Abstract
An active miniature inverted repeat transposable element (MITE), mPing, was discovered by computer-assisted analysis of rice genome sequence. The mPing element is mobile in rice cell culture and in a few rice strains where it has been amplified to >1,000 copies during recent domestication. However, determination of the transposase source and characterization of the mechanism of transposition have been hampered by the high copy number of mPing and the presence of several candidate autonomous elements in the rice genome. Here, we report that mPing is active in Arabidopsis thaliana, where its transposition is catalyzed by three sources of transposase from rice: the autonomous Ping and Pong elements and by a cDNA derived from a Ping transcript. In addition to transposase, the product of a second element-encoded ORF of unknown function is also required for mPing transposition. Excision of mPing in A. thaliana is usually precise, and transposed copies usually insert into unlinked sites in the genome that are preferentially in or near genes. As such, this will be a valuable assay system for the dissection of MITE transposition and a potentially powerful tagging system for gene discovery in eukaryotes.
Keywords: autonomous element, PIF/Harbinger, transgenic
Transposable elements (TEs) comprise the largest fraction of eukaryotic genomes, where they often account for at least half of total content. Two broad classes of eukaryotic TEs have been recognized: Type I (RNA) elements use an RNA-mediated mechanism for amplification, whereas type II (DNA) elements use a DNA-mediated mechanism for transposition. Whereas type I elements can reach copy numbers in the thousands and tens of thousands, type II elements rarely attain these levels (1, 2). One exception is miniature inverted repeat TEs (MITEs), which are found in both prokaryotic and eukaryotic genomes, where their copy numbers can exceed several thousand (3–5).
MITEs are short (<500 bp), noncoding elements that are prevalent near genes (6–8). Most type II elements are classified by their encoded transposase, the enzyme that catalyzes element excision from one chromosomal locus and reinsertion elsewhere. However, because MITEs have no coding capacity, the classification of two MITE superfamilies, Stowaway and Tourist, was initially based on structural features including the sequence of the short terminal inverted repeat (TIR) and the target site duplication (TSD) that flanks the element as a direct repeat (4, 5). The TSD of Stowaway MITEs is TA, whereas that of Tourist MITEs is TAA. Similarity between the Stowaway TIRs and TSDs and those of members of the Tc1/mariner superfamily led to the hypothesis that transposase encoded by this superfamily catalyzes Stowaway transposition (9). In contrast, as described below, the association between Tourist MITEs and members of the PIF/Harbinger superfamily is based on both functional criteria and sequence similarity.
The first active MITE, the Tourist mPing element from rice, was recently reported to transpose in three different assay systems: long-term rice cell culture (10), short-term anther culture (11), and plants derived from γ-irradiation of seed from the cultivar Gimbozu (12). Further analysis of mPing in Gimbozu and related landraces that were not exposed to irradiation revealed mPing to be actively transposing (13). Unfortunately, these strains are not suitable for the analysis of transposition behavior because each contained ≥1,000 mPing copies. Such high copy numbers preclude the determination, for example, of whether elements preferentially transpose to linked or unlinked sites in the genome.
Another important question that is difficult to address in the high copy number strains is the identity of the transposase source. Prior studies have furnished circumstantial evidence that two elements, Ping and Pong, encode potential sources of transposase (10, 11). As the name suggests, mPing is derived by deletion from the larger Ping element (Fig. 1), and, as such, Ping is a likely transposase source. However, Ping is not present in the genome of cultured rice cells where mPing is transposing. Instead, a closely related element called Pong was identified in the sequenced rice genomes and subsequently shown to be transposing with mPing in the same cultured rice cells (10).
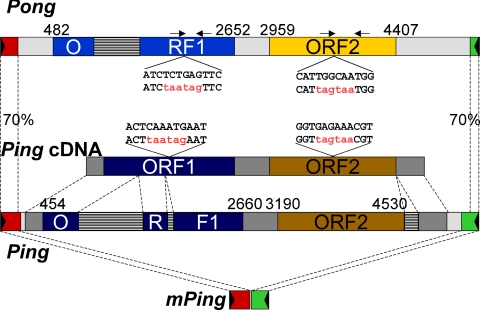
Fig. 1.
