Abstract
Background
Paulinella chromatophora is a freshwater filose amoeba with photosynthetic endosymbionts (chromatophores) of cyanobacterial origin that are closely related to free-living Prochlorococcus and Synechococcus species (PS-clade). Members of the PS-clade of cyanobacteria contain a proteobacterial form 1A RubisCO (ribulose-1,5-bisphosphate carboxylase/oxygenase) that was acquired by horizontal gene transfer (HGT) of a carboxysomal operon. In rDNA-phylogenies, the Paulinella chromatophore diverged basal to the PS-clade, raising the question whether the HGT occurred before or after the split of the chromatophore ancestor.
Results
Phylogenetic analyses of the almost complete rDNA operon with an improved taxon sampling containing most known cyanobacterial lineages recovered the Paulinella chromatophore as sister to the complete PS-clade. The sequence of the complete carboxysomal operon of Paulinella was determined. Analysis of RubisCO large subunit (rbcL) sequences revealed that Paulinella shares the proteobacterial form 1A RubisCO with the PS-clade. The γ-proteobacterium Nitrococcus mobilis was identified as sister of the Paulinella chromatophore and the PS-clade in the RubisCO phylogeny. Gene content and order in the carboxysomal operon correlates well with the RubisCO phylogeny demonstrating that the complete carboxysomal operon was acquired by the common ancestor of the Paulinella chromatophore and the PS-clade through HGT. The carboxysomal operon shows a significantly elevated AT content in Paulinella, which in the rbcL gene is confined to third codon positions. Combined phylogenies using rbcL and the rDNA-operon resulted in a nearly fully resolved tree of the PS-clade.
Conclusion
The HGT of the carboxysomal operon predated the divergence of the chromatophore ancestor from the PS-clade. Following HGT and divergence of the chromatophore ancestor, diversification of the PS-clade into at least three subclades occurred. The γ-proteobacterium Nitrococcus mobilis represents the closest known relative to the donor of the carboxysomal operon. The isolated position of the Paulinella chromatophore in molecular phylogenies as well as its elevated AT content suggests that the Paulinella chromatophore has already undergone typical steps in the reductive evolution of an endosymbiont.
Background
Paulinella chromatophora is a thecate filose amoeba of the Rhizaria that contains a photosynthetic entity of cyanobacterial origin termed chromatophore. A similar process initiated the evolution of plastids likely more than a billion years ago. It has previously been shown that neither the Paulinella host cell nor the chromatophores are related to the eukaryotic lineage containing primary plastids. Instