Abstract
The ability to infect macrophages is a common characteristic shared among many mycobacterial species. Mycobacterium avium, Mycobacterium tuberculosis, and Mycobacterium kansasii enter macrophages, using the complement receptors CR1, CR3, CR4, and the mannose receptor. To identify M. avium genes and host cell pathways involved in the bacterial uptake by macrophages, we screened a M. avium transposon mutant library for the inability to enter macrophages. Uptake-impaired clones were selected. Sequence of six M. avium clones identified one gene involved in glycopeptidolipid biosynthesis, one gene encoding the conserved membrane protein homologue to the M. avium subsp. paratuberculosis MAP2446c gene and four others belonging to the same region of the chromosome. Analysis of the chromosome region revealed a pathogenicity island inserted between two tRNA sequences with 58% of G+C content versus 69% in the M. avium genome. The region is unique for M. avium and is not present in M. tuberculosis or M. paratuberculosis. Although the mutants did not differ from the WT bacterium regarding the binding to macrophage cell membrane, analysis of macrophage proteins after 1 h infection revealed a deficiency in the mutant to phosphorylate certain proteins on uptake. To understand M. avium interaction with two evolutionarily distinct hosts, the mutants were evaluated for Acanthamoeba castellanii invasion. The defect in the ability of the mutants to invade both cells was highly similar, suggesting that M. avium might have evolved mechanisms that are used to enter amoebas and human macrophages.
Keywords: uptake
Mycobacterium avium complex is an intracellular pathogen that can infect a variety of host cells (1–4). M. avium, like the majority of pathogenic mycobacteria, infects and replicates within macrophages (5, 6), which are the phagocytic cells where M. avium establishes a long-term infection (7).
A number of studies in vitro have determined that M. avium, in a manner similar to Mycobacterium tuberculosis, is taken up by macrophages using the complement receptors CR3, CR4, and CR1 (8–10), and/or the mannose receptor (8, 11). M. tuberculosis has been found to bind to the CD11b chain of the β-integrin of the CR3 and CR4 receptors, which happened without participation of the serum complement proteins (10). Other studies demonstrated that the mannose capped lipoarabinomannan antigen of virulent M. tuberculosis cell wall is capable to recognize and bind to the mannose receptors on the macrophage membrane (11).
It has been hypothesized that virulent mycobacteria bind to many receptors because pathogenic mycobacteria would use several different receptors on the macrophage membrane to be internalized, as a strategy to guarantee phagocytosis, even when a site-specific macrophage would not contain one or more membrane receptors. In addition, this property would address the possibility that mycobacteria may not express all of the ligands during all phases of infection. Most mycobacteria evolved to survive inside phagocytic cells, although they can enter and replicate in other host cells (2). The reason some pathogens adapted to the intracellular environment in the first place is unknown, but presumably it is related to the availability of specific nutrients and because of the protection that the intracellular environment provides against host defense mechanisms. It seems to be clear that both M. avium and M. tuberculosis can alter their environment in the macrophage, very likely creating the right condition for gene expression associated with the natural progression of the disease (12).
It is possible that M. avium and M. tuberculosis differ regarding the primary pathways by which they enter or are ingested by macrophages. Although both bacteria have a common ancestor source, they evolved in a diverse manner, allowing for important differences to take place in their genome. To obtain new insight into the mechanism by which M. avium is ingested by macrophages, it was decided to screen a transposon mutant bank for phagocytosis in vitro.
Results
Identification of M. avium Mutants Associated With Low Invasion of Macrophages.
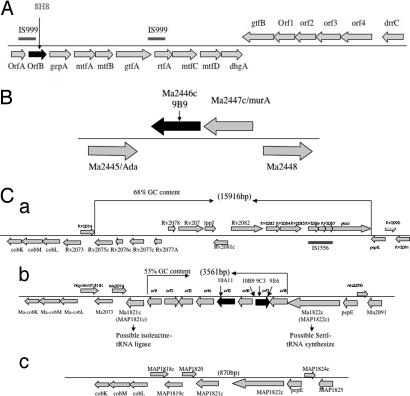
A transposon library was partially screened for invasion of human monocyte cell line and the phenotype of the selected clones was confirmed by using human monocyte-derived macrophages. Three groups of mutants were identified: (i) six clones with uptake of <5% in 1 h, compared with 14 ± 0.5% of the WT bacterium; (ii) 17 clones with uptake between 5% and 8%; and (iii) 39 mutants between 8.1% and 12%. We decided to concentrate initially on the group with <5% uptake. As shown in Table 1, six clones were identified. Among them, the 8H8 clone had an inactivated gene belonging to the glycopeptolipid synthesis pathway (Fig. 1A); the 9B9 clone had a conserved membrane protein inactivated (Fig. 1B); and the other four clones (9C3, 9E6, 10A11, and 10B9) had genes in a unique region of the M. avium chromosome (Fig. 1C). Among the mutants identified, 9C3, 9E6, and 10B9 had mutation in the different locations of ORF7 of this region (Fig. 1C). Although complemented 9C3 strain restored the same phenotype of the WT bacterium, this was not observed to complemented 8H8 mutant, possibly because of the downstream polar effect (Table 1).
Table 1.