Structures of Pong, Ping, and mPing. Predicted ORFs are shown for Pong, Ping, and Ping cDNA. The dark gray regions in Ping stand for exons, and the striped regions stand for introns. The black triangles represent TIRs. The numbers mark the start and end positions of ORFs. Sites and sequences for ORF mutagenesis are shown. The original top strand sequences are shown above the mutated sequences with the introduced nonsense mutations in red. The arrowheads above Pong indicate positions of PCR primers for verifying the presence of mutated ORFs. Amplicon sizes of 346 and 485 bp are expected for ORF1 and ORF2, respectively (see Fig. 5).
Ping and Pong are members of the PIF/Harbinger superfamily that is widespread in both plants and animals (14–16). Plants and insect genomes are particularly rich in family members where the vast majority contains two ORFs (ORF1 and ORF2) (16). ORF2 probably encodes the transposase because it contains a putative DDE motif, a signature for transposase catalytic centers (10, 17, 18). The function of the ORF1 product is unknown although a role in DNA binding has been proposed based on the identification of a myb-like domain in several family members (10). The sequenced rice genome (from Oryza sativa, Nipponbare) has >200 family members including five nearly identical Pong elements and a single Ping element. Of all of the elements in rice, Ping and Pong are most closely related with identical 13-bp TIRs, ≈70% DNA sequence identity in the subterminal regions, and 60% (74% positive) and 84% (90% positive) amino acid sequence identity in ORF1 and ORF2, respectively (16). Whether Ping or Pong or both elements encode functional transposase and what is the function of ORF1 are additional questions that cannot be easily addressed in rice, because strains with actively transposing mPing elements also contain multiple copies of Ping and Pong elements. Here, we report the successful deployment of a heterologous transposition system for mPing in Arabidopsis thaliana. By using this assay, we have addressed and answered several questions that could not be answered in rice concerning the movement of mPing and the source of transposase.
Results
Transposition Assay System.
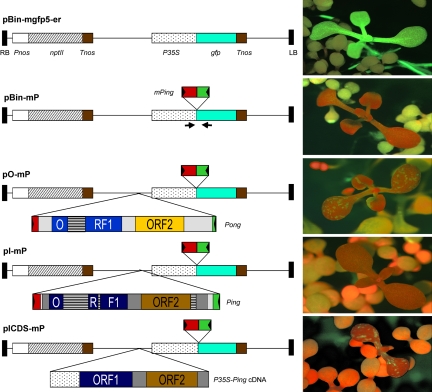
Transposition assays in eukaryotes usually consist of two components: a transposase source and a reporter for TE excision. In this study, both components were engineered into a single T-DNA that was then transferred into the A. thaliana genome via Agrobacteria-mediated transformation. The starting point for all constructs was a T-DNA-containing vector with a selectable marker (neomycin phosphotransferase, npt II) and a reporter gene composed of the green fluorescent protein (gfp) coding region fused to the cauliflower mosaic virus (CaMV) 35S promoter (Fig. 2, pBin-mgfp5-er) (19). This construct was modified by the insertion of a mPing element flanked by a trinucleotide TSD TAA at the junction of the 5′-UTR and the translation start codon (Fig. 2, pBin-mP). This construct served as a negative control because A. thaliana transformants with pBin-mP displayed no GFP expression (Fig. 2).
Fig. 2.
T-DNA constructs transformed into A. thaliana. The arrowheads indicate PCR primer locations for mPing excision analysis. See the legend for Fig. 1 for annotation of Ping, Pong, and mPing. The positive control is pBin-mgfp5-er, and the negative control is pBin-mP. To the right of each construct are representative images of transformants with red fluorescence from chlorophyll and green fluorescence from GFP. The dark red seedlings with true leaves are transgenic, and the yellowish seedlings without true leaves are nontransgenic. npt II, neomycin phosphate transferase; RB and LB, right and left borders of T-DNA; Pnos, promoter of nopaline synthase gene; Tnos, terminator of nopaline synthase gene; P35S, promoter of CaMV 35S gene.
To test for transposition activity, three different sources of tranposase were engineered into pBin-mP: the native rice Pong and Ping elements (Fig. 2, pO-mP and pI-mP) and a cDNA (encoded by Ping) fused to the CaMV 35S promoter (Fig. 2, pICDS-mP). The full-length cDNA contains the predicted ORF1 and ORF2 with an intergenic spacer sequence of 527 bp.
