 5′ and 3′ sequences via double-stranded oligonucleotides. The complete cDNA was then cloned into pcDNA3.1/V5–His–TOPO (Invitrogen, Carlsbad, CA) linking the V5–His epitope to the C-terminal end. A truncation mutant, WNK2-195, encodes amino acids 1–195 of human WNK2 and was cloned into the BamHI and XbaI sites of pcDNA3.1/V5–His (Invitrogen) to generate pcDNA3.1-195/V5–His. WNK2–S352A, S356A (S), WNK2–K207A (K), and the lung cancer mutant WNK2–G1619E (G) were generated by using the site-directed mutagenesis method. Each cDNA was verified by sequencing (see SI Table 6).
Reporter Gene Assays.
Five promoter–reporter constructs were made by cloning different regions of the WNK2 CpG island into BglII and HindIII restriction sites in the pGL3-Basic luciferase vector (Promega, Madison, WI) (see SI Table 7). The pGL3 basic vector was used as a basal level of luciferase activity and the vector pGL3 promoter containing the simian virus 40 promoter was used as a positive control. One microgram of each vector was cotransfected with 10 ng (1:100) of pRL–TK (Promega) expressing Renilla luciferase by using FuGENE 6 (Roche, Indianapolis, IN). After 48 h, firefly luciferase activity was measured by using the Dual-Luciferase reporter assay system (Promega) and normalized against Renilla luciferase activity.
In Vitro Kinase Assays.
Nondenatured whole-cell protein extracts were prepared from control and V5-tagged WNK2-transfected HEK293 cells and were incubated with 3 μg of an antibody recognizing the V5 epitope tag (anti-V5; Invitrogen) for 1 h at 4°C with gentle rocking. Antigen–antibody complexes were precipitated by incubation with a 1:100 dilution of protein-A/G PLUS agarose for 1 h at 4 °C with gentle rocking. After three washes with 1 M NaCl/20 mM Tris·HCl (pH 7.4), immunoprecipitated proteins were washed once with 10 mM MgCl2/10 mM Hepes (pH 8.0). In vitro kinase reactions were performed as previously described by Lenertz et al. (19).
Colony-Formation Assay.
Cells were seeded in six-well tissue culture plates and transfected with equal molar amounts of either WNK2–V5–His–TOPO WT (3 μg) or pcDNA3.1/V5–His–TOPO control vector (1 μg). After 24 h, cells were split at 1:100 and selected with Geneticin (G418) (GIBCO, Carlsbad, CA) at 800 μg/ml for 10–14 days. Cells were fixed in 20% methanol and stained with crystal violet, and colonies with >50 cells were counted and expressed as a percentage of empty vector controls for each cell line.
For additional information, see the SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Dr. Jane Fridlyand for generating array CGH heat maps and statistical analyses and Dr. Justin Smith for critical comments on the manuscript. This work was supported by grants from the American Brain Tumor Association (to C.H.), the Howard Hughes Medical Institute (to P.J.), the American Cancer Society (to J.F.C.), and the National Institutes of Health (to J.F.C.).
Abbreviations
- CGH
comparative genomic hybridization
- RLGS
restriction landmark genome scanning.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700683104/DC1.
