Abstract
MicroRNAs (miRNAs) are noncoding small RNA of ≈22 bases, which suppress expression of target genes through translational block or degradation of a target's transcript. Recent studies uncovered specific miRNA expression profiles in human malignancies. Nevertheless, the mechanisms underlying cancer-specific miRNA expression are largely unknown. miRNA biogenesis consists of a series of steps beginning with generation of a primary transcript, termed pri-miRNA, and continuing into excision of a stem-loop hairpin structure within pri-miRNA by the nuclear RNaseIII enzyme Drosha, transportation to the cytoplasm, and further processing by a second RNaseIII enzyme Dicer, into a 22-base mature duplex RNA. In principle, alteration in any step during this maturation process could affect miRNA production. The ALL-1 (also termed MLL) gene was originally isolated by virtue of its involvement in recurrent chromosome translocations associated with acute leukemias, particularly in infant and therapy-related leukemias. These translocations result in the fusion of ALL-1 with partner genes and the consequent production of chimeric leukemogenic proteins. Here, we identify specific miRNAs up-regulated in leukemias triggered by All1 fusions. Further, we demonstrate coimmunoprecipitation of the All1/Af4 and All1/Af9 fusions with Drosha, disrupted by treatment with DNase I. Finally, we present evidence from ChIP experiments for All1 fusion protein-mediated recruitment of Drosha to target genes encoding miRNAs. Such recruitment may underlie the enhanced expression of the relevant miRNAs.
The ALL-1 gene, also termed MLL, has been cloned from chromosome band 11q23, a site involved in multiple chromosome abnormalities associated with both acute lymphoblastic leukemia (ALL) and acute myeloblastic leukemia (1, 2). The chromosome translocations result in the fusion of the ALL-1 gene with one of >50 different partner genes and the production of leukemogenic proteins composed of the N-terminal All1 sequence and a portion of the partner protein encoded by the segment of the gene positioned 3′ to the breakpoint (1, 2). The most prevalent ALL-1 rearrangement involves recombination with AF4 and production of the All1/Af4 protein. This rearrangement is associated with very poor prognosis in infants and adults (3). The molecular pathways deregulated by All1 fusion proteins are still largely unknown.
MicroRNAs (miRNAs) are short 20- to 22-nt RNAs that negatively regulate gene expression at the posttranscriptional level by base-pairing to the 3′ UTR of target messenger RNAs. More than 400 miRNAs have been identified in human, and they are evolutionarily conserved. It has been shown that miRNAs regulate various physiological and pathological pathways, such as cell differentiation, cell proliferation, and tumorigenesis (reviewed in ref. 4). Extensive expression profiling studies revealed cell-type-specific miRNA fingerprints in B cell chronic lymphocytic leukemia, breast cancer, colon cancer, gastric cancer, glioblastoma, hepatocellular carcinoma, papillary thyroid cancer, and endocrine pancreatic tumors (reviewed in ref. 5). Calin et al. (6) showed that although miRNA genes represent only 1% of the mammalian genome, >50% of miRNA genes are located within regions associated with amplifications, deletions, and translocations in cancer. These somatic mutations of miRNA genes underlie some of the specific miRNA expression patterns in cancer. Additional factors contributing to deregulation of miRNAs are unknown, but could act at the level of transcription or miRNA maturation. miRNA biogenesis begins with a primary transcript, termed pri-miRNA, which is generated by RNA polymerase II (reviewed in ref. 7). Within the pri-miRNA, the miRNA itself is contained within ≈60–80 nt that can fold back on itself to form a stem–loop hairpin structure. This hairpin structure is recognized and excised from pri-miRNA by the microprocessor complex composed of the nuclear RNase III enzyme, Drosha, and its binding partner DGCR8. The excised miRNA hairpin, referred to as pre-miRNA, is transported to the cytoplasm in association with RAN-GTP and Exportin 5, where it is further processed by a second RNase III enzyme, Dicer, which releases a 22-nt mature duplex RNA with 5′ phosphate and 2-nt 3′ overhang. The antisense RNA strand is incorporated into the RISC complex, which targets it to mRNA(s) by base-pairing. This miRNA interferes with translation of the mRNA or cleaves it.
Here, we present evidence for All1 fusion protein-mediated recruitment of Drosha to target genes encoding specific miRNAs. This recruitment might be the cause for the enhanced expression of the relevant miRNAs.
Results
Identification of miRNAs Deregulated in Leukemic Cell Lines Harboring ALL-1 Rearrangements.