For each construct, primary transformants were selected on Murashige and Skoog medium (20) containing kanamycin (50 mg/liter). Seedlings harboring pO-mP (9 of 22 transformants) and pICDS-mP (6 of 9 transformants) showed GFP expression in cotyledons (as sectors), true leaves (as stripes), hypocotyls (as stripes), or roots (sectors or stripes) when observed with a fluorescence dissection microscope. In contrast, no GFP expression was visualized in 156 seedlings transformed with pI-mP (Fig. 2).
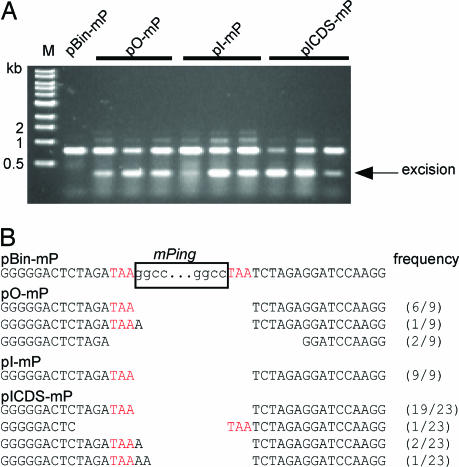
To verify that mPing excision was responsible for GFP expression, PCR analysis was performed on DNA samples extracted from cotyledons by using PCR primers flanking the mPing donor site (primer positions shown in Fig. 2). PCR products consistent with mPing excision were seen after amplification of DNA from primary transformants of pO-mP and pICDS-mP (Fig. 3A). For pI-mP, an mPing excision band was not apparent in the PCR products from primary transformants at the same stage of development (four true leaves; data not shown). However, an apparent excision product was detected when DNA was isolated from older plants containing pI-mP (≈12 leaves) (Fig. 3A).
Fig. 3.
Analysis of mPing excision. (A) Agarose gel of PCR products of mPing donor sites (see Fig. 2 for primer positions). The size marker is a 1-kb ladder (New England Biolabs). Three independent transformants are shown for each construct. (B) Sequences of mPing donor sites after excision events catalyzed by different transposase sources with their frequency in parentheses. The sequence before excision of mPing is shown at the top along with the remainder of the 3-bp TSD sequence (in red). The results are combined from different plants (Ping, 2; Pong, 4; ICDS, 3).
Several putative PCR excision products were cloned and sequenced. The vast majority of excision sites (83%; 34 of 41) contained only a single copy of the TAA trinucleotide; both mPing and the other copy of the TSD were deleted (Fig. 3B). These products are referred to as precise excision events, because under normal circumstances, such an event would restore the locus to the sequence before mPing insertion. In this experiment, “precise” excision of mPing results in a gfp gene with a 3-bp insertion (TAA) because two copies of TAA plus mPing were originally inserted into the 5′-UTR upstream of gfp during plasmid construction. The presence of TAA at the site of excision rules out the possibility of contamination by the gfp gene from the original plasmid (pBin-mgfp5-er). The remaining 17% of the excision sites contain deletions beyond the TSDs or insertions of “A” or “AA” following the retained TSD (Fig. 3B).
Preferential Insertion of mPing into Unlinked Sites.
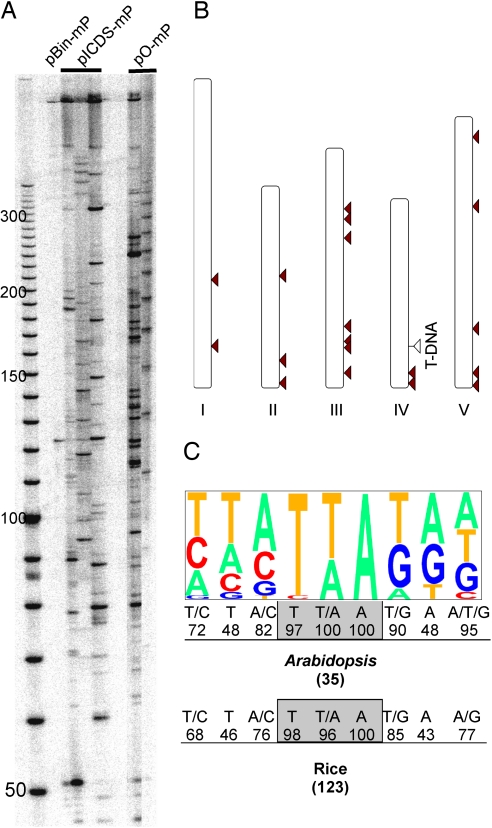
Transposition involves both excision and reinsertion. The rice mPing element, like other Tourist-MITEs, inserts at the trinucleotide TAA/TTA that is duplicated on insertion. In addition, mPing has a strong preference for insertion into or near rice genes (13). To isolate the sites of mPing insertion in A. thaliana, two different strategies were used. For one approach, transposon display analysis [a modification of amplified fragment-length polymorphism technique (21)] was used with DNA isolated from leaves of primary transformants (T1 plants) harboring pICDS-mP or pO-mP T-DNA (see Materials and Methods). All samples tested showed multiple bands on transposon display gels (Fig. 4A), with each sample displaying distinct banding patterns suggestive of independent insertion events. A total of 14 sequences flanking mPing insertion sites were identified in the A. thaliana genome by this approach [supporting information (SI) Table 1, insertions 1–14].
