Abstract
Inactivation of the p53 pathway represents the most common molecular defect of human cancer. But in the setting of melanoma, a highly aggressive and invariably fatal malignancy in its advanced disseminated form, mutation/deletion of p53 is relatively rare, whereas its positive regulator ARF is often lost. Here, we show that genetic deficiency in Arf but not p53 facilitates rapid development of melanoma in a genetically engineered mouse model. This difference is accounted for, at least in part, by the unanticipated observation that, unlike fibroblasts, senescence control in melanocytes is strongly regulated by Arf and not p53. Moreover, oncogenic NRAS collaborates with deficiency in Arf, but not p53, to fully transform melanocytes. Our data demonstrate that ARF and p53, although linked in a common pathway, suppress tumorigenesis through distinct, lineage-dependent mechanisms and suggest that ARF helps restrict melanoma progression by executing the oncogene-induced senescence program in benign nevi. Thus, therapeutics designed to restore wild-type p53 function may be insufficient to counter melanoma and other malignancies in which ARF holds p53-independent tumor suppressor activity.
Keywords: genetically engineered mice, MET, nevi, p16INK4A, rhabdomyosarcoma
Cutaneous malignant melanoma is an extremely aggressive, often fatal disease that has proven to be largely resistant to current therapeutic approaches. Melanoma incidence has increased steadily for several decades, underscoring the need to understand the genetic and environmental mechanisms driving disease risk, initiation, and progression. Although significant progress in our understanding of the etiologies and genetic underpinnings of melanoma has recently been made (1), this knowledge has yet to be translated into effective treatment strategies. The INK4A-ARF (CDKN2A) locus on chromosome 9p21, often deleted or otherwise inactivated in heritable and sporadic melanoma, is strongly implicated in melanoma suppression (1). INK4A-ARF encodes two distinct tumor suppressors, transcriptionally initiated from separate promoters and read in two different reading frames: p16INK4A, referred to hereafter as INK4A, and p14ARF (p19Arf in mouse), referred to hereafter as ARF. INK4A positively regulates the pRB tumor suppressor by inhibiting CDK4, and numerous studies have documented inactivation of INK4A in human melanoma, often sparing ARF (1). But the relevance of loss of ARF function as a distinct event in melanoma has been demonstrated by reports of ARF-specific, INK4A-independent mutations and ARF-specific promoter methylation (1). In contrast, mutation of TP53 is infrequent in primary human melanoma (2), and, when TP53 mutations do occur, the mutant p53 tends to retain transactivation function (3). ARF had been thought to function predominantly as a positive regulator of the p53 tumor suppressor through inhibition of MDM2 (4). However, experimental evidence has recently emerged, mostly through an analysis of fibroblasts, demonstrating roles for ARF distinct from its ability to regulate p53 stability. Accordingly, ARF- and p53-deficient mice do not exhibit identical tumor phenotypes, and ARF interacts with a variety of other proteins, including E2F1, Myc, NFκB, and CtBP, and can function independently of p53 in ribosome biosynthesis, DNA-damage response, apoptosis, and autophagy (4).
Previous studies have shown that the INK4A/RB pathway plays a significant role in human and mouse melanocyte senescence (5–7). Here, we show that, in addition to INK4A, ARF strongly regulates melanocyte proliferation, underscoring the detrimental impact of loss of the INK4A-ARF locus so characteristic of human melanoma (1). Moreover, we demonstrate that ARF, and not p53, efficiently suppresses early melanoma genesis and operates independently of p53 to induce senescence in melanocytes. This functional distinction provides a mechanistic explanation for the common inactivation of ARF but only rare disruption of p53 in melanoma.
Results and Discussion
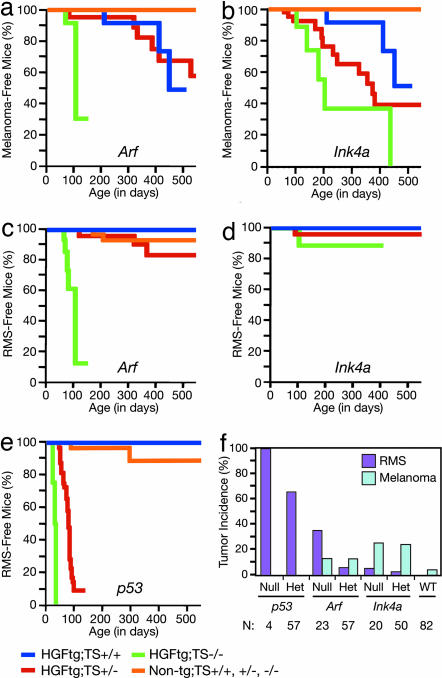
The genetic and molecular pliability of the mouse make it an exceptional model system to dissect the complex genetic principles of human cancer. Previously, we demonstrated that transgenic mice expressing the Met ligand hepatocyte growth factor/scatter factor (HGF/SF) from the metallothionein promoter were prone to both embryonal rhabdomyosarcoma (RMS) and UV-induced cutaneous malignant melanoma that developed in stages highly reminiscent of their human counterparts (8, 9). The risk of developing these two malignancies was significantly enhanced when HGF/SF-transgenic mice were placed on a genetic background in which the Ink4a-Arf locus was genetically inactivated (10, 11). To elaborate the specific roles of these tumor suppressors in oncogenesis, the HGF/SF transgene was bred onto a background deficient in either Arf or Ink4a alone. After neonatal UV irradiation, HGF/SF-transgenic mice devoid of either Arf or Ink4a exhibited premalignant melanocytic lesions by as early as 2 months of age and developed cutaneous melanoma with a significantly reduced mean onset age of ≈3.5 months and 7 months, respectively (Fig. 1 a and b). The mean onset age for melanoma in HGF/SF-transgenic Ink4a+/− mice fell between the values for the Ink4a+/+ and Ink4a−/− mice, and the survival of the heterozygotes was significantly reduced relative to Ink4a+/+ mice. Somewhat unexpectedly, the mean onset age for HGF/SF-transgenic Arf+/− mice was not significantly different from Arf+/+ controls (Fig. 1a). Our mouse model therefore readily confirmed the importance of these tumor suppressors in melanoma (12, 13).