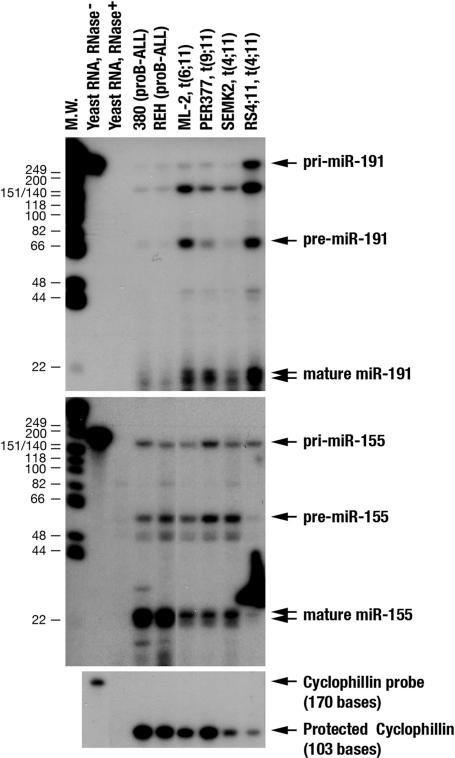
Applying miRNA microarray analysis, we determined the miRNA expression profiles of human leukemic cell lines harboring ALL-1 rearrangements. A total of 18 miRNAs were found to be up-regulated at statistical significance in cell lines with rearranged ALL-1, including SEMK2 and RS4;11 pro-B cells with the t(4;11) and PER377 cells with the t(9;11). Two pro-B cell lines with no ALL-1 abnormalities, 380 and REH, did not show up-regulation (Table 1). Northern analysis supported and expanded these findings [supporting information (SI) Fig. 6]. To confirm and extend some of the results of the microarrays, the expression of miR-191, ranked top in the analysis, and miR-155, which did not show differential expression in lines with ALL-1 gene rearrangements, were determined by applying RNase protection assay (Fig. 1). This assay enabled resolution of the three forms of miRNA (for the identification of protected species corresponding to pri-, pre-, and mature miR-191 and miR-155, see SI Fig. 7). Although miR-191 mature species could hardly be detected in REH and 380 cells, it was abundant in lines expressing All1 fusion proteins, including ML-2 with the t(6;11) chromosome translocation, PER377, SEMK2, and RS4;11 cells. In contrast, mature miR-155 was expressed in 380 and REH cells to considerably higher levels compared with the other cells. This assay also showed that the degree of the pri-miR-191 protection (expression) was similar in all leukemic cells (except for RS4;11) regardless of the expression level of the mature species. This finding suggests that the higher abundance of mature miR-191 in ALL-1-associated leukemias is not caused by overproduction of the pri-miRNA.
Table 1.
Comparison of miRNA expression profiles of cells with and without ALL-1 rearrangement
| miRNA | SAM score* | False discovery rate, %† |
|---|---|---|
| Up-regulated miRNAs | ||
| hsa-mir-191 | 4.84 | 0 |
| hsa-mir-24-1 | 4.42 | 0 |
| hsa-mir-221 | 4.27 | 0 |
| hsa-mir-24-2 | 3.91 | 0 |
| hsa-mir-192 | 3.84 | 0 |
| hsa-mir-222 | 3.75 | 0 |
| hsa-mir-196a-1 | 3.59 | 0 |
| hsa-mir-023b | 3.27 | 0 |
| hsa-mir-146a | 3.26 | 0 |
| hsa-mir-023a | 3.10 | 0 |
| hsa-mir-128b | 2.83 | 0 |
| hsa-mir-128a | 2.69 | 0 |
| hsa-mir-220 | 2.54 | 0 |
| hsa-mir-196b | 2.39 | 0 |
| hsa-mir-223 | 2.26 | 0 |
| hsa-mir-146b | 2.20 | 0 |
| hsa-mir-214 | 1.90 | 2.46 |
| hsa-mir-135a-1 | 1.90 | 2.46 |
| Down-regulated miRNAs | ||
| hsa-mir-125b-1 | −3.86 | 0 |
| hsa-mir-125b-2 | −3.19 | 0 |
| hsa-mir-100 | −2.45 | 2.97 |
miRNA expression profiles were determined in triplicate by probing miRNA-chip with total RNAs from three cell lines expressing All1 fusion protein and two cell lines bearing similar phenotype but lacking ALL-1 abnormalities. Genomic loci of bold-typed miRs have been previously identified as binding sites for normal ALL1 (11).
*SAM identifies genes with statistically significant scores (i.e. paired t tests). Each gene is assigned a score on the basis of its change in gene expression relative to the standard deviation of repeated measurements for that gene. Genes with scores greater than a threshold are deemed potentially significant.