Percent of uptake and intracellular growth of M. avium 109 and clones in U937 and monocyte-derived macrophages
| Clone | Uptake U937 (1 h), % | Uptake hM0 (1 h), % | Growth in Mo U937 (4-day) |
|---|---|---|---|
| WT bacterium | 14 ± 0.4 | 16 ± 0.8 | log 1.8 |
| 8H8 | 2 ± 0.1 | 2.2 ± 0.1 | log 0.1 |
| 9B9 | 2 ± 0.3 | 2.1 ± 0.4 | log 2.7 |
| 9C3 | 1 ± 0.2 | 1.0 ± 0.3 | log 2.8 |
| 9E6 | 3 ± 0.6 | 2.8 ± 0.2 | log 2.5 |
| 10A11 | 1 ± 0.3 | 1.3 ± 0.1 | log 2.8 |
| 10B9 | 3 ± 0.4 | 3.4 ± 0.3 | log 1.3 |
| Complemented 9C3 | 13.8 ± 0.5 | 14.9 ± 0.9 | log 1.8 |
| Complemented 8H8 | 1.6 ± 0.3 | 1.5 ± 0.4 | log 0.5 |
Percent of uptake was calculated as the ratio between the number of bacteria added to the tissue culture well and the number of bacteria recovered from the lysed macrophages. Average cfu of the bacterium strains: 2.1 ± 0.8 × 106. The assays were repeated at least 3 times, and the results represent the mean ± SD. All of the clones shown in the table had statistically significant decrease of uptake when compared with the wild-type strain.
Fig. 1.
Chromosome regions. (A) Organization of the chromosome region inactivated in the 8H8 clone of M. avium involved in the glycosylation of the lipopeptide core. (B) Organization of the chromosome region inactivated in the M. avium 9B9 clone. The M. avium gene names correspond to MAP numbers from M. avium subsp. paratuberculosis genome sequence. (C) Genetic organization of M. avium 104 PI associated with low invasion of macrophages and virulence in mice. The M. avium 104 (b) sequence and gene organization of this region is presented in the comparison with M. tuberculosis H37Rv (a) and M. avium subsp. paratuberculosis (c) similar loci. Numbers in parentheses indicate the approximate size of the different regions in above-mentioned mycobacterial species. The gene name or corresponding Rv or MAP numbers from M. tuberculosis H37Rv and M. avium subsp. paratuberculosis genome sequence is shown above the construct. Unknown M. avium genes are presented as ORFs 1–8. The arrows on the genes indicates the location of disrupted genes by the insertion of Tn5367 transposon.
Genetic Organization of M. avium Pathogenicity Island (PI).
The alignments of the three genetic maps of an ≈12 kb genome segment from M. avium 104, M. tuberculosis H37Rv, and M. avium subsp. paratuberculosis region are shown in Fig. 1C. A comparison of M. avium PI region with upstream and downstream 4.5 kb to the same region of above-mentioned mycobacterial species revealed differences in the gene organization. Eight putative ORFs were identified in the PI, using the ORF-predicting software (http://cgrb.orst.edu/). Four ORFs share some homology with products of M. tuberculosis Rv2074 and Rv3902c and/or M. avium subsp. paratuberculosis MAP1820 and MAP1821c. The remaining ORFs in this region do not have similarity to any bacterial genome. Sequencing of this region showed that it is composed of eight ORFs with a G+C content of 58%, in contrast with the G+C content of M. avium (from 69% to 72%). Locus organization in M. paratuberculosis and M. tuberculosis indicates that this region is unique for M. avium, and it is not presented in the other two mycobacterial species. Also, flanking this region are the genes Ma1821c, a possible isoleucine tRNA ligase, and Ma1822c, a possible seril-tRNA, which supports the hypothesis that the region has been acquired during evolution. The protein sequence analysis of upstream or downstream region of the PI revealed a high level of protein identity between M. avium and selected mycobacterial regions. This observation suggests that the region has been acquired horizontally and corresponds to a PI. Analysis of four other M. avium strains (100, 101, 104, and 109) by DNA microarray and real-time PCR confirmed the presence of the PI region (data not shown).
Bacterial Survival in Macrophages.
To determine whether the identified mutants were able to grow inside macrophages, U937 mononuclear cells were infected and bacterial survival was monitored after 4 days. None of the mutants or WT bacterium were cytotoxic to the monolayers (data not shown). As shown in Table 1, with the exception of the 8H8 mutant (attenuated mutant), all others were able to grow inside cells in a way comparable with the WT bacterium, indicating that the defect acquired was linked to the uptake.
In addition, the evaluation of 9C3, 9B9, and 9E6 in U937 cells, human monocyte-derived macrophages, and RAW 267.4 cells (J. Early, M.W., L.E.B., and B. Yuan, unpublished data) have shown that all mutants can grow intracellularly in a way comparable with the WT bacterium. In mice, the mutants also showed decreased ability to enter spleen macrophages [Table 1 and supporting information (SI) Fig. 6].
The Ability of M. avium Mutants to Invade the Amoeba.
M. avium is an environmental microorganism that infects amoeba (13). Because it is possible that M. avium evolved this mechanism to infect macrophages from the ability to be ingested by amoeba, we tested whether the mutants 8H8, 9C3, 9E6, and 9B9 could be taken up by Acanthamoeba castellanii. As shown in Table 2, in contrast to the mutation in the glycopeptidolipid component of the cell wall (8H8 mutant), the mutations in the PI significantly inhibited uptake during the incubation, suggesting that the genes in this island were acquired to give M. avium the ability to be ingested by environmental amoeba. The complementation of the mutant 9C3 recovered the WT phenotype.
Table 2.
Invasion of Acanthamoeba castellanii by M. avium
| Bacteria strain | Invasion at 1 h, % |
|---|---|
| WT | 28.6 ± 6 |
| 8H8 | 24.7 ± 4 |
| 9C3 | 3.3 ± 1* |
| 9E6 | 3.1 ± 2* |
| 9B9 | 2.7 ± 0.5* |
| Complemented 9C3 | 27.6 ± 4.2 |
*, P < 0.05 compared with the WT bacterium.