Fig. 4.
Insertion sites of transposed mPing elements in the A. thaliana genome. (A) Autoradiograph of a transposon display gel of DNA from primary transformants. Samples are from those shown in Fig. 3A. (B) Distribution of mPing insertions on A. thaliana chromosomes. The white triangle indicates the single T-DNA insertion locus; red arrowheads indicate somatic mPing insertions identified from the single-copy T-DNA line. (C) Target site preference of mPing. Pictogram of the extended 9-bp target site based on 35 mPing insertions. The letter sizes are proportional to their frequencies. The rice data are from Naito et al. (13).
To determine whether mPing inserts into linked or unlinked sites in A. thaliana, we first identified a transformant that harbored a single copy of the pICDS-mP T-DNA by DNA gel blot analysis among the GFP-expressing T1 plants (data not shown). Additionally, a segregation ratio of 3:1 (48 kanamycin resistant vs. 14 susceptible; χ2 = 0.16, P > 0.65) was observed in the T2 generation, confirming the presence of a single-locus T-DNA insertion in the T1 line. The position of the single-copy T-DNA insertion was determined by thermal asymmetric interlaced PCR and subsequent sequencing to be on chromosome 4 (see Materials and Methods). Of 21 mPing insertions isolated by inverse PCR (from nine resistant T2 plants), 20 inserted into TTA or TAA, whereas 1 inserted into CTA (SI Table 1, insertions 15–35). Insertion sites were distributed on all five chromosomes, with only two insertions located on chromosome 4 (Fig. 4B). These data suggest that mPing has no preference for insertion into linked sites.
The genomic “neighborhoods” of 35 A. thaliana mPing insertions were examined and summarized in SI Table 1. Of the 35 insertions, 33 (94%) are in single-copy sequence with 24 (68.6%) within 1 kb of a gene: 3 in exons, 2 in introns, and 19 within 1 kb of a coding sequence. In a computerized simulation, only 69% of random insertions occur in single-copy sequence and 56% within 1 kb of a gene in the Arabidopsis genome. In rice, an extended 9-bp target site preference has been reported for mPing (13). A similar consensus was derived from the 35 A. thaliana insertions (Fig. 4C).
Requirement of ORF1 and ORF2 for mPing Excision.
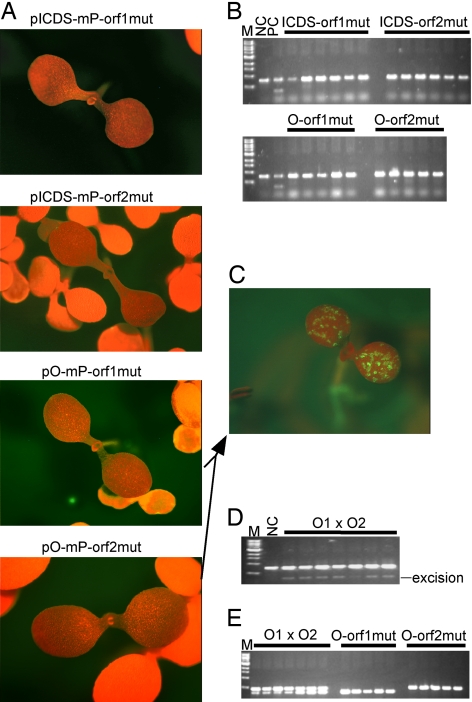
Like most members of the PIF/Harbinger superfamily, Ping and Pong have two ORFs (10, 16). To determine whether both ORFs are required for mPing excision, consecutive nonsense mutations were engineered in the middle of each ORF (Fig. 1). When these constructs replaced their progenitor constructs in the excision assay, no GFP expression was observed in transgenic plants (Pong orf1, n = 18; Pong orf2, n = 27; ICDS orf1, n = 25; ICDS orf2, n = 34) (Fig. 5A). Furthermore, PCR analysis of DNA extracted from the transformants did not reveal any excision products (Fig. 5B).