Fig. 1.
Consequences of the loss of Arf, Ink4a, and p53 on development of melanoma and RMS in HGF/SF-transgenic mice. (a) Melanoma Kaplan–Meier (K–M) survival analysis of UV-irradiated Arf-deficient mice. Key is below the figure: HGF/SF-transgenic (HGFtg), nontransgenic (Non-tg), tumor suppressor (TS), wild type (+/+), heterozygote (+/−), or homozygote (−/−). Differences between Arf homozygote and heterozygote, and Arf homozygote and wild type were statistically significant (P = 0.0053 and P < 0.0001, respectively; Tarone–Ware). (b) Melanoma K–M survival analysis of UV-irradiated Ink4a-deficient mice. Differences between Ink4a wild type and heterozygote, and Ink4a wild type and homozygote were statistically significant (P = 0.023 and P = 0.0006, respectively; Tarone–Ware). (c) RMS K–M survival analysis of Arf-deficient mice. (d) RMS K–M survival analysis of Ink4a-deficient mice. (e) RMS K–M survival analysis of p53-deficient mice. (f) Lifetime incidence of RMS and melanoma in HGF/SF-transgenic mouse population with loss of various tumor suppressors. Number (N) of mice in each group is below the graph.
RMS was very common in HGF/SF-transgenic Arf−/− mice, irrespective of UV irradiation, with a mean age of onset of ≈3.7 months (Fig. 1c), and, again, the loss of one Arf allele did not significantly affect RMS genesis. RMS was relatively rare in HGF/SF-transgenic Ink4a−/− mice (Fig. 1d), suggesting that the Arf-p53 axis plays a critical role in suppression of embryonal RMS. Indeed, this hypothesis was confirmed by generating HGF/SF p53 mutant cohorts, showing that Tp53−/− and Tp53+/− mice rapidly developed RMS (Fig. 1e). Additionally, 14 of 14 RMS tumors arising in Tp53+/− mice sustained loss of the wild-type Tp53 locus (data not shown). Nonetheless, the Rb pathway was typically disrupted in RMS arising in Tp53+/− mice, through either decreased pRb or Ink4a expression or overexpression of Cdk4 or Cdk6 (data not shown). These results suggest that, although p53 pathway disruption is a preferential, critical early step in RMS genesis, Rb pathway disruption still plays an important role later in progression.
Remarkably, HGF/SF-transgenic mice in which either one or both Tp53 alleles were inactivated failed to develop melanocytic lesions of any kind, while succumbing to RMS by a mean age of onset of 2.7 or 1.4 months, respectively (summarized in Fig. 1f). This was in clear contrast to HGF/SF-transgenic mice deficient in Arf, which experienced the same risk of developing either melanoma or RMS with a mean onset age of ≈3.6 months (Fig. 1 a and c). The age at which the Arf-deficient mice developed melanoma overlapped with the life expectancy of the Tp53+/− mice: for example, one Arf−/− mouse died at 72 days bearing a melanoma of 120 mm3 as its only tumor. Based on the fact that melanoma developed in p53-deficient mice expressing mutant HRAS with a mean age of onset of >6 months (14; L. Chin and N. Bardeesy, personal communication), it is very likely that melanomas would have eventually developed in the HGF/SF-transgenic p53-deficient mice had they not succumbed to RMS, but at an age much later than the Arf-deficient mice. To overcome the short lifespan of our p53-deficient cohorts, we crossed the HGF/SF transgene onto a C57BL/6 background, which resulted in more viable and long-lived transgenic mice (HGF/SF-BL6); p53-deficient HGF/SF-BL6 mice live 50% longer than their FVB/N counterparts. Of six such p53-deficient HGF/SF-BL6 mice living an average age of 113 days, only one (16.7%) developed a small 2-mm3 melanoma, and no melanocytic lesions developed in the only available HGF/SF-BL6 Tp53−/− mouse, which lived 129 days. Similarly, 15% of HGF/SF-BL6 mice with wild-type p53 developed melanoma at 113 days (n = 24). Together, these data emphasize the relative importance of Arf in melanocytes and raise the intriguing possibility that Arf possesses melanoma tumor suppressor activity that is independent of p53. This notion is supported by our earlier work showing that melanomas in the UV-irradiated HGF/SF-transgenic mouse often experience loss of exon 2 of the Ink4a-Arf locus, but rarely harbored Tp53 mutations (11).