Macrophage Uptake During Mycobacterium smegmatis Infection Expressing M. avium PI.
To investigate whether the M. avium-identified 3.5-kb region had any effect on the ability of M. smegmatis to be phagocytized by the macrophage, 1,931-bp (clone 1 containing ORF1, ORF2, ORF3, ORF4) and 1,630-bp (clone 2 containing ORF5, ORF6, ORF7, ORF8) fragments of the PI were constructed in the shuttle expression vector pJAM2, resulting in pLDPI-Reg1 and pLDPI-Reg2 plasmid constituents, electroporated into M. smegmatis, and maintained as described in ref. 14. The transformants were screened for invasion in parallel with M. smegmatis WT strain.
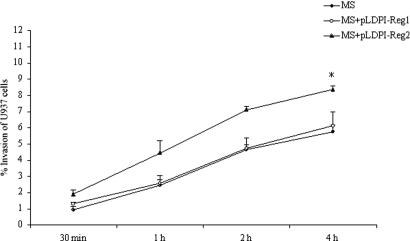
Fig. 2 shows that M. smegmatis clone 1 had similar invasion to the WT strain mc2 155 after 30 min, 1 h, 2 h, and 4 h of exposure at multiplicity of infection 10:1. In contrast, in three independent experiments, M. smegmatis clone 2, expressing the downstream portion of M. avium PI region, showed increased invasion in a time dependent manner and a statistically significant difference at the 4-h time point (P < 0.05) compared with WT bacterium.
Fig. 2.
Invasion of U937 cells by M. smegmatis clones containing M. avium PI region 1 (clone 1) and 2 (clone 2). M. smegmatis with no plasmid was used as control. P < 0.05 for the comparison between control M. smegmatis and clone 1 to clone 2 point invasion.
Binding of Biotinylated M. avium Extracts to Macrophages.
SI Fig. 7A shows five bacterial ligand molecules of M. avium membrane proteins with size range 35–150 kDa that interact with macrophage surface proteins. Comparison of M. avium 109 WT and mutants regarding the binding to macrophage receptors showed no difference.
Identification of Cell-Surface Proteins in Macrophages.
Three macrophage proteins with molecular masses of ≈29 kDa, 52 kDa, and 56 kDa were shown to bind M. avium (SI Fig. 7B). All bacterial clones showed similar results suggesting that both M. avium mutants and WT interact with the same surface proteins of macrophage.
Phosphorylation of Macrophage Proteins.
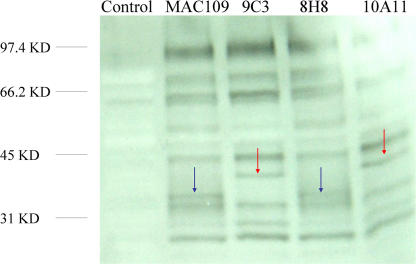
The importance of tyrosine kinase protein in signal transduction pathways is well established. Analysis of phosphorylated proteins of macrophages after 1 h of infection with mutants and WT bacteria, revealed a 36-kDa phosphorylated protein upon uptake of WT bacterium and 8H8 mutant, and a 41-kDa phosphorylated protein upon infection with 9C3 and 10A11 mutants. A differential level of phosphorylation of several proteins was also observed (Fig. 3).
Fig. 3.
Western blot analysis of macrophage phosphorylated proteins after 1 h infection with M. avium WT and transposon mutants.
Amino Acid Sequence of Phosphorylated Host Cell Proteins.
A search from Mascot (Matrix Science, London, U.K.) and IPI human database (PEAKS Studio; Bioinformatics Solutions, Waterloo, ON, Canada) softwares for the 36-kDa phosphorylated protein from M. avium WT and 8H8 infected macrophages revealed the amino acid sequence with 92% identity to the 36-kDa glyceraldehyde-3-phosphate dehydrogenase (GeneBank accession no. NP_002037). The 41-kDa phosphorylated protein sequence from M. avium 9C3- and 10A11-infected macrophage gels identified the skeletal muscle and kidney inositol phosphate (SKIP1) with 81% identity (GenBank accession no. AAK58174) (Table 3).
Table 3.
Amino acid sequences of macrophage phosphorylated proteins
| Protein | GenBank accession no. | Sequence | Identity |
|---|---|---|---|
| 36-kDa protein from macrophage GAPDH | NP_002037 | galqniipastgaakavxkvixel | 22/24 (92%) |
| galqniipastgaakavgkvipel | |||
| 41-kDa protein SKIP1 | AAK58174 |
savxhcxqsaivlxdlklrkfvxste savahchqsaivlgdlklrkfvfste |
22/26 (81%) |
SKIP1, skeletal muscle and kidney inositol phosphate 1. The WT bacterium and 8H8 mutant prosphorylate the 36-kDa protein upon entry, whereas the protein is not activated by the mutants 9C3 and 10A11. In contrast, these mutants phoshorylate the 41-kDa protein SKIP1, whereas the WT bacterium and 8H8 mutant do not.
Actin Polymerization upon Uptake.