†The percentage of such genes identified by chance is the q value of false discovery rate. miR-155 and 27a, investigated in this article, are not up-regulated in the cell lines with ALL-1 translocations.
Fig. 1.
RNase protection assay for the detection of miR-191 and miR-155 in leukemic cell lines with ALL-1 rearrangements. Twenty micrograms of total RNAs was hybridized overnight at 43°C with in vitro-transcribed antisense probe of miR-191 or miR-155 and subsequently treated with RNase. Protected probe fragments were resolved on a 15% polyacrylamide gel containing 8 M urea. End-labeled φX174/HinfI was used as a molecular weight marker. As a loading control, 10 μg of total RNA was also subjected to hybridization with a Cyclophillin probe.
All1 Fusion Proteins, All1/Af4 and All1/Af9, Physically Interact with Drosha in Vivo.
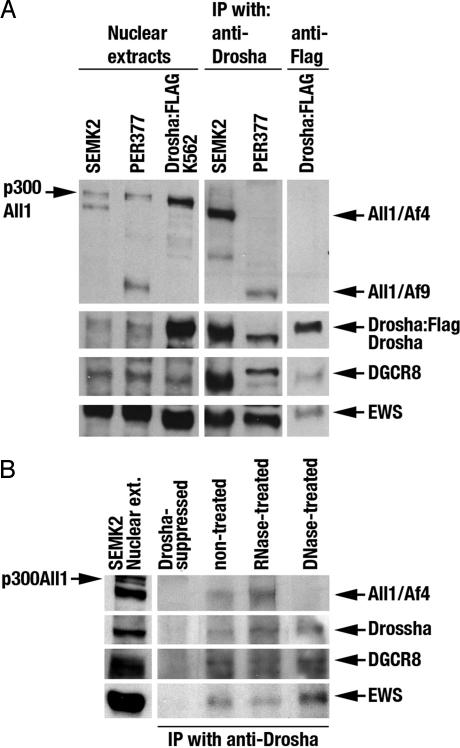
The localization of both Drosha and All1 fusion proteins to the cell nuclei raised the possibility that the latter affects Drosha-mediated miR-191 processing. To test the possibility of physical interaction between Drosha and All1 fusion proteins, we applied co-immunoprecipitation (IP) methodology. It has been previously reported that the exogenously expressed Drosha:FLAG assembles a complex, termed the microprocessor complex (8–10). In addition, Drosha:FLAG was found to assemble a second and larger multiprotein complex of >2 MDa, which contained many RNA binding proteins including EWS (Ewing sarcoma) (10). We used anti-Drosha Ab to precipitate endogenous Drosha produced in SEMK2 and PER377 cell nuclei. In parallel, Drosha:FLAG was precipitated with anti-Flag mAb from whole-cell lysates of transfected K562 cells. Drosha in the immunoprecipitates was eluted by adding an excess amount of the synthetic peptide previously used to generate the Ab. The eluates were subjected to Western blot analysis (Fig. 2) and in vitro cleavage assays to measure processing of miR-191 and miR-155 probes (Fig. 3). The Western blot analysis demonstrated co-IP of two known Drosha-associated proteins, DGCR8 and EWS (Fig. 2A). Strikingly, the fusion proteins All1/Af4 and All1/Af9 coprecipitated with Drosha (Fig. 2A). In contrast, normal p300 All1 did not coprecipitate. Reciprocal IP directed against All1/Af4 using anti-Af4C-terminal Ab failed to coimmunoprecipitate Drosha, although this Ab effectively precipitated the fusion protein (data not shown). The co-IP of the All1 fusion proteins with Drosha is not caused by cross-reaction, because the anti-Drosha Ab did not precipitate All1/Af4 from SEMK2 cells in which the Drosha protein was down-regulated by interference RNA (Fig. 2B). The failure of anti-Af4 Ab to coprecipitate Drosha suggests that only a small portion of All1/Af4 is associated with Drosha or that the association masks the relevant epitope on the Af4 C-terminal region. We next sought to determine whether the association between All1/Af4 and Drosha is RNA- or DNA-dependent. To this end, SEMK2 nuclear extracts were treated extensively with either RNase or DNase and subjected to IP with anti-Drosha Ab. Western blot analysis showed the presence of the All1/Af4, Drosha, DGCR8, and EWS proteins in the immunoprecipitate of RNase-treated nuclear extracts (Fig. 2B). Significantly, DNase treatment abrogated the association of Drosha with All1/Af4, while the association with other proteins was sustained (Fig. 2B). These results suggest that (genomic) DNA is involved in the physical interaction between All1/Af4 and the Drosha complex.