What ARF function then is critical for the suppression of melanoma? An important process in which both RB and p53 pathways have been implicated is cellular senescence, thought to constitute a physiological response to a variety of stressors long evident in cultured cells (15, 16). Replicative senescence is triggered by telomere exhaustion; however, senescence can be prematurely induced by the expression of oncogenes such as mutant BRAF and NRAS (17), commonly found in human nevi and melanoma (1). It is now well appreciated that senescence represents a potent anticancer mechanism, although the genetic wiring of the response appears to be context dependent (17). In benign nevi, which consist of senescent melanocytes, oncogene-induced senescence represents a critical block of progression to melanoma (5, 17, 18). INK4A has been implicated in melanocyte senescence (5–7); however, recent evidence suggests that INK4A alone may not be sufficient, and another collaborating factor or factors may be required to execute the melanocyte senescence program (17, 18).
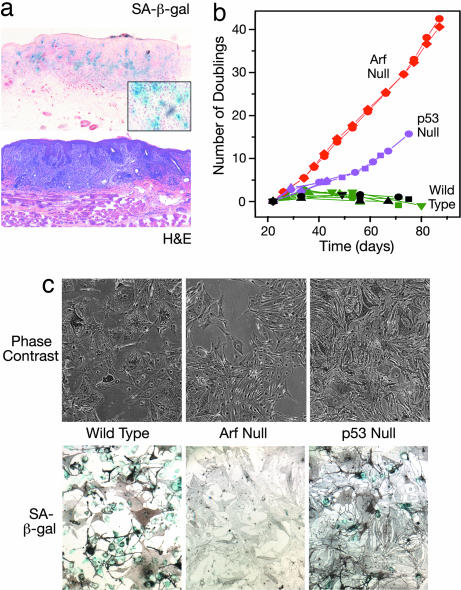
We identified a senescent subset of lesional cells in early melanomas arising from our UV-irradiated HGF/SF-transgenic mice, as evidenced by positive staining with senescence-associated acidic β-galactosidase (SA-β-gal) (Fig. 2a). Based on our in vivo data, we hypothesized that, unlike senescence in the well studied fibroblast model system Arf, but not p53, may play a key role in the regulation of melanocyte senescence. To test this notion, primary melanocytes were isolated from the neonatal skin of mice deficient in Arf, p53, or Ink4a. Fig. 2b shows that, although wild-type melanocytes senesced stably at ≈4 weeks, cultured primary Arf−/− melanocytes showed no detectable growth arrest over 3 months. Cultured Ink4a−/− melanocytes could also grow without signs of senescing but required the presence of survival factors such as endothelin-1 and stem cell factor [supporting information (SI) Fig. 6a]. In contrast, Tp53−/− melanocytes on the same genetic background initially exhibited slowed proliferation and the large size, hyperpigmentation, and positive SA-β-gal staining indicative of the senescence observed in wild-type melanocytes (Fig. 2c and SI Fig. 6b). After ≈50 days in culture, some p53-deficient melanocytes began to recover, eventually growing at a rate similar to that of Arf-null melanocytes (Fig. 2b). Notably, successful establishment of a Tp53−/− melanocyte cell line (melan-p53–1) coincided initially with full suppression of Ink4a expression (SI Fig. 7a). Subsequently, in later passages, melan-p53–1 exhibited enhanced growth associated with diminished expression of Arf (SI Fig. 7 b and c). These data substantiate the idea that loss of p53 was insufficient to permit melanocytes to escape senescence. Additional support for a reduced role for p53 in senescence in mouse melanocytes comes from mice designed to express a hyperactive form of p53, which exhibit accelerated cellular aging in various tissues but normal pigmentation of hair and skin (19). In human melanocytes, p53 appears to contribute only subtly to replicative senescence (7, 20), suggesting that the senescence molecular wiring may not be very different between mouse and human melanocytes.
Fig. 2.
Loss of Arf function bypasses senescence in primary mouse melanocytes. (a) SA-β-gal staining in an early cutaneous melanoma arising in a UV-irradiated HGF/SF-transgenic mouse. (Inset) Higher magnification of blue-stained cells. (b) Growth analyses of primary mouse melanocytes isolated and cultured from wild-type, Arf-deficient, and p53-deficient neonatal mouse skins. Each curve represents a separate primary line from one mouse. For each data point (i.e., each passage), melanocytes were collected from triplicate 3-cm dishes by trypsinization, each suspension was counted in triplicate, and cells were replated at 3 × 104 cells per ml. The mean relative population increase was calculated and converted to number of population doublings. SEMs are no bigger than the symbols, and no more than 7% of the mean. (c) Microscopic image of melanocytes by using phase contrast (Upper) and bright field microscopy after staining for SA-β-gal (Lower). Blue indicates senescent cells; stain is masked by melanin in some cells.