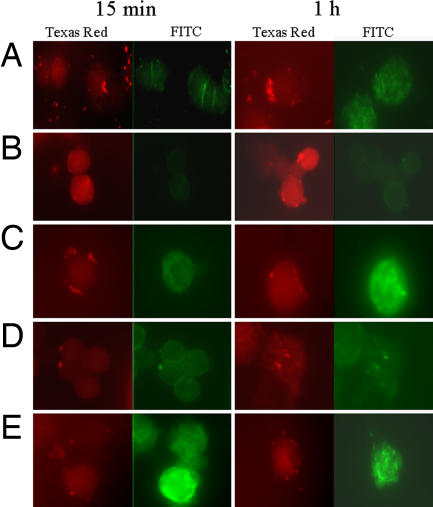
To identify changes in actin cytoskeleton reorganization after short-time infection, we examined the interaction of U937 macrophages with M. avium WT versus 9C3 mutant and complemented strain, as well as M. smegmatis WT versus M. smegmatis clone 2 containing pLDPI-Reg2. The 9C3 mutant had reduced ability to induce actin polymerization, compared with the WT bacterium, and the cytoskeleton rearrangements were localized rather than diffused (Fig. 4 A and B). Over time, actin reorganization increased during 9C3 strain infection. Complemented mutant restored the ability of bacteria to induce actin polymerization with comparable efficiency as WT (Fig. 4C) at both time points. In addition, infected macrophages with M. smegmatis WT and containing clone 2 showed clear difference in actin rearrangement at 15-min and 1-h time infections (Fig. 4 D and E). The results indicate that ORF7 is involved in inducing actin polymerization.
Fig. 4.
Mycobacterium induced actin cytoskeleton rearrangements in U937 cells. Macrophages were infected with M. avium WT (A), 9C3 mutant (B), 9C3 complemented strain (C), M. smegmatis WT (D), or M. smegmatis containing pLDPI-Reg2 (E) for 15 min and 1 h. Bacteria (red) were labeled with rhodamine (Left; Texas red channel) and the actin (green) was visualized by fluorescein-phalloidin staining (Right; FITC channel). Slides were analyzed by using a DM400B coded fluorescent microscope (Leica, Wetzlar, Germany).
Host Protein Interacting with ORF7.
To investigate the cellular proteins interacting with ORF7, U937 cell lysates were incubated with His-tag-ORF7 protein or His-tag alone (control) that had been immobilized on agarose beads. The major His-tag-ORF7 binding protein was actin (Fig. 5). No host proteins bound to His-tagged beads (data not shown).
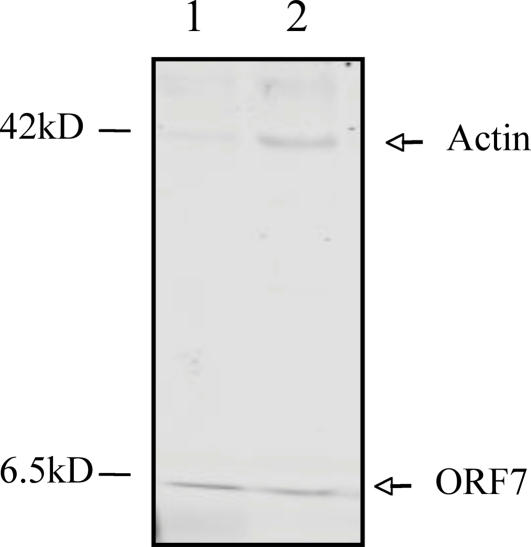
Fig. 5.
In vitro interaction of ORF7 protein with actin. ORF7 protein interaction with host proteins was examined with His-tagged pull-down assay as described in Materials and Methods. Lane 1, purified His-tagged ORF7 protein; lane 2, His-tagged ORF7 bound to actin.
Discussion
Uptake of M. avium by host macrophages has been the subject of a number of studies. M. avium enters macrophages in vitro using similar receptors used by M. tuberculosis, i.e., the complement receptors CR1, CR3, and the mannose receptors (2, 10, 11). In addition, other macrophage receptors have been suggested as entry pathways for virulent mycobacteria, such as the scavenger receptor and CD36 (9, 15). It seems clear, however, that virulent mycobacteria have alternative pathways to be taken by macrophages, which suggests that the process is mainly proactive.
Our current study demonstrates that M. avium phagocytosis by host macrophages is significantly more complex than previously presumed. Screening of an M. avium transposon library for mutants deficient in entry, identified a PI not present in either M. tuberculosis or M. paratuberculosis. The expression of the operon in M. smegmatis with consequent change of phenotype indicates that the proteins are likely expressed on the bacterial surface. The G+C content of this chromosomal region suggests horizontal acquisition from a microorganism probably different from mycobacteria. Also, the fact that it is not present in M. tuberculosis indicates that M. avium probably acquired the region to interact with the environment.
M. avium and other environmental mycobacteria, can infect amoeba (13, 16). The M. avium mutants' ability to infect macrophages and amoeba with comparable deficiency raises the hypothesis that the PI might have been acquired to facilitate M. avium interaction with environmental protozoa. It is plausible to hypothesize that, by using the PI, M. avium avoids macrophage-killing mechanisms and/or lives in a vacuole that is more amenable to a less virulent bacterium, when compared with M. tuberculosis. Other pathogens, like Legionella pneumophila, also infect amoeba in the environment (17). Genes required for Legionella to survive within macrophages are quite similar to the ones required to live inside amoeba (18), indicating the ability to infect and survive in protozoa might have adapted Legionella to the mammalian cell environment.