Fig. 2.
Purification of Drosha protein from ALL-1-rearranged leukemic cells by IP with anti-Drosha Ab. (A) Western blot detection of immunoprecipitated proteins. Ab 169 reacting with All1 N-terminal epitope was used for the detection of All1/Af4 and of All1/Af9. For unambiguous identification of Drosha, recombinant Drosha exogenously expressed in K562 cells transfected with pCK-Drosha-Flag plasmid was purified by IP (Drosha:FLAG). Twenty micrograms of nuclear extracts of leukemic cells or ≈2.5 μg of immuno-purified Drosha was used in the analysis. Note that endogenous Drosha was purified from nuclear extracts, whereas Drosha:FLAG was from whole-cell lyzate. (B) Western blot detection of proteins immunoprecipitated with anti-Drosha Ab from SEMK2 nuclear extracts treated either with RNase or DNase.
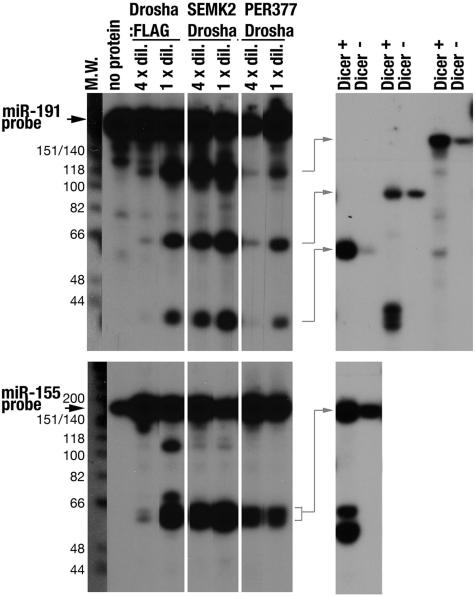
Fig. 3.
In vitro cleavage assays of Drosha immunopurified from plasmid-transfected cells and ALL-1-associated leukemic cell lines. Equal amounts of Drosha, determined by Western analysis (Fig. 2A), were used in all reactions; the corresponding volumes of the nondiluted samples were 10 μl of Drosha:FLAG, 5 μl of SEMK2 Drosha, and 20 μl of PER377 Drosha. (Left) Cleaved products of miR-191 and miR-155 were resolved on denaturing 9% polyacrylamide gel. The cleaved products were excised from the gel, electro-eluted, and subjected to further cleavage with recombinant Dicer enzyme. (Right) Fifteen percent denaturing gel was used to resolve and identify the 22-nt mature products.
The in vitro cleavage assays showed that all Drosha preparations generated three species of miR-191 cleavage products. Of these, the species of ≈66 nt was identified as pre-miR-191 because of its cleavage by recombinant Dicer enzyme (Fig. 3 Upper Right). Similarly, the mixture of miR-155-processed products, surmised to be composed of three species of 55, 59, and 65 nt, was shown to be further cleaved by Dicer, resulting in generation of 22-base products (Fig. 3 Lower Right). These results indicated that the three affinity-purified Drosha preparations were functionally active with both miR-191 and miR-155 templates. Drosha containing All1/Af4 exhibited the strongest processing activity, whereas Drosha containing All1/Af9 had less processing activity, similar to that of the Drosha:FLAG preparation.
All1/Af4-Mediated Drosha Recruitment to miRNA Loci.