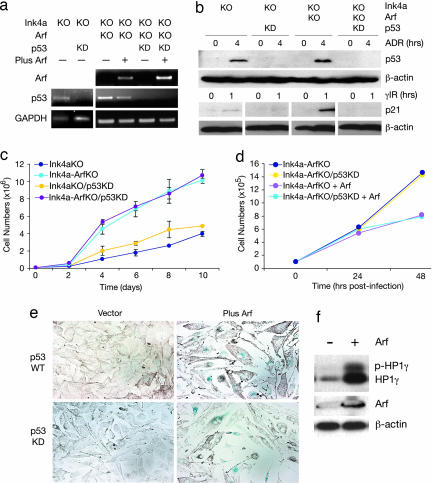
Cultures of early passage mouse melanocytes were then used to study p53-independent Arf activity in melanocytes. To avoid the rapid senescence that characterizes cultured wild-type melanocytes, subsequent experiments were performed with melanocytes whose growth was enabled by germ-line deficiency of Ink4a. This is a relevant genetic context for studying the consequences of Arf deficiency because, in human melanoma genesis, ARF is frequently lost in conjunction with loss of INK4A. Expression of p53 was knocked down in the presence and absence of Arf by using a retroviral vector containing a p53-specific shRNA-mir (LUMP-p53.1224) (21). This retroviral vector efficiently knocked down p53 RNA and protein levels (Fig. 3 a and b) and blocked its ability to induce p21 expression after γ-irradiation (Fig. 3b). The following melanocyte lines were used: Ink4a−/−, Ink4a-Arf−/−, Ink4a−/− with p53 knockdown, and Ink4a-Arf−/− with p53 knockdown. Growth analysis of these four cell lines in vitro showed that melanocytes deficient in Arf exhibited a strong growth advantage relative to those carrying wild-type Arf, a phenotypic difference not overtly affected by the functional status of p53 (Fig. 3c). This result was confirmed in melanocyte line melan-p53–1, derived from Tp53−/− mice (SI Fig. 7c). These data indicate that Arf regulates melanocyte proliferation in a p53-independent manner.
Fig. 3.
Proliferation of mouse melanocytes is regulated by Arf but not p53. (a) To avoid studying senescence-prone melanocytes, all cells were genetically Ink4a-deficient. p53 expression was stably knocked down (KD) in Ink4a−/− and Ink4a-Arf−/− (KO) melanocytes by infection with retrovirus LUMP-p53.1224, a shRNA-mir (21). Arf was reexpressed in Ink4a-Arf−/− melanocytes by infection with a pBabe retrovirus containing Arf cDNA (+) (23). Melanocytes are wild type for a given allele unless otherwise indicated. Expression of Arf (top blot) and p53 (middle blot) RNA was assessed by RT-PCR; GAPDH RNA expression was used as control (bottom blot). (b) p53 activity was blocked by expression of the shRNA-mir. Ink4a−/−, Ink4a−/− with p53KD, Ink4a-Arf−/−, and Ink4a-Arf−/− with p53KD melanocytes were treated with either 0 or 1 μM ADR (upper set of blots), or 0 or 20 Gy γ-IR (lower set of blots). Expression of p53 and p21 was detected by immunoblotting and β-actin antibody used to confirm equal loading. (c) Growth analysis of established p53KD melanocytes. Ink4a−/−, Ink4a−/− with p53KD, Ink4a-Arf−/−, and Ink4a-Arf−/− with p53KD melanocytes were seeded at 1 × 105 cells per dish, and cell numbers were counted between 2 and 10 days. Data points represent means ± SE of three independent experiments. Growth of all cells expressing Arf was significantly different from cells without Arf (P < 0.001). (d) Ink4a-Arf−/− and Ink4a-Arf−/− with p53KD melanocytes were infected with the pBabe retrovirus containing Arf cDNA or empty vector, as shown in a, and the number of cells were counted at 24 and 48 h after infection. (e) Cells were stained with SA-β-gal at 48 h after retrovirus infection. Blue indicates senescent cells, as in 2c. (f) Overexpression of Arf induces phosphorylated HP1γ. The Tp53−/− melanocyte line melan-p53–1 was infected with pBabe retrovirus containing Arf cDNA or empty vector. Forty-eight hours after infection, HP1γ was analyzed by immunoblotting (22). Total and phosphorylated HP1γ are indicated, as are Arf and a β-actin loading control.
To further evaluate the role of Arf in melanocyte senescence, a retroviral vector was used to restore expression of Arf in Ink4a-Arf−/− melanocytes, with and without p53 knockdown (Fig. 3a). Forty-eight hours after infection, Arf expression inhibited cell proliferation (Fig. 3d) and induced morphological senescence as evidenced by SA-β-gal staining, irrespective of the presence of functional p53 (Fig. 3e). This result was confirmed in Arf-infected melan-p53–1 melanocytes (SI Fig. 8a). Moreover, the ability of Arf to induce senescence in this Tp53−/− cell line was corroborated through analysis of phosphorylation of heterochromatin protein-1γ (HP1γ) (Fig. 3f) and its association with prominent senescence-associated heterochromatin foci (SI Fig. 8b) (22). In contrast, Arf reexpression in Ink4a-Arf−/− melanocytes did not induce an overt apoptotic response over this time period, irrespective of p53 expression (SI Fig. 9a).
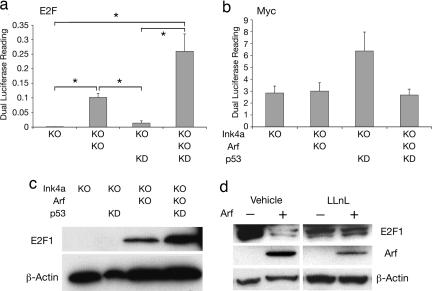
To determine possible targets of p53-independent Arf control over melanocyte cell proliferation, luciferase reporter assays were used. Although found to be a target of Arf regulation in other cell types (4), Myc activity was not significantly affected by the loss of Arf in melanocytes (Fig. 4b). E2F1 has also been reported to be a direct, p53-independent target of ARF (4, 23). We discovered that E2F activity was significantly elevated in melanocytes devoid of Arf (Fig. 4a). Knockdown of p53 had no overt effect on E2F activity by itself but did enhance the activity achieved through loss of Arf alone, although this change did not reach significance. Subsequent analysis revealed that levels of E2F1 were greatly enhanced in Arf-deficient melanocytes relative to wild type (Fig. 4c). The consequence of p53 loss mirrored the luciferase reporter assay in that no effect was observed unless Arf expression was also lost (Fig. 4c). To further investigate the mechanism of Arf-regulated E2F1 expression, Arf was reexpressed in Arf-deficient melanocytes in the presence and absence of a proteasomal inhibitor. Arf reexpression decreased the level of E2F1; however, this effect was abrogated in the presence of the proteasomal inhibitor, LLnL (Fig. 4d). These data demonstrate that Arf down-regulates melanocyte E2F1 levels through a proteasomal-degradation mechanism that could account, at least in part, for its ability to trigger melanocyte growth arrest. Importantly, E2F activity has been associated with senescence in mouse and human melanocytes and has been implicated in human melanoma (24–26). However, because Arf has many binding partners and other activities (4), additional mechanisms may be involved in Arf-mediated senescence.
Fig. 4.
E2F1 level and activity are significantly enhanced upon inactivation of Arf. (a) Ink4a−/−, Ink4a−/− with p53KD, Ink4a-Arf−/−, and Ink4a-Arf−/− with p53KD melanocytes were transfected with a luciferase reporter plasmid for E2F activity (E2F-luc). Luciferase activity, normalized to readings from a cotransfected pRL-TK plasmid, was measured after 16 h. Data represent means ± SE of three independent experiments of three replicates each. Asterisks indicate a statistically significant difference between samples (P < 0.05). (b) Same as a, but here, a reporter for Myc activity was used (Myc-luc). (c) Western blot showing expression of E2F1 in Ink4a−/−, Ink4a−/− with p53KD, Ink4a-Arf−/−, and Ink4a-Arf−/− with p53KD melanocytes (upper blot). β-Actin was used as control for protein loading (lower blot). (d) Arf can induce E2F1 degradation. Arf was reexpressed in Ink4a-Arf−/− melanocytes by infection with a retrovirus containing Arf cDNA. Melanocytes were treated with proteasomal inhibitor 50 μM LLnL or vehicle for 16 h at 24 h after infection. Western blots depict expression of E2F1 (top blot), Arf (middle blot), and β-actin as loading control (bottom blot).
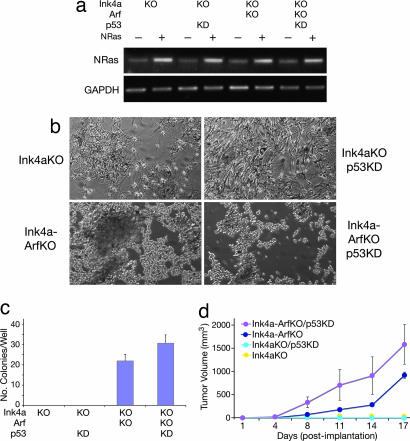
Having demonstrated the importance of Arf in the regulation of melanocyte senescence, we next determined whether Arf was a barrier to melanocyte transformation by NRAS, frequently mutated in human melanoma (1). We again used the set of four mouse melanocyte lines growth-enabled through deficiency of Ink4a; all cells were subjected to infection by a retrovirus encoding mutant NRAS. Enhanced expression of NRAS was confirmed by RT-PCR (Fig. 5a). We found that, in the context of NRAS activation (and Ink4a-deficiency), loss of Arf, but not p53, was sufficient to confer both a transformed morphology (Fig. 5b) and the capability for anchorage-independent growth (Fig. 5c and SI Fig. 9b). This result was confirmed in melan-p53–1 melanocytes, which were not transformed by NRAS (SI Fig. 8c). The role of Arf in melanocyte transformation was further characterized by in vivo growth analysis in nude mice. The results showed that, in concert with mutant NRAS, the loss of Arf, but not p53 alone, allowed melanocytes to form tumors readily in vivo (Fig. 5d). Interestingly, we did note an effect of p53 loss with respect to E2F activity, soft agar growth, and tumorigenesis, but only in melanocytes already deficient in Arf, reinforcing the concept that Arf and p53 possess discrete functions in melanocytes and melanoma. These data corroborate our initial findings in the transgenic mouse model and support a prominent, distinct role for ARF and a muted or delayed role for p53 in suppressing melanoma.