The difference in macrophage protein phosphorylation upon entry by mutants with genes inactivated in the PI region of the chromosome compared with either the WT bacterium and mutants with gene inactivated in different chromosomal sites (8H8 clone), suggests that the WT bacterium bypasses a serine-threonine kinase phosphorylation. This skeletal muscle and kidney inositol phosphate, SKIP, is an inositol polyphosphate 5-phosphatase that hydrolyzes phosphatidylinositol 3,4,5-triphosphate ([PHI3,4,5]P3) (19). The [PHI3,4,5]P3 is second manager that mobilizes Ca2+ stores. It also binds to several actin-regulating proteins, such as vinculin, profilin, and gelosin (20). Therefore, by hydrolizing PHI-phosphates SKIP plays a negative role in regulating the actin cytoskeleton. SKIP is also 54% similar to the yeast protein ATG1. ATG1 is a serine/threonine protein kinase involved in cytoplasm-to-vacuole transport in yeast and found to be essential in autophagy, where it is required for the formation of autophagosomes (21, 22). M. tuberculosis has been shown recently to inhibit autophagy in macrophages (23). Whether this same characteristic is also found with M. avium is currently unknown, but the answer is being actively pursued in the laboratory. Recently, Boucrot and colleagues demonstrated that Salmonella enterica, a pathogen that survives within vacuoles in macrophages, recruits the plus-end-directed motor kinesin (24). In fact, Salmonella effector proteins translocate into the host cell, and bacterial SifA protein targets SKIP, a host protein that down-regulates the recruitment of kinesin on the bacterial vacuole, and therefore controls vacuolar membrane dynamics. Because SKIP is inversely related with kinesin recruitment to the vacuole, and consequently impacts the motor activity of the vacuole, it would suggest that the WT bacterium is located in a vacuole that actively interacts with kinesin and microtubules (25). Recent work in the laboratory (Jha et al., submitted for publication) sheds light on the complex steps of vacuole membrane dynamics in M. avium-infected macrophages. The activation of some host cell proteins may specifically recruit other proteins to the vacuole membrane, creating the conditions for the uptake of needed nutrients for bacterial replication and virulence gene regulation (12).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) has been described on the surface of phagocytic cells and has a role in signal transduction (26–28). More recently, GAPDH has been found to associate with β-tubulin recruitment to membrane (26, 28). Thus, an association appears to exist between GAPDH and microtubule remodeling. GAPDH has also been shown to bind F-actin and participate in the cytoskeleton rearrangement, because it is necessary for energy production (29, 30). Our observation, both that the ORF7 binds specifically with actin and that the presence of PI in M. avium leads to cytoskeleton rearrangement and GAPDH phosphorylation, suggests a pathogen strategy to command the vacuole trafficking.
The mutants in the PI do not bind to different receptors on macrophage membrane, based on the binding to the biotinylated proteins, suggesting that effects downstream of the receptor also have significant impact on the uptake. Many of the enteric pathogens have been shown to interfere with steps downstream of the membrane receptor, and inactivation of bacterial genes involved in these interactions impair invasion (27).
The PI region contains possible transport proteins or chaperones. The expression of ORFs 1 to 4 in M. smegmatis did not alter the uptake by macrophages; however, the expression of ORFs 5 to 8 significantly increased bacterial invasion, suggesting that some of these genes may be linked to the transport of ORF7. Actin polymerization study also confirms our observation. In fact, ORF8 promoter overlaps with the promoter of ORF7, and ORF8 protein sequence matches significantly with some oxireductases that have been described to participate in protein folding before transport to the membrane (31). Because the inactivation of ORF7 and ORF5 gives similar phenotype, it is plausible to assume that ORFs 5 – 8 function would be to transport actin-interacting protein ORF7. Our results indicate that ORF7 is involved in inducing actin polymerization and is required for efficient M. avium entry into macrophages. The function of ORFs 1 – 4 is unknown at this time.
Mutations in the PI also impact the ability of the bacterium to be taken up by macrophages in vivo, indicating that M. avium uses the same pathway in the host. All M. avium mutants in the PI region did multiply like the WT bacterium, supporting the role of the PI only in macrophage uptake.
Our findings raised a number of important questions. Why does M. avium use a pathway, likely acquired to enter amoeba, to interact with macrophages? Which kind of advantages does it offer?
The future dissection of the pathways of phagocytosis by macrophages should help in the understanding of this complex interaction.
Materials and Methods
Bacterial Strains, Growth Conditions, and DNA Manipulations.
M. avium 109 strain isolated from the blood of an AIDS patient was grown on Middlebrook 7H11 agar (Difco, Detroit, MI) containing 10% oleic acid-albumin-glucose-catalase for 8–10 days and used as the source for macrophage or mouse infection experiments and transposon library preparation. M. smegmatis mc2 155 competent cells were used for screening of pLDPI-Reg1 and pLDPI-Reg2 plasmids containing 1,931-bp (ORF1, ORF2, ORF3, ORF4) and 1,630-bp (ORF5, ORF6, ORF7, ORF8) regions (respectively) of M. avium PI for infection assay in macrophages. Acetamidase promoter expression vector pJAM2, kindly provided by J. A. Triccas and B. Gicquel (Pasteur Institute, Paris, France) (14), was used to construct the pLDPI-Reg1 and pLDPI-Reg2 plasmids. M. avium chromosomal DNA was extracted from mutants as described in ref. 32, and used for sequencing of genes interrupted by transposon.
Macrophages.
U937 human monocytic cell line (ATCC CRL-1593–2) and human monocyte derived macrophages were cultured as reported in ref. 33. Human monocytes were obtained from the blood of volunteers, using protocols approved by the Institutional Human Subject committee. The purification, culture, and maturation have been described in refs. 33 and 34.
Screening of M. avium Transposon Library.