Fig. 5.
ORF1 and ORF2 are required for mPing transposition. (A) Primary transformants bearing mutated ORFs. Representative images of transformants are shown for each construct. (B) Agarose gels of PCR products of the mPing donor site in transformants containing a single ORF of Ping or Pong. (C) Representative image of the GFP-expressing seedlings from Pong complementation analysis. (D) Agarose gel of PCR products of the mPing donor site in GFP-expressing plants represented by C. Each lane represents an individual plant. (E) Agarose gel of PCR products of mutated ORFs using mutation-specific primers for samples shown in D and their parents. See Fig. 1 for primer positions. Each PCR contains primers for both mutated ORFs. The upper bands are from mutated ORF2, and the lower bands are from mutated ORF1. O1xO2 are plants derived from crossing parents with mutated ORF1 and mutated ORF2. Samples from independent plants are shown. M, size marker (same as that in Fig. 3); NC, negative control (pBin-mP transformant); PC, positive control (pICDS-mP transformant).
Although these data suggest that excision of mPing requires the product of both ORFs, it does not address the question of whether both ORFs need be on the same element. To address this issue, we performed complementation analysis where transformants containing nonsense mutations in ORF1 or ORF2 were crossed to bring a functional copy of each ORF together in the progeny. Twelve GFP-expressing progenies were obtained when parental plants bearing mutated Pong constructs were crossed (Fig. 5C). PCR analysis using genomic DNA prepared from these seedlings revealed excision of mPing (Fig. 5D). When PCR primers specific for the mutated ORFs were used (see Fig. 1 for primer positions), these plants were confirmed to contain both T-DNAs (they contain both mutated ORFs, whereas their parental plants contain only one mutated ORF) (Fig. 5E).
Transcription of the ORFs from Ping and Pong.
Complementation by mutagenized constructs indicates that excision of mPing requires both ORFs and that they need not be produced by a single element (construct). However, most PIF/Harbinger members have two ORFs, and, as such, we were interested in determining how the two ORFs are expressed in rice. To this end, we performed database searches to identify full-length cDNA sequences or ESTs corresponding to the ends (5′ or 3′) of full-length cDNAs. BLAST searches against the database in GenBank using Ping and Pong as queries suggested the presence of transcription start sites in the spacer sequence between ORF1 and ORF2. In addition, when the elements were used for promoter prediction, promoters with high scores (1.00 or 0.99 on a scale of 0 to 1) were predicted upstream of ORF2 and (with lower scores, 0.65 or 0.61) upstream of ORF1.
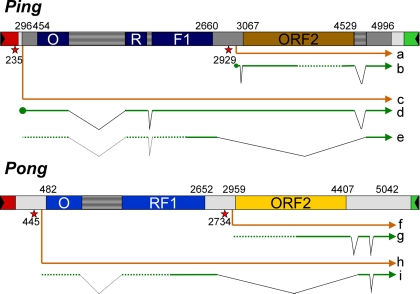
Gene models based on these data for the Ping and Pong elements are shown in Fig. 6(and also in SI Text). Promoters predicted near the 5′-ends of Ping and Pong are proposed to initiate premRNAs “c” and “h,” respectively, and to contain both ORFs. Transcript “c” is further spliced to mRNA “e” which encodes ORF1. The cDNA used in this study (“d”) is probably an intermediate in the production of the fully spliced transcript “e.”
Fig. 6.
Gene models of Ping and Pong. Primary transcripts “a,” “c,” “f,” and “h” are shown in orange. Splicing products are shown as green arrows. Filled circles represent mRNA caps, wedges represent spliced introns, and dotted regions represent unfinished part of full-length cDNAs. Transcription start sites and ORF boundaries are indicated as numbers. Red stars with positions represent predicted promoters. The full-length Ping cDNA used for pICDS-mp is from mRNA “d.” mRNA “b” is supported by GenBank accession nos. CI669389, CI446806, CI696632, CI475894 3′, CI672819, CI450618, and the 5′-RACE cDNA is supported by EF441275 (see Results); mRNA “d” is supported by AK068363; mRNA “e” is supported by CI161443 and CI244010; mRNA “g” is supported by AK068654, CI122968, CI089660, and CI337546; and mRNA “i” is supported by CI080985.