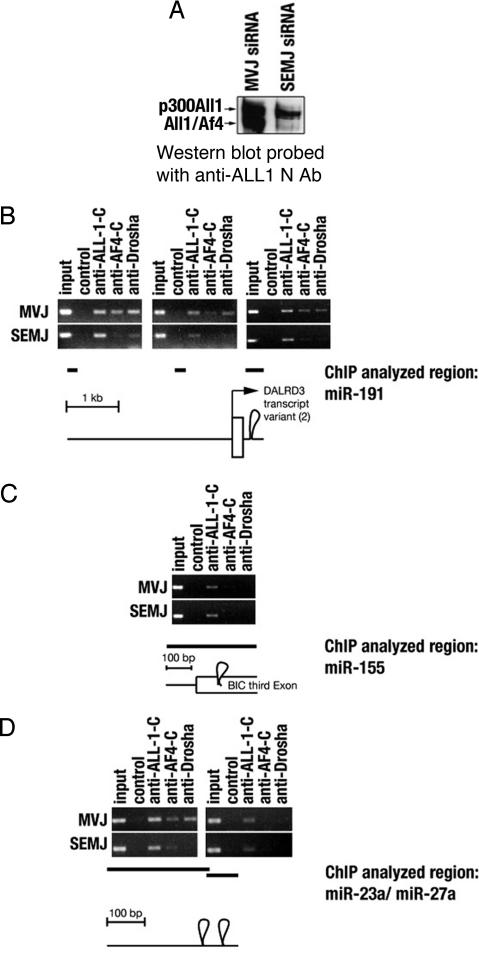
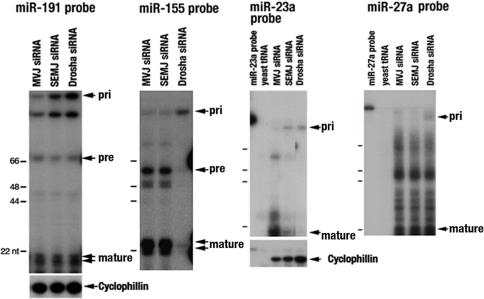
The dependency of the physical interaction between All1/Af4 and Drosha on cellular DNA prompted us to investigate the occupancy of the two proteins on the miR-191 gene. Here, we took advantage of our previous finding that normal All1 binds to DNA regions located 3.5 and 1.5 kb upstream of miR-191 hairpin as well as to the region spanning the hairpin sequence itself (11). ChIP analysis was done on: (i) SEMK2 cells transfected with SEMK2-fusion junction-specific siRNA; the latter down-regulates the All1/Af4 protein at an efficiency of >90%; and (ii) SEMK2 cells expressing siRNA that targets a different ALL-1/AF4 junction and therefore does not affect the level of the fusion protein in the cells (the two siRNAs are referred to as SEMJ and MVJ siRNA, respectively; the amount of All1/Af4 protein in the transfectants is shown in Fig. 4A). The analysis of chromatin of cells containing MVJ siRNA and intact SEMK2 cells (data not shown) showed co-occupancy of normal All1, All1/Af4, and Drosha proteins on the three regions within the miR-191 locus (Fig. 4B). In contrast, no occupancy of All1/Af4 and Drosha on miR-155 hairpin was detected (Fig. 4C). Knockdown of All1/Af4 by treatment with SEMJ siRNA resulted in reduced occupancy of the fusion protein on the three sites within the miR-191 gene and a concurrent loss of Drosha binding (Fig. 4B). This finding suggests All1/Af4-mediated Drosha recruitment onto the miR-191 locus. The investigation was further extended to two additional miRNA loci. The miR-23a and miR-27a genes are aligned in 5′ to 3′ configuration and are spaced by an interval of 84 nt. The expression microarray analysis showed miR-23a, but not miR-27a, to be up-regulated in leukemic cells expressing All1 fusion proteins (Table 1). The protein binding profiles of normal All1, All1/Af4, and Drosha within the miR-23a and 27a regions spanning the hairpin sequences resembled the profiles of miR-191 and miR-155, respectively (Fig. 4D, MVJ). The binding of both All1/Af4 and Drosha to the miR-23a gene is reduced or eliminated in SEMK2 cells knocked out for All1/Af4 (Fig. 4D, SEMJ).
Fig. 4.
All1/Af4-dependent localization of Drosha in miR-191 and miR-23a genomic loci and the effect of All1/Af4 knockdown on Drosha recruitment. (A) Elimination of most of the All1/Af4 protein from SEMK2 cells treated with SEMJ siRNA. (B–D) ChIP analysis for determination of recruitment of normal All1, All1/Af4, and Drosha proteins to genomic loci encoding miR-191, miR-155, miR-23a, and miR-27a. Chromatins tested were from SEMK2 cells treated with the nonfunctional siRNA MVJ or from cells treated with the SEMJ siRNA, which knocks down most of All1/Af4.
All1/Af4 Knockdown Causes Accumulation of Specific Pri-miRNAs.
To investigate the consequence of the reduction in amounts of All1/Af4 and Drosha bound to the genomic regions encoding miR-191 and miR-23a, we determined the expression level of primary and processed RNA products of the loci in comparison to those encoded by the miR-155 and miR-27a genes. The products from SEMK2 cells treated with MVJ siRNA, SEMJ siRNA, or Drosha-specific siRNA were analyzed by RNase protection assay (Fig. 5; for the miR-23a and miR-27a probes, see SI Fig. 8). Both All1/Af4 and Drosha knockdown resulted in accumulation of the primary transcript of miR-191 and miR-23a, indicating impairment of Drosha function by either manipulation. The apparent impairment caused by both knockdown of Drosha and All1/Af4 is reflected in reduced abundance of the 22-base mature miR-23a. In contrast, knockdown of All1/Af4 in cells treated with SEMJ siRNA did not increase the abundance of pri-miR-155 or pri-miR-27a, compared with cells treated with the inert MVJ siRNA (as expected, knockdown of Drosha brought about accumulation of pri-miR-155 and pri-miR-27a). This finding suggests that elimination of All1/Af4 impairs processing of pri-miR-191 and pri-miR-23a, but not of pri-miR-155 or pri-miR-27a.