Five thousand clones of M. avium 109 transposon library were constructed as described in ref. 35, and a total of 2,000 clones were screened individually. U937 macrophages were seeded (2 × 105 cells per well) in 24-well tissue culture plates at 80% confluence and treated with 500 ng/ml of Phorbol 12-Myristate 13-Acetate (PMA; Sigma, St. Louis, MO). Macrophages were infected with WT M. avium or transposon mutants (multiplicity of infection of 10) for 2 h at 37°C. The wells were washed 3 times with Hanks's Balanced Salt solution (HBSS) (GIBCO, Grand Island, NY) to remove extracellular bacteria and monolayers were lysed with sterile water for 15 min, as described (35). The percentage of the inoculum that was taken up by macrophages infected with the transposon mutants was compared with the percentage of uptake of the WT (M. avium 109) bacterium. The invasion experiments were performed at least 3 times. Uptake-deficient clones were then tested for phagocytosis five times, using human monocyte-derived macrophages, as described above. The method for complementation is described in SI Text.
Sequencing of Transposon Mutants and Data Analysis.
Nonspecific nested suppression PCR method was used for identification of M. avium genes interrupted by the Tn5367 transposon as reported in refs. 35 and 36. Database search of the sequenced fragments were performed at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov), using BLAST network service, and at the database at The Institute for Genomic Research Institute (www.tigr.org). Sequence homology between M. avium and M. tuberculosis and/or M. paratuberculosis was used to determine the M. avium gene interrupted with transposon.
The identified M. avium unique sequences that did not show any homology to other mycobacterial genomes were retrieved from the CGRB Biocomputing and Bioinformatics database (http://cgrb.orst.edu), and ORFs were detected by using the EMBOSS program Getorf. The G+C content of mycobacterial DNA regions were analyzed with the oligonucteotide properties calculator from Basic Sciences website (www.basic.northwestern.edu/biotools/OligoCalc.html) at the Northwestern University Medical School.
To verify whether this region was present in M. avium strains, the strains 100, 101, 104, and 109 were tested in a custom-made DNA array as part of a large evaluation of common genes (M.H., M.W., L.E.B., and M. Kent, unpublished data).
Infection of Acanthamoeba castellanii.
To examine whether the M. avium clones were able to infect amoeba, A. castellanii was cultured in 712 peptone yeast glucose media and then transferred to a 24-well tissue culture plate. A monolayer of 1 × 105 amoeba in HBSS was infected with 105 M. avium WT or mutants for 1 h. The amoeba were lysed with 0.1% Triton X-100, and the lysate was diluted and plated onto 7H10 agar as described (13).
Expression of M. avium PI Region in M. smegmatis and Screening for Invasion Assay.
U937 macrophages (1 × 105 cells) were seeded overnight in 24-well plates and infected with M. smegmatis mc2 155, containing pLDPI-Reg1 (clone 1), pLDPI-Reg2 (clone 2), or no plasmids (106 bacteria) for 30 min, 1 h, 2 h, and 4 h. U937 cells in triplicate wells were lysed at each time point, and the number of viable intracellular mycobacteria was determined by plating the serial diluted lysates onto 7H10 agar plates. cfus were determined after incubation at 37°C for 3 days. Actual inoculum concentration were determined by plating serial dilutions of M. smegmatis clones and the WT bacterium on 7H10 agar plates, either containing kanamycin or no antibiotic. The experiments were repeated 3 times.
Biotinylation of Surface Proteins on M. avium and Macrophages.
Bacterial and macrophage surface proteins were biotinylated with ECL protein biotinylation module (Amersham Biosciences, Piscataway, NJ) according manufacture's instructions. M. avium mutants (8H8, 9C3, 10A11) and WT (109) were grown to mid-log phase. Bacteria were harvested by centrifugation at 3,000 × g in a Beckman Allegra-6 (Beckman Coulter, Fullerton, CA) for 15 min and washed twice in HBSS. Pellets were resuspended in ice-cold 40 mM bicarbonate buffer, incubated with biotinylation reagent (biotinamidocoproate N-hydroxysuccinamide ester in dimethylformamide) at a concentration of ≈6 × 107 per ml of cells and gently rotated for 30 min at a room temperature. At the end of incubation bacteria were washed twice by resuspending in PBS and spinning at 3,500 rpm for 10 min to remove unbound biotin.
U937 cells were washed in HBSS, resuspended to a density of 2 × 108 cells/ml in bicarbonate buffer, mixed with biotinylation reagent and incubated on a roller mixer for 30 min. Cells were harvested by centrifugation 500 rpm for 10 min at 4°C and washed twice with PBS.
Preparation of Bacterial and Macrophage Extracts.
The biotinylated bacteria were lysed with rapid mechanical disruption with bead-beater in a hypotonic buffer containing 250 mM NaCH, 25 mM Tris·HCl (pH 7.5), 5 mM EDTA (pH 8.0), 1% detergent Nonidet P-40, and a protease inhibitory mixture. The extract was centrifuged, and supernatant were used for the isolation of bacterial ligands that bind to macrophages. The extract of macrophages was prepared as reported in ref. 38. Briefly, biotinylated macrophages were resuspended in 50 mM Tris·HCl buffer, pH 7.5, containing protease inhibitory mixture, 100 mM octyl-β-d-glucopyranoside, 1 mM phenylmethylsulfanyl fluoride, 1 mM CaCl2, and 1 mM MgCl2 (extraction buffer) and incubated on a roller mixer for 1 h at 4°C. The extract was centrifuged and supernatant were used for the isolation of macrophage receptors that bind to M. avium. The extracts of the biotinylated bacteria and macrophages were analyzed by Western blot.
Evaluation of Macrophage Surface Proteins and Bacterial Ligands.