Fig. 5.
NRAS collaborates with Arf deficiency to transform melanocytes. (a) Ink4a−/−, Ink4a−/− with p53KD, Ink4a-Arf−/−, and Ink4a-Arf−/− with p53KD melanocytes were infected with vector LZRS containing mutant NRAS (G12V) (36). RT-PCR demonstrated NRAS expression levels with and without infection (upper blot). GAPDH served as a control (lower blot). (b) Microscopic image of the phenotypic consequences of mutant NRAS expression in melanocytes. All images show NRAS-infected cells. (c) Quantification of the ability of melanocytes to grow under anchorage-independent conditions after NRAS infection (in colonies per well). All experiments were in triplicate; bars represent mean ± SE of three wells. P = 0.057 for Ink4a/Arf−/− with and without p53KD. (d) In vivo tumor growth analysis of NRAS-transformed melanocytes. NRAS-transformed melanocytes were collected and s.c. inoculated at 0.5 × 106 cells per injection site per nude mouse. Tumor volume was measured every 3 days after injection. Data points represent mean tumor volume ±SE (error bars for some points are too small to see); five mice in each group.
Melanocytes are highly specialized cells built to absorb and survive significant stress associated with melanin production and UV exposure and can be intermittently induced into temporary states of active proliferation. Here, we show that, unlike fibroblasts, where p53 inactivation is sufficient to prevent fibroblast senescence and ARF expression can only trigger fibroblast growth arrest and senescence in the presence of functional p53 (15, 27–30), Arf can regulate melanocyte senescence in a manner independent of p53 and distinct from Ink4a. Loss of Arf in melanocytes results in a significant growth advantage relative to p53 or Ink4a deficiency, whereas restoration of Arf induces cellular growth arrest, senescent morphological alterations, SA-β-gal expression, and phosphorylation of HP1γ, irrespective of p53 status. Moreover, data derived from transgenic mice and NRAS-transformed melanocytes validate the relevance in melanoma genesis of loss of ARF vis-à-vis p53.
If indeed the molecular wiring of senescence is sufficiently similar in mouse and human, then the ability of ARF to regulate melanocyte senescence and influence melanoma genesis in a p53-independent manner has clinical significance. Deletions at the CDKN2A locus are currently the second most frequent genetic event in cancer, second only to TP53 alterations. ARF/p53 pathway members would seem then to comprise very promising candidates for broadly effective targeted therapy. Development of novel, rational therapies based on p53 is ongoing and has focused mainly on targeting MDM2/p53 interactions and the use of small molecules to alter the conformation of mutant p53 to restore wild-type function and not on ARF (31). However, our data demonstrate that the function of ARF and p53 in melanocytes and their roles in melanoma are not redundant. Saadatmandi et al. (32) showed that ARF expression can suppress the growth and viability of a variety of human tumor cells both with and without functional p53. In the future, it may be prudent to determine the presence and influence of alterations in ARF separately from those in p53 for individual melanoma cases and stratify patients for treatment accordingly.
Materials and Methods
Details of some materials and methods are available in the SI Text.
Genetically Engineered Mice.
Mice carrying a HGF/SF transgene whose expression is regulated by a metallothionein gene promoter and flanking regions were made as described (8). Mice carrying specific inactivating mutations in Arf or Ink4a were generated as described (33). Tp53−/− mice (34) were obtained from The Jackson Laboratories (Bar Harbor, ME). Unless otherwise indicated, all mice were on a genetic background consisting of ≈90% FVB/N:10% C57BL/6. All melanomas were initiated by a single dose of UV radiation at 3.5 days of age as described (9). RMS development was unaffected by UV irradiation, so UV irradiated and untreated cohorts were combined. Primers used for genotyping are listed in SI Table 1. All mouse work was performed with the approval of the National Cancer Institute Animal Care and Use Committee, in accordance with American Association for the Accreditation of Laboratory Animal Care guidelines and policies established by the National Institutes of Health.
Melanocyte Derivation, Culture, and Growth Curves.
All melanocytes were derived from mouse stocks on a C57BL/6J background, including: Arf−/− mice [a gift from F. Zindy and C. Sherr (St. Jude Children's Research Hospital)], Ink4a−/− mice (Ink4a*/*) (12) and Tp53−/− mice (34) (The Jackson Laboratories). Primary melanocyte cultures were prepared from neonatal skins as described by using a feeder layer of mitomycin-treated immortal murine XB2 keratinocytes for the first two passages only (35). Cultures were grown at 37°C in RPMI medium 1640 with 10% FBS, 200 nM 12-O-tetradecanoyl phorbol 13-acetate, and 200 pM cholera toxin (35), hereafter referred to as complete melanocyte growth medium (CMGM). They were gassed with 10% CO2 in humidified air, final pH 6.9–7.0. Ink4a-deficient cultures were additionally supplemented with 10 ng/ml recombinant mouse stem cell factor (SCF; R & D Systems, Minneapolis, MN) and 10 nM endothelin-1 (Sigma–Aldrich, St. Louis, MO). Medium was changed twice weekly.