Fig. 5.
Effect of All1/Af4 knockdown on accumulation of pri miR-191. Abundance of the precursors pri-miR-191, pri-miR-155, pri-miR-23a, and pri-miR-27a, as well as of their processed products, was tested in SEMK2 cells treated with the nonactive MVJ siRNA or knocked down for either All1/Af4 (SEMJ) or Drosha. The RNAs were identified by RNase protection assay. Note that Drosha knockdown increased the abundance of all primary transcripts. In contrast, knockdown of All1/Af4 (SEMJ) was associated with higher abundance of pri-miR-191 and pri-miR-23a.
Discussion
In the present study we identified several miRNAs up-regulated in ALL-1-associated leukemias. Further, we showed that leukemogenic All1 fusion proteins, All1/Af4 and All1/Af9, physically interact with Drosha, the nuclear RNase III enzyme essential for miRNA biogenesis. The notion that alterations in nuclear pri-miRNA processing mediated by Drosha and its associated protein(s) affect miRNA production in vivo originated previously because of some discrepancies between the levels of primary transcripts, precursors, and mature miRNA species. Thus, human embryonic stem cells express measurable amount of the primary transcript encoding let-7a-1 but lack mature species (12). Similarly, the level of miR-155 in diffuse large B cell lymphoma showed only a weak correlation with the level of BIC RNA spanning miR-155 (13). Further, a recent study determining expression of all three molecular forms of let-7g in mouse embryo showed that the mature species is detectable at 10.5 days gestation and is high at 14.5, whereas the primary transcript is highly expressed throughout development (14). Similar discrepancies were also found in several miRNAs known to be associated with mouse development (14). The fact that no accumulation of the precursor species was detected led to the suggestion that the differentiation events occurring during embryonic development activate Drosha processing of specific miRNA. In the same study, Thomson et al. further extended their findings to primary human tumors and provided evidence supporting the Drosha processing block proposed to cause the down-regulation of specific miRNAs (14). Thus, regulation of Drosha activity might have an important physiological role. By applying ChIP analysis and RNase protection assay to leukemic cells expressing All1/Af4, or impaired in this expression because of enforced taking in of siRNA directed against the latter, we could show recruitment of both All1/Af4 and Drosha to specific miRNA genomic loci, correlated with augmentation of processing of the primary transcript. We suggest that the apparent enhanced production of the mature miRNA in cell lines producing All1 fusion proteins could be caused by Drosha binding to the corresponding locus.
It still remains to be determined whether the interaction with Drosha is general for All1 fusion proteins, whether the association between these proteins is direct, and if so if it involves the partner polypeptides. Our findings point to a mechanism by which miRNAs may be regulated and a function for All1 leukemic proteins. It is noteworthy that up-regulation of miR-191 was recently found to be associated with poor prognosis in acute myeloid leukemias (R. Garzon, S. Volinia, C.-G. Liu, C. Fernandez-Cymering, T. Palumbo, F. Pichiorri, M. Fabbri, K. Coombes, H. Alder, T.N., N. Flomenberg, G. Marcucci, G. A. Calin, S. M. Kornblau, C. D. Bloomfield, M. Andreeff, and C.M.C., unpublished work). Up-regulation of miR-191 was also observed in study of six different types of solid tumors, including colon, breast, and lung cancer (15). Further studies are needed to determine the mechanisms associated with miR-191 up-regulation in these tumors.
Materials and Methods
Cell Culture and Antibodies.
Human pro-B cell ALL 380, pre-B cell ALL 697, ML-2 with the t(6;11), and SEMK2 and MV4;11 with the t(4;11) were obtained from DSMZ, Braunschweig, Germany. REH pro-B cell ALL, pro-B cell RS4;11 with the t(4;11), and K562 were purchased from the American Type Culture Collection, Manassas, VA. PER377 with the t(9;11) was obtained from Ursula Kees (University of Western Australia, Perth, Australia). All cell lines were maintained in RPMI medium 1640 supplemented with 10% FBS. Antibodies against Drosha (ab12286), DGCR8 (ab24162), and a Drosha synthetic peptide (ab12307) were purchased from Abcam, Cambridge, MA. Ab against EWS (A300–308A) was made by Bethyl Laboratories, Montgomery, TX. Anti-FLAG M2 mAb and 3× FLAG peptide were obtained from Sigma, St. Louis, MO. Ab 169 directed against ALL-1 N terminus was as described (16). Ab against AF4 C terminus was generated in rabbit by using bacterially synthesized polypeptide spanning AF4 residues 2323–2886.