Eight- to ten-day-old cultures of M. avium mutants and WT (≈ 5 × 108 per ml) were incubated with biotinylated macrophage extract (1 mg of protein per ml) at room temperature under continual agitation. Two hours later, suspensions were centrifuged at 250 × g, unbound macrophage extract was removed, and bacteria were collected from supernatant with centrifugation at 3,000 × g. M. avium pellets were washed three times with PBS, resuspended in 1 ml of extraction buffer containing 20 mM EDTA and 25 mM octyl-β-d-glucopyranoside and incubated for 2 h at room temperature on rotator. Macrophage bound protein(s) elutes were collected by centrifugation and processed for Western blot analysis.
Freshly harvested 7.3 × 108 cells/ml U937 macrophages were incubated with biotinylated M. avium mutants and WT extracts (1 mg of protein per ml) for 2 h at room temperature with constant agitation. Suspensions were centrifuged at 250 × g for 10 min and pellets were washed 3 times to remove unbound bacterial extracts. The bound bacterial proteins were eluted from macrophages with extraction buffer rotating for 2 h at room temperature. Elutes were collected for further analysis by Western blot.
Analysis of Macrophage Phosphorylated Proteins.
U937 macrophages were infected with M. avium 8H8, 9C3,10A11 mutants, and WT for 15 min and 1 h (multiplicity of infection 10 bacteria:1 cell). Both infected and uninfected cell cultures were lysed with 0.25% SDS and subjected to immunoprecipitation. Supernatants were precleared with protein A-Sepharose (45 μl) for 1 h at 4°C. Mouse monoclonal agarose conjugate antibody to phosphotyrosine pTyr (Santa Cruz Biotechnology, Santa Cruz, CA) was added to the samples and incubated for overnight at 4°C with agitation. Immune precipitates were separated on a 12.5% (wt/vol) SDS-acrylamide gels. One gel was examined for Western blot analysis, and duplicate gel was stained with Coomassie blue staining and used for In-Gel Tryptic Digestion (Pierce, Rockford, IL) of selected phosphorylated proteins. The phosphorylated proteins were developed with ECL chemiluminescence detection kit (Amersham Parmacia).
The selected phosphorylated protein spots were excised from gels, diced into 2 × 2 mm pieces, and distained twice in distaining solution (80 mg of ammonium bicarbonate with 20 ml of acetonitrite and 20 ml of ultra pure water), using In-Gel Tryptic Digestion kit (Pierce) according to the manufacturer's protocol. The samples were digested in activated Tripsin solution and used for further analysis.
Mass Spectrometric Sequencing and Database Search.
Digested proteins were analyzed at Environmental Health Science Center of Mass Spectrometry facility, Oregon State University, Corvallis, by chromatography and electrospray ionization mass spectrometry. Database search and sequence comparisons were performed by using Mascot (Matrix Science) and IPI human (Bioinformatics Solutions) softwares. The resulting MS/MS spectra were automatically batch-analyzed for each peptide, and successful identifications were based on the number of matching peptide masses and the percent sequence coverage by those matches.
Actin Staining.
To label bacteria, M. avium 109 (WT), 9C3 mutant, a 9C3-complemented strain, and M. smegmatis mc2 155 (WT) and M. smegmatis containing pLDPI-Reg2 were incubated with 200 μg/ml rhodamine (Molecular Probes, Eugene, OR) diluted in PBS containing 3% FBS. After 1 h of incubation, bacteria were washed several times with PBS containing 1% FBS and used to infect U937 cells for 15 min and 1 h. Macrophages were then washed with PBS, fixed with 4% paraformaldehyde for 1 h, and permeabilized with 0.1% Triton X-100 for 10 min. Infected cells were incubated for 30 min with fluorescein-phalloidin (Molecular Probes) at a final concentration of 100 units per ml in PBS containing 1% BSA, to label F-actin.
His-Tagged Pull-Down Assay.
The macrophage extract was prepared as described above. Alternatively, Escherichia coli expressing His-tag-ORF7 protein were harvested in PBS and disrupted with bead-beater, and bacterial lysate was cleared by centrifugation at 15,000 × g for 10 min. Expressed protein were purified according the manufacturer's protocol (Novagen, Madison, WI). U937 cell lysates were mixed with 1 mg of purified His-tag-ORF7 protein and 60 μl of His-tag agarose conjugate rabbit polyclonal IgG beads (500 g/ml with 25% agarose conjugate). The mixture was incubated at 4°C for overnight. Samples were centrifuged, washed 3 times with PBS and resuspended and boiled for 5 min in Laemmli sample buffer containing β-mercaptoethanol. Proteins were stained with Coomassie blue on 12% SDS-polyacrylamide gel, excised from gel as described above and sequenced by electrospray ionization mass spectrometry.
Statistical Analyses.
All in vitro experiments were carried out in duplicate and repeated at least 3 times. The in vivo experiments were carried out with seven mice per experimental group and repeated at least twice. Significance of the differences between experimental groups and control groups was analyzed by Student's t test or ANOVA. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Denny Weber for the editing and preparation of the manuscript. The chromatography and mass spectrometric sequencing was supported in part by the National Institute of Environmental Health Science, National Institutes of Health Grant P30 ES00210. This work was supported by National Institute of Allergy and Infectious Diseases Grant AI-43199.
Abbreviation
- PI
pathogenicity island.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610746104/DC1.