Long-term growth curves were generated from several independent primary lines (each from one skin) per genotype. Number of lines generated: three for Tp53−/−, two for Arf−/−, four for Ink4a−/−, and eight for wild-type melanocytes. Cells were collected from triplicate 3-cm dishes per line by trypsinization and each suspension counted in triplicate by hemocytometer and replated at 3 × 104 cells per ml, 2 ml per dish. The mean relative population increase was calculated and converted to number of population doublings.
For other experiments, melanocytes were grown as above except with 5% CO2, the pH being adjusted with HCl to reach pH 7.1–7.2 on incubation. Medium was replaced every 48 h. For short-term growth curves, Ink4a−/−, Ink4a−/− with p53 knockdown, Ink4a-Arf−/−, Ink4a-Arf−/− with p53 knockdown and melan-p53–1 melanocytes were plated at 1 × 105 cells per 10-cm dish. Cells were collected by trypsinization and counted by using a Coulter counter (Beckman, Fullerton, CA) every 2 days. The experiment was performed three times with triplicate dishes.
Adriamycin (ADR) (Sigma–Aldrich) was dissolved in deionized H2O and sterilized by passage through a 0.2-μm filter before use. Cells were treated with a final concentration of 1 μM ADR for 4 h in CMGM. Twenty grays of γ-irradiation was used for p21 induction. Cells were incubated at 37°C for 1 h after irradiation. LLnL (Sigma–Aldrich) was dissolved in 95% ethanol. Cells were treated with a final concentration of 50 μM LLnL for 16 h in CMGM. Cells were rinsed with PBS and collected for analysis after individual treatment.
Vectors and Generation of Modified Cell Lines.
Melanocytes with p53 knockdown were generated by infection with the retrovirus LUMP-p53.1224, which contains a p53-specific shRNA-mir (21). Briefly, EcoPack 293 cells were transfected with 10 μg of DNA, and viral supernatant was collected after 2 days and used to infect melanocytes. Cells were selected in 1.5 μg/ml puromycin. Expression of Arf and NRAS in cultured melanocytes was carried out by infection with the retrovirus pBabe containing Arf or the vector LZRS containing NRAS (G12V) (36), respectively, by using the same procedure. The Arf cDNA retrovirus was constructed as described (23), and the pBabe vector was used as a control.
SA-β-Gal Staining.
SA-β-gal staining was performed by using a senescence-detection kit (Biovision Research Products, Mountain View, CA) according to the manufacturer's instructions. Briefly, mouse melanocytes were fixed 48 h after infection with Arf retrovirus or control and incubated overnight at 37°C with staining solution. For tumor samples, fresh-frozen tissues were fixed in formaldehyde/glutaraldehyde for 10 min at room temperature. Slides were rinsed with PBS and incubated in staining solution [1 mg/ml β-galactosidase, 40 mM citric acid/sodium phosphate (pH 6.0), 5 mM potassium ferrocyanide, 0.15 M sodium chloride, and 2 mM magnesium chloride] at 31°C overnight. Slides were thoroughly rinsed and counterstained with 0.1% neutral red. For primary melanocytes, staining was performed between passages 3–5.
Statistical Analysis.
The differences in growth in vitro and in vivo among the four cell lines were tested by using the analysis of covariance (ANCOVA). In the model, time was included as a covariate to reflect cell proliferation or tumor growth. Analysis was done by using SAS PROC GLM. For Kaplan–Meier survival curves, the Tarone–Ware method was used, and for analysis of luciferase activity, Student's t test was used.
Supplementary Material
Acknowledgments
We acknowledge Dr. Paul Khavari (Stanford University, Stanford, CA) for the NRAS retroviral vector, Dr. Huei-Min Lin (National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD) for E2F-luciferase and Myc-luciferase reporters, and Drs. Frederique Zindy and Charles Sherr for Arf KO mouse skins. We thank Dr. Frances Noonan (George Washington University, Washington, DC) for stimulating discussions and useful advice, Cari Graff-Cherry for superb animal care and technical support, and Dr. Binbing Yu for assistance with statistical analyses. This work was supported in part by the Intramural Research Program of the National Cancer Institute (NCI), National Institutes of Health, in part by NCI Contract N01-CO-12400, and in part by Wellcome Trust Program Grants 064583 and 078327 (to E.V.S.).
Abbreviations
- CMGM
complete melanocyte growth medium
- HGF/SF
hepatocyte growth factor/scatter factor
- HP1γ
heterochromatin protein-1γ
- RMS
rhabdomyosarcoma
- SA-β-gal
senescence-associated acidic β-galactosidase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611638104/DC1.