References
- 1.Cavenee WK, Dryja TP, Phillips RA, Benedict WF, Godbout R, Gallie BL, Murphree AL, Strong LC, White RL. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 2.Knudson AG., Jr Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, et al. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 5.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber T, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 6.Baylin SB, Ohm JE. Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA. Semin Hematol. 2005;42:S3–S8. doi: 10.1053/j.seminhematol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Rush LJ, Dai Z, Smiraglia DJ, Gao X, Wright FA, Fruhwald M, Costello JF, Held WA, Yu L, Krahe R, et al. Blood. 2001;97:3226–3233. doi: 10.1182/blood.v97.10.3226. [DOI] [PubMed] [Google Scholar]
- 9.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, et al. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 10.Zardo G, Tiirikainen MI, Hong C, Misra A, Feuerstein BG, Volik S, Collins CC, Lamborn KR, Bollen A, Pinkel D, et al. Nat Genet. 2002;32:453–458. doi: 10.1038/ng1007. [DOI] [PubMed] [Google Scholar]
- 11.Hatada I, Hayashizaki Y, Hirotsune S, Komatsubara H, Mukai T. Proc Natl Acad Sci USA. 1991;88:9523–9527. doi: 10.1073/pnas.88.21.9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong C, Maunakea A, Jun P, Bollen AW, Hodgson JG, Goldenberg DD, Weiss WA, Costello JF. Cancer Res. 2005;65:3617–3623. doi: 10.1158/0008-5472.CAN-05-0048. [DOI] [PubMed] [Google Scholar]
- 13.Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH. J Biol Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- 14.Verissimo F, Jordan P. Oncogene. 2001;20:5562–5569. doi: 10.1038/sj.onc.1204726. [DOI] [PubMed] [Google Scholar]
- 15.Kahle KT, Wilson FH, Lalioti M, Toka H, Qin H, Lifton RP. Curr Opin Nephrol Hypertens. 2004;13:557–562. doi: 10.1097/00041552-200409000-00012. [DOI] [PubMed] [Google Scholar]
- 16.Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, et al. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 17.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu BE, Min X, Stippec S, Lee BH, Goldsmith EJ, Cobb MH. J Biol Chem. 2002;277:48456–48462. doi: 10.1074/jbc.M207917200. [DOI] [PubMed] [Google Scholar]
- 19.Lenertz LY, Lee BH, Min X, Xu BE, Wedin K, Earnest S, Goldsmith EJ, Cobb MH. J Biol Chem. 2005;280:26653–26658. doi: 10.1074/jbc.M502598200. [DOI] [PubMed] [Google Scholar]
- 20.Smith JS, Jenkins RB. Front Biosci. 2000;5:D213–D231. doi: 10.2741/smith. [DOI] [PubMed] [Google Scholar]
- 21.Smith JS, Perry A, Borell TJ, Lee HK, O'Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, et al. J Clin Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki H, Zlatescu MC, Betensky RA, Johnk LB, Cutone AN, Cairncross JG, Louis DN. J Neuropathol Exp Neurol. 2002;61:58–63. doi: 10.1093/jnen/61.1.58. [DOI] [PubMed] [Google Scholar]
- 23.Mueller W, Hartmann C, Hoffmann A, Lanksch W, Kiwit J, Tonn J, Veelken J, Schramm J, Weller M, Wiestler OD, et al. Am J Pathol. 2002;161:313–319. doi: 10.1016/S0002-9440(10)64183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker C, du Plessis DG, Joyce KA, Fildes D, Gee A, Haylock B, Husband D, Smith T, Broome J, Warnke PC. Ann Neurol. 2005;57:855–865. doi: 10.1002/ana.20496. [DOI] [PubMed] [Google Scholar]
- 25.Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, et al. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 26.Fallon KB, Palmer CA, Roth KA, Nabors LB, Wang W, Carpenter M, Banerjee R, Forsyth P, Rich K, Perry A. J Neuropathol Exp Neurol. 2004;63:314–322. doi: 10.1093/jnen/63.4.314. [DOI] [PubMed] [Google Scholar]
- 27.McDonald JM, See SJ, Tremont IW, Colman H, Gilbert MR, Groves M, Burger PC, Louis DN, Giannini C, Fuller G, et al. Cancer. 2005;104:1468–1477. doi: 10.1002/cncr.21338. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, Flynn H, Passe S, Felten S, Brown PD, et al. Cancer Res. 2006;66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 29.Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, Mohapatra G, Hosek SM, Kimmel D, O'Fallon J, Yates A, et al. Oncogene. 1999;18:4144–4152. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- 30.Kitange G, Misra A, Law M, Passe S, Kollmeyer TM, Maurer M, Ballman K, Feuerstein BG, Jenkins RB. Genes Chromosomes Cancer. 2005;42:68–77. doi: 10.1002/gcc.20108. [DOI] [PubMed] [Google Scholar]
- 31.Jones PA, Martienssen R. Cancer Res. 2005;65:11241–11246. doi: 10.1158/0008-5472.CAN-05-3865. [DOI] [PubMed] [Google Scholar]
- 32.Davuluri RV, Grosse I, Zhang MQ. Nat Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- 33.Xu BE, Stippec S, Lazrak A, Huang CL, Cobb MH. J Biol Chem. 2005;280:34218–34223. doi: 10.1074/jbc.M505735200. [DOI] [PubMed] [Google Scholar]
- 34.Nagase T, Kikuno R, Hattori A, Kondo Y, Okumura K, Ohara O. DNA Res. 2000;7:347–355. doi: 10.1093/dnares/7.6.347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.