Although mRNA “b” is supported by EST sequences corresponding to both ends of the full-length cDNA clones in GenBank (Fig. 6, “b”), the sequence in the middle is missing. To confirm the integrity of mRNA “b,” a cDNA was cloned from a rice strain with actively transposing mPing elements (O. sativa cv Aikoku123) by using 5′-RACE (13) (see Materials and Methods). The sequence of this cDNA (GenBank accession no. EF441275) supports the transcription start site, the splicing pattern, and an intact ORF2 for mRNA “b.”
Discussion
Demonstration that the rice MITE mPing can transpose in the model plant A. thaliana provides a powerful assay system to address several questions about the transposition mechanism of Tourist-like MITEs. The conclusions from this study and directions of future studies are summarized and discussed below.
TEs related to Ping and Pong were identified in numerous plant and animal genomes with the vast majority containing two ORFs (ORF1 and ORF2) (16). In this study, we demonstrated that functional copies of both ORFs are necessary for transposition of mPing. Transgenic A. thaliana containing a single functional ORF (one or two) cannot promote excision; however, activity is restored when these strains are crossed, but only in progeny containing functional copies of both ORFs. With regard to ORF function, ORF2 is probably the transposase because it contains the DDE motif. However, ORF2 does not encode a recognizable DNA binding domain (10, 16). To function, DNA transposases bind near the ends of the element, positioning the catalytic domain for the excision reaction. It was previously noted that ORF1 encodes a weak myb-like domain, suggesting a role for the ORF1 product in DNA binding (10). As such, the ORF1 protein may bind to the element ends where it could serve as a platform for transposase (ORF2) binding. Alternatively, it could bind to the intergenic region and regulate ORF2 transcription initiation. If this were the case, overexpression of ORF2 should bypass the requirement for the ORF1 product and promote mPing transposition. This scenario is unlikely because mPing transposition was not detected in plants overexpressing Ping ORF2 from a CaMV 35S promoter (C.N.H. and S.R.W., unpublished data). With a requirement for both ORFs for mPing transposition, the A. thaliana assay should be valuable in the design of experiments to understand what the ORFs encode, especially a role for the ORF1 product.
The results of this study, combined with those of prior reports, provide a strong case for the development of mPing into a potent gene tagging tool. Here, we demonstrate that, regardless of the source of transposase tested, excision of mPing is usually precise. As such, this is only the second characterized plant transposon with a preference for precise excision (22). Our data also indicate that mPing, unlike most (but not all) DNA transposons, does not have a preference for insertion into sites linked to the donor. Thus, the features of mPing transposition, including the ability to attain high copy numbers, widespread species distribution, precise excision, and transposition to unlinked sites, make it an extremely attractive system to deploy for gene tagging protocols in both plants and animals.
Materials and Methods
Plasmid Construction.
Full-length Pong, Ping, and mPing were amplified from genomic DNA isolated from O. sativa (Nipponbare) by using pfu DNA polymerase (Stratagene, La Jolla, CA). The Ping cDNA clone was obtained from the Rice Genome Resource Center of National Institute of Agrobiological Sciences of Japan (accession no. AK068363). mPing was cloned into the XbaI site on binary vector pBin-mgfp5-er to obtain pBin-mP, into which Pong was cloned into the SbfI site (pO-mP), Ping was cloned between HindIII and SbfI (pI-mP). To construct pICDS-mP, PCR products of the full-length Ping cDNA were digested with NcoI. The gel-purified larger fragment (containing the coding region) was cloned between NcoI and PmlI sites on pCambia1305.2, resulting in pCambia-ICDS. The fragment containing the 35S promoter and Ping cDNA on this plasmid was amplified by using pfu DNA polymerase using primers tagged with HindIII and SbfI sites. After digestion and gel purification, the fragment was cloned between the HindIII and SbfI sites of pBin-mP, resulting in pICDS-mP. Primer sequences are available on request.
A. thaliana Transformation and Selection.
A. thaliana ecotype Columbia was transformed with Agrobacterium tumorfacience (GV3103) bearing binary vectors according to Bechtold et al. (23). Seeds of transformed plants were germinated on Murashige and Skoog solid medium (0.2% phytagel) containing 150 mg/liter timentene and 50 mg/liter kanamycin. Transformants were distinguishable after 7–10 days of incubation at 26°C (16:8 day/night cycle). Transgenic plants are observed and imaged under blue and white light by using fluorescence stereoscope Leica MZ10 F (Leica, Wetzlar, Germany).