References
- 1.Chin L, Garraway LA, Fisher DE. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 2.Hussein MR, Haemel AK, Wood GS. Eur J Cancer Prev. 2003;12:93–100. doi: 10.1097/00008469-200304000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Soussi T, Kato S, Levy PP, Ishioka C. Hum Mutat. 2005;25:6–17. doi: 10.1002/humu.20114. [DOI] [PubMed] [Google Scholar]
- 4.Sherr CJ. Nat Rev Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 5.Gray-Schopfer VC, Cheong SC, Chong H, Chow J, Moss T, Abdel-Malek ZA, Marais R, Wynford-Thomas D, Bennett DC. Br J Cancer. 2006;95:496–505. doi: 10.1038/sj.bjc.6603283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sviderskaya EV, Hill SP, Evans-Whipp TJ, Chin L, Orlow SJ, Easty DJ, Cheong SC, Beach D, DePinho RA, Bennett DC. J Natl Cancer Inst. 2002;94:446–454. doi: 10.1093/jnci/94.6.446. [DOI] [PubMed] [Google Scholar]
- 7.Sviderskaya EV, Gray-Schopfer VC, Hill SP, Smit NP, Evans-Whipp TJ, Bond J, Hill L, Bataille V, Peters G, Kipling D, et al. J Natl Cancer Inst. 2003;95:723–732. doi: 10.1093/jnci/95.10.723. [DOI] [PubMed] [Google Scholar]
- 8.Takayama H, LaRochelle WJ, Sharp R, Otsuka T, Kriebel P, Anver M, Aaronson SA, Merlino G. Proc Natl Acad Sci USA. 1997;94:701–706. doi: 10.1073/pnas.94.2.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noonan FP, Recio JA, Takayama H, Duray P, Anver MR, Rush WL, De Fabo EC, Merlino G. Nature. 2001;413:271–272. doi: 10.1038/35095108. [DOI] [PubMed] [Google Scholar]
- 10.Sharp R, Recio JA, Jhappan C, Otsuka T, Liu S, Yu Y, Liu W, Anver M, Navid F, Helman LJ, et al. Nat Med. 2002;8:1276–1280. doi: 10.1038/nm787. [DOI] [PubMed] [Google Scholar]
- 11.Recio JA, Noonan FP, Takayama H, Anver MR, Duray P, Rush WL, Lindner G, De Fabo EC, DePinho RA, Merlino G. Cancer Res. 2002;62:6724–6730. [PubMed] [Google Scholar]
- 12.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 13.Sharpless NE, Kannan K, Xu J, Bosenberg MW, Chin L. Oncogene. 2003;22:5055–5059. doi: 10.1038/sj.onc.1206809. [DOI] [PubMed] [Google Scholar]
- 14.Bardeesy N, Bastian BC, Hezel A, Pinkel D, DePinho RA, Chin L. Mol Cell Biol. 2001;21:2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben-Porath I, Weinberg RA. Int J Biochem Cell Biol. 2005;37:961–976. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Sherr CJ, DePinho RA. Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- 17.Mooi WJ, Peeper DS. N Engl J Med. 2006;355:1037–1046. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- 18.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 19.Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, et al. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 20.Bandyopadhyay D, Timchenko N, Suwa T, Hornsby PJ, Campisi J, Medrano EE. Exp Gerontol. 2001;36:1265–1275. doi: 10.1016/s0531-5565(01)00098-5. [DOI] [PubMed] [Google Scholar]
- 21.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 22.Zhang R, Chen W, Adams PD. Mol Cell Biol. 2007;27:2342–2358. doi: 10.1128/MCB.02019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martelli F, Hamilton T, Silver DP, Sharpless NE, Bardeesy N, Rokas M, DePinho RA, Livingston DM, Grossman SR. Proc Natl Acad Sci USA. 2001;98:4455–4460. doi: 10.1073/pnas.081061398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 25.Halaban R. Cancer Metastasis Rev. 2005;24:339–356. doi: 10.1007/s10555-005-1582-z. [DOI] [PubMed] [Google Scholar]
- 26.Nelson MA, Reynolds SH, Rao UN, Goulet AC, Feng Y, Beas A, Honchak B, Averill J, Lowry DT, Senft JR, et al. Cancer Biol Ther. 2006;5:407–412. doi: 10.4161/cbt.5.4.2512. [DOI] [PubMed] [Google Scholar]
- 27.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 28.Dimri GP, Itahana K, Acosta M, Campisi J. Mol Cell Biol. 2000;20:273–285. doi: 10.1128/mcb.20.1.273-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei W, Hemmer RM, Sedivy JM. Mol Cell Biol. 2001;21:6748–6757. doi: 10.1128/MCB.21.20.6748-6757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dirac AM, Bernards R. J Biol Chem. 2003;278:11731–11734. doi: 10.1074/jbc.C300023200. [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Rastinejad F, El-Deiry WS. Cancer Biol Ther. 2003;2:S55–S63. [PubMed] [Google Scholar]
- 32.Saadatmandi N, Tyler T, Huang Y, Haghighi A, Frost G, Borgstrom P, Gjerset RA. Cancer Gene Ther. 2002;9:830–839. doi: 10.1038/sj.cgt.7700505. [DOI] [PubMed] [Google Scholar]
- 33.Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 34.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Butel JS, Bradley A. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 35.Sviderskaya EV, Wakeling WF, Bennett DC. Development (Cambridge, UK) 1995;121:1547–1557. doi: 10.1242/dev.121.5.1547. [DOI] [PubMed] [Google Scholar]
- 36.Chudnovsky Y, Adams AE, Robbins PB, Lin Q, Khavari PA. Nat Genet. 2005;37:745–749. doi: 10.1038/ng1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.