References
- 1.Bermudez LE. Clin Immunol Immunopathol. 1991;61:225–235. doi: 10.1016/s0090-1229(05)80026-1. [DOI] [PubMed] [Google Scholar]
- 2.Bermudez LE, Young LS. Infect Immun. 1994;62:2021–2026. doi: 10.1128/iai.62.5.2021-2026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGarvey J, Bermudez LE. Clin Chest Med. 2002;23:569–583. doi: 10.1016/s0272-5231(02)00012-6. [DOI] [PubMed] [Google Scholar]
- 4.Wu HS, Kolonoski P, Chang YY, Bermudez LE. Infect Immun. 2000;68:2979–2984. doi: 10.1128/iai.68.5.2979-2984.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowle AJ, Dahl R, Ross E, May MH. Infect Immun. 1991;59:1823–1831. doi: 10.1128/iai.59.5.1823-1831.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inderlied CB, Kemper CA, Bermudez LE. Clin Microbiol Rev. 1993;6:266–310. doi: 10.1128/cmr.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrofsky M, Bermudez LE. Infect Immun. 2005;73:2621–2627. doi: 10.1128/IAI.73.5.2621-2627.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bermudez LE, Young LS, Enkel H. Infect Immun. 1991;59:1697–1702. doi: 10.1128/iai.59.5.1697-1702.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst JD. Infect Immun. 1998;66:1277–1281. doi: 10.1128/iai.66.4.1277-1281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlesinger LS, Bellinger-Kawahara CG, Payne NR, Horwitz MA. J Immunol. 1990;144:2771–2780. [PubMed] [Google Scholar]
- 11.Schlesinger LS. J Immunol. 1993;150:2920–2930. [PubMed] [Google Scholar]
- 12.Wagner D, Maser J, Lai B, Cai Z, Barry CE, III, Honer Zu Bentrup K, Russell DG, Bermudez LE. J Immunol. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- 13.Cirillo JD, Falkow S, Tompkins LS, Bermudez LE. Infect Immun. 1997;65:3759–3767. doi: 10.1128/iai.65.9.3759-3767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Triccas JA, Parish T, Britton WJ, Gicquel B. FEMS Microbiol Lett. 1998;167:151–156. doi: 10.1111/j.1574-6968.1998.tb13221.x. [DOI] [PubMed] [Google Scholar]
- 15.Philips JA, Rubin EJ, Perrimon N. Science. 2005;309:1251–1253. doi: 10.1126/science.1116006. [DOI] [PubMed] [Google Scholar]
- 16.Tenant R, Bermudez LE. Curr Microbiol. 2006;52:128–133. doi: 10.1007/s00284-005-0218-4. [DOI] [PubMed] [Google Scholar]
- 17.Cirillo JD, Cirillo SL, Yan L, Bermudez LE, Falkow S, Tompkins LS. Infect Immun. 1999;67:4427–4434. doi: 10.1128/iai.67.9.4427-4434.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molmeret M, Alli OA, Zink S, Flieger A, Cianciotto NP, Kwaik YA. Infect Immun. 2002;70:69–78. doi: 10.1128/IAI.70.1.69-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ijuin T, Takenawa T. Mol Cell Biol. 2003;23:1209–1220. doi: 10.1128/MCB.23.4.1209-1220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ijuin T, Mochizuki Y, Fukami K, Funaki M, Asano T, Takenawa T. J Biol Chem. 2000;275:10870–10875. doi: 10.1074/jbc.275.15.10870. [DOI] [PubMed] [Google Scholar]
- 21.Straub M, Bredschneider M, Thumm M. J Bacteriol. 1997;179:3875–3883. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirkegaard K, Taylor MP, Jackson WT. Nat Rev Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 24.Boucrot E, Henry T, Borg JP, Gorvel JP, Meresse S. Science. 2005;308:1174–1178. doi: 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- 25.Karcher RL, Deacon SW, Gelfand VI. Trends Cell Biol. 2002;12:21–27. doi: 10.1016/s0962-8924(01)02184-5. [DOI] [PubMed] [Google Scholar]
- 26.Huitorel P, Pantaloni D. Eur J Biochem. 1985;150:265–269. doi: 10.1111/j.1432-1033.1985.tb09016.x. [DOI] [PubMed] [Google Scholar]
- 27.Meyer-Siegler K, Mauro DJ, Seal G, Wurzer J, deRiel JK, Sirover MA. Proc Natl Acad Sci USA. 1991;88:8460–8464. doi: 10.1073/pnas.88.19.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tisdale EJ. J Biol Chem. 2002;277:3334–3341. doi: 10.1074/jbc.M109744200. [DOI] [PubMed] [Google Scholar]
- 29.Cao F, Yanagihara N, Burke JM. Cell Motil Cytoskeleton. 1999;44:133–142. doi: 10.1002/(SICI)1097-0169(199910)44:2<133::AID-CM5>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 30.Waingeh VF, Gustafson CD, Kozliak EI, Lowe SL, Knull HR, Thomasson KA. Biophys J. 2006;90:1371–1384. doi: 10.1529/biophysj.105.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watarai M, Tobe T, Yoshikawa M, Sasakawa C. Proc Natl Acad Sci USA. 1995;92:4927–4931. doi: 10.1073/pnas.92.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danelishvili L, Poort MJ, Bermudez LE. FEMS Microbiol Lett. 2004;239:41–49. doi: 10.1016/j.femsle.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 33.Danelishvili L, McGarvey J, Li YJ, Bermudez LE. Cell Microbiol. 2003;5:649–660. doi: 10.1046/j.1462-5822.2003.00312.x. [DOI] [PubMed] [Google Scholar]
- 34.Bermudez LE, Parker A, Petrofsky M. Clin Exp Immunol. 1999;116:94–99. doi: 10.1046/j.1365-2249.1999.00852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Miltner E, Wu M, Petrofsky M, Bermudez LE. Cell Microbiol. 2005;7:539–548. doi: 10.1111/j.1462-5822.2004.00484.x. [DOI] [PubMed] [Google Scholar]
- 36.Winkler MA, Hickman RK, Golden A, Aboleneen H. Biotechniques. 2000;28:890–895. doi: 10.2144/00285st01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.