Microarray Analysis.
Microarray analysis was performed as described (17). Raw data were normalized and analyzed in GeneSpring 7.2 software (Silicon Genetics, Redwood City, CA). Expression data were median-centered by using both the GeneSpring normalization option and the global median normalization of the Bioconductor package (www.bioconductor.org) with similar results. Statistical comparisons were done by using the GeneSpring ANOVA tool and the significance analysis of microarray (SAM) software (www-stat.stanford.edu/∼tibs/SAM/index.html).
miRNA Detection.
RNase Protection assays (RPA) were performed by using the RPA III kit from Ambion, Austin, TX, according to the manufacturer's instructions. Five to 20 μg of total RNA extracted with TRIZOL reagent (Invitrogen, Carlsbad, CA) was used per reaction. Cyclophillin antisense control template was obtained from Ambion and labeled by using T7 RNA polymerase.
Vector Construction and Probe Preparation.
A genomic fragment spanning miR-191 hairpin was prepared by digesting BAC clone RP13–131K19 with PflMI-Bsu36I, blunt-ending, and subcloning into the SmaI site of pGEM-3Z (Promega, Madison, WI) in both orientations. These constructs were linearlized with BamHI and used as templates for generating RNA probes by using the Riboprobe in vitro transcription kit with T7 RNA polymerase (Promega). Probes with sense and antisense orientation were purified on a denaturing gel and used in in vitro cleavage assay and RNase protection assay, respectively. The miR-155 hairpin region, embedded within the third exon of the BIC gene, was PCR-amplified from the human IMAGE cDNA clone 5176657. The forward primer ATGCCTCATCCTCTGAGTGCT tethered with EcoRI site and the reverse primer CTCCCACGGCAGCAATTTGTT tethered with HindIII site, corresponding to nucleotides 261–281 and 401–421 (13) respectively, were used for amplification. Subsequently, the PCR product was cloned into the EcoRI—HindIII sites of the pGEM-3Z vector. Sense and antisense RNA probes were synthesized by using T7 RNA polymerase and SP6 RNA polymerase, respectively. Genomic regions spanning miR-23a and miR-27a hairpin sequences were PCR-amplified as shown in SI Fig. 8 and cloned into the HindIII–EcoRI sites of the pGEM3Z vector. Antisense probes of miR-23a and miR-27a were prepared by digesting the recombinants with NaeI and Bsu36I, respectively, followed by in vitro transcription with Sp6 RNA polymerase.
IP.
K562 cells were transfected with pCK-drosha-flag by using a Nucleofector apparatus according to the manufacturer's instructions (Amaxa, Gaithersburg, MD). Transfected cells (2 × 108) were lysed and subjected to IP with anti-Flag M2 mAb as described (8). Briefly, 25 mg of whole-cell lysate was incubated with 500 μg of mAb after preclearing with protein G Sepharose (GE Healthcare, Piscataway, NJ) at 4°C overnight. Immunocomplex was precipitated with protein G Sepharose and washed, and the Drosha:FLAG in the precipitate was eluted by adding 3× FLAG peptide at a concentration of 0.4 mg/ml in a buffer containing 30 mM Hepes (pH 7.4), 100 mM KCl, 5% glycerol, 0.2 mM EDTA, 7.5 mM MgCl2, and 2 mM DTT. Elution was repeated three times, each time for 30 min at room temperature, and eluates were combined. For IP of endogenous Drosha, 50 mg of nuclear extracts from SEMK2 or PER377 cells prepared by the method of Dignam et al. (18) was subjected to IP with 300 μg of anti-Drosha Ab. The anti-Drosha Ab, purchased from Abcam, was generated in rabbit by immunizing with a synthetic peptide derived from the N-terminal region of Drosha. The peptide is commercially available. Our preliminary study with small-scale IP showed that the addition of excess Drosha peptide to anti-Drosha immunoprecipitate releases Drosha; this procedure enabled purification of the Drosha complex in a native form. The peptide was used for the elution of Drosha at a concentration of 0.4 mg/ml. In some IPs, as shown in Fig. 2B, 250 μg of SEMK2 nuclear extacts was mixed either with 50 μl of DNase-free RNase (Roche, Indianapolis, IN) or 50 units of RQ1 DNase (Promega) and subjected to preclearing with protein A Sepharose (GE Healthcare) at room temperature for 60 min. This process was followed by IP with 10 μg of anti-Drosha Ab.