Sequencing of Excision Sites.
Genomic DNA was extracted from cotyledon leaves expressing GFP and used for PCR analysis of mPing excision sites. PCR master mix (Promega, Madison, WI) was used with the following primers: 5′-agacgttccaaccacgtcttcaaagcaag-3′ and 5′-cctctccactgacagaaaatttgtgccca-3′. PCR products were cloned into TOPO vector (Invitrogen, Carlsbad, CA) for sequencing.
Analysis of mPing Insertion Sites.
mPing insertions in A. thaliana were detected by transposon display. Distinct DNA bands from transposon display gels were excised, reamplified, and cloned as described (24, 25). Sequences of cloned fragments were determined at the Molecular Genetics Instrumentation Facility (University of Georgia). Position of flanking sequences were mapped by BLAST search against the annotated A. thaliana genome database of Gramene (www.gramene.org; TAIR, version 6).
Mapping of T-DNA Insertion Sites.
Genomic DNA preparation and DNA blot hybridization were as described (26). A total of 5 μg of A. thaliana genomic DNA digested with NcoI was used for DNA gel blot analysis. GFP coding sequence was used as probes. Sequences flanking the T-DNA inserts were amplified by thermal asymmetric interlaced PCR as described (27). Two rounds of amplification were performed with the following nested T-DNA primers from the left border: 5′-ctggaacaacactcaaccctatctc-3′ and 5′-gatttcggaaccaccatcaaacag-3′. Thermal asymmetric interlaced PCR products were cloned into TOPO cloning vector (Invitrogen) for sequencing.
Genomic DNA Gel Blot Analysis and Inverse PCR.
A. thaliana genomic DNA (1 μg) was digested with DraI in 100 μl at 37°C for 6 h and then purified (PCR clean up kit; Qiagen, Valencia, CA). Approximately 200 ng of DNA was self-ligated overnight at 4°C in 50 μl by using T4 DNA ligase (Invitrogen) and 4 μl was used for inverse PCR. Inverse PCR was performed with the mPing-specific primers (available on request) using phusion DNA polymerase (New England Biolabs, Beverly, MA). Products were cloned into TOPO PCR 2.0 vector (Invitrogen) for sequencing.
Site-Directed Mutagenesis and Complementation Analysis.
Nonsense mutations were introduced into the middle of ORF1 and ORF2 of Pong and Ping cDNA with QuikChange Multi Site-Directed Mutagenesis kit (Stratagene, CA) using pO-mP and pICDS-mP as templates. Primers for Pong ORF1 and ORF2 were 5′-gattactggactaaggtaacttaatagaatacaagcggatatgacgatga-3′ and 5′-cggcttctccaacttggttagtaacgtggttttcctggaatgttcggcag-3′, respectively. Primers for Ping cDNA ORF1 and ORF2 were 5′-tggtcaaggttgaagtcagcgatctaatagttcaatgactattggagtac-3′ and 5′-ttcggtagcattgactgtatgcattagtaatgggaaaggtgcccaactgc-3′, respectively. Mutations were confirmed by sequencing the target regions. The mutated binary vectors (pO-mP-orf1mut, pO-mP-orf2mut, pICDS-mP-orf1mut, and pICDS-mP-orf2mut) were used in the mPing excision assay.
For complementation analysis, transformants bearing Ping and Pong ORF1 and ORF2 mutations were reciprocally crossed, and hybrids were screened for GFP sectors. If found, genomic DNA was used for PCR analysis to detect mPing excision products. To verify the presence of both mutated ORFs of Pong, PCR analysis was performed by using the following primers: 5′-gattggagaattcaatgattactggactaaggtaacttaata-3′ and 5′-gcgtcactttgataattttttcaatattgtcccctaggatg-3′ for ORF1; and 5′-ccgaacggcttctccaacttggttagt-3′ and 5′-cgttttaagatgcaaaatcttcgctgcaatacacc-3′ for ORF2.
5′-RACE.
mRNA was purified by using the RNeasy Plant Mini kit and Oligotex mRNA Mini kit (Qiagen). The SMART RACE cDNA Amplification kit (Clontech, Palo Alto, CA) was used with the Ping-specific primer 5′-gtgatctgatagaagcaact-3′. The resulting PCR products were cloned into pCR 2.1-TOPO (Invitrogen) for sequencing.