In Vitro Processing of pri-miRNAs.
In vitro processing assay was done essentially as described (8). Amounts to be added of Drosha:FLAG and two Drosha preparations were determined by measuring the content of Drosha by Western blot analysis. Briefly, 20 μl of reaction mixtures containing immuno-purified Drosha, 7.5 mM MgCl2, 20 units of RNase inhibitor (RNasin, Promega), 2 mM ATP, 2 mM DTT, and 1 × 105 cpm of the labeled probe was incubated at 37°C for 90 min. The reactions were terminated by adding 20 μl of buffer containing 20 mM Tris (pH 8.0), 10 mM EDTA, 1% SDS, and 2 μg of proteinase K (Roche), followed by incubation at 45°C for 30 min. After extraction with phenol/chloroform and chloroform, the processed products were ethanol-precipitated and resolved on a polyacrylamide gel containing 8 M urea.
RNA Interference.
siRNA duplexes targeting ALL-1/AF4 and Drosha mRNAs in SEMK2 cells were transfected by applying Amaxa Nucleofector using kit V and program T-20. Twenty-four hours after transfection, cells were harvested, subjected to a second transfection, and subsequently grown in culture for an additional 48 h. Target sequences of SEMJ siRNA and MVJ siRNA were 5′-AAGAAAAGCAGACCUACUCCA-3′ and 5′-AAGAAAAGGAAAUGACCCATT-3′, respectively. The former siRNA targets ALL-1/AF4 mRNA produced in SEMK2 cells, whereas the latter targets ALL-1/AF4 mRNA produced in MV4;11 cells. Note that the first 8 nt in both siRNAs correspond to ALL-1 mRNA sequence immediately 5′ of the fusion point and are identical, whereas the following 13 nt correspond to AF4 sequences that vary between the fusions and accordingly between the siRNAs; thus, MVJ siRNA will be inactive in SEMK2 cells. The sequence of Drosha siRNA is from ref. 10. The siRNAs were synthesized by Dharmacon, Lafayette, CO.
ChIP Assay.
ChIP assays were performed by using the ChIP assay kit from Upstate Biotechnology (Lake Placid, NY) with minor modifications. Briefly, 5 × 107 formaldehyde-treated SEMK2 cells were lysed in 1 ml of buffer containing 50 mM Hepes (pH 7.4), 140 mM NaCl, 1% Triton X, 0.1% Na-deoxycholate, and 1× Complete protease inhibitor (Roche). A 50-μl aliquot of the preparation was treated to reverse the cross-linking, deproteinized with proteinase K, extracted with phenol chloroform, and determined for DNA concentration. An aliquot of chromatin preparation containing 25 μg DNA was used per ChIP. DNase-free RNase (Roche) was added at a concentration of 200 μg/ml during reverse cross-linking. After deproteiniztion with proteinase K, DNA was purified in 50 μl of Tris-EDTA with a PCR purification kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. A 1-μl aliquot was used for PCR. Primer sequences are listed in SI Table 2.
Supplementary Material
Acknowledgments
We thank V. N. Kim (Seoul National University, Seoul, Korea) for pCK-Drosha-flag plasmid, C. Liu for miRNA chip analysis, S. Volinia for statistical analysis of the chip results, and R. Garzon for Northern blot analysis. This research is supported by National Institutes of Health Grant CA81534-06 and U.S.-Israel Binational Grant 2003223.
Abbreviations
- miRNA
microRNA
- ALL
acute lymphoblastic leukemia
- IP
immunoprecipitation
- EWS
Ewing sarcoma
- SAM
significance analysis of microarray.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704559104/DC1.
References
- 1.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E. Cell. 1992;71:701–709. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 2.Tkachuk DC, Kohler S, Cleary ML. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 3.Johansson B, Moorman AV, Haas OA, Watmore AE, Cheung KL, Swanton S, Secker-Walker LM. Leukemia. 1998;12:779–787. doi: 10.1038/sj.leu.2401012. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamur S, Shimizu M, Rattan S, Bulrich F, Negrini M, et al. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim VN. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 8.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landthaler M, Yalcin A, Tuschl T. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Gregory RL, Yan K-P, Amuthan G, Chendrimada T, Doratotaji B, Cooch N, Shiekhattar R. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 11.Guenther MG, Jenner RG, Chevailer B, Nakamura T, Croce CM, Canaani E, Young RA. Proc Natl Acad Sci USA. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 17.Liu C-G, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru DC, Shimizu M, Zupo S, Dono M, et al. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.