Supplementary Material
Acknowledgments
We thank Lisa Kanizay for technical assistance, the laboratories of Zhenghua Ye for help with A. thaliana transformation, Richard Meagher for help with fluorescence microscopy, Timothy C. Hall and James Haseloff for providing vectors, and Jeff Bennetzen for insightful comments. This work was supported by a grant from the National Institutes of Health (to S.R.W.), by a grant from the National Science Foundation (to S.R.W.), and by the University of Georgia Office of the Vice President for Research.
Abbreviations
- TE
transposable element
- MITE
miniature inverted repeat transposable element
- TIR
terminal inverted repeat
- TSD
target site duplication.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.D. is a guest editor invited by the Editorial Board.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. EF441275).
This article contains supporting information online at www.pnas.org/cgi/content/full/0702080104/DC1.
References
- 1.Bennetzen JL. Plant Mol Biol. 2000;42:251–269. [PubMed] [Google Scholar]
- 2.Deininger PL, Roy-Engel AM. In: Mobile DNA II. Craig N, Craigie R, Geller M, Lambowitz A, editors. Washington, DC: Am Soc Microbiol; 2002. pp. 1074–1092. [Google Scholar]
- 3.Liu SV, Saunders NJ, Jeffries A, Rest RF. J Bacteriol. 2002;184:6163–6173. doi: 10.1128/JB.184.22.6163-6173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bureau TE, Ronald PC, Wessler SR. Proc Natl Acad Sci USA. 1996;93:8524–8529. doi: 10.1073/pnas.93.16.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bureau TE, Wessler SR. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bureau TE, Wessler SR. Proc Natl Acad Sci USA. 1994;91:1411–1415. doi: 10.1073/pnas.91.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Arbuckle J, Wessler SR. Proc Natl Acad Sci USA. 2000;97:1160–1165. doi: 10.1073/pnas.97.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang G, Dong J, Chandrasekharan MB, Hall TC. Mol Genet Genomics. 2001;266:417–424. doi: 10.1007/s004380100530. [DOI] [PubMed] [Google Scholar]
- 9.Feschotte C, Mouches C. Mol Biol Evol. 2000;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- 10.Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- 11.Kikuchi K, Terauchi K, Wada M, Hirano HY. Nature. 2003;421:167–170. doi: 10.1038/nature01218. [DOI] [PubMed] [Google Scholar]
- 12.Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, Inoue H, Tanisaka T. Nature. 2003;421:170–172. doi: 10.1038/nature01219. [DOI] [PubMed] [Google Scholar]
- 13.Naito K, Cho E-Y, Yang G, Campbell M, Yano K, Okumoto Y, Tanisaka T, Wessler SR. Proc Natl Acad Sci USA. 2006;103:17620–17625. doi: 10.1073/pnas.0605421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jurka J, Kapitonov VV. Proc Natl Acad Sci USA. 2001;98:12315–12316. doi: 10.1073/pnas.231490598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Feschotte C, Zhang Q, Jiang N, Eggleston WB, Wessler SR. Proc Natl Acad Sci USA. 2001;98:12572–12577. doi: 10.1073/pnas.211442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Jiang N, Feschotte C, Wessler SR. Genetics. 2004;166:971–986. doi: 10.1534/genetics.166.2.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rezsohazy R, Hallet B, Delcour J, Mahillon J. Mol Microbiol. 1993;9:1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 18.Mahillon J, Chandler M. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haseloff J, Siemering KR, Prasher DC, Hodge S. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 21.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Z, Yan X, Maurais S, Fu H, O'Brien DG, Mottinger J, Dooner HK. Plant Cell. 2004;16:1105–1114. doi: 10.1105/tpc.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bechtold N, Pelletier G. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 24.Casa AM, Brouwer C, Nagel A, Wang L, Zhang Q, Kresovich S, Wessler SR. Proc Natl Acad Sci USA. 2000;97:10083–10089. doi: 10.1073/pnas.97.18.10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casa AM, Nagel A, Wessler SR. Methods Mol Biol. 2004;260:175–188. doi: 10.1385/1-59259-755-6:175. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F, Peterson T. Plant Cell. 2005;17:903–914. doi: 10.1105/tpc.104.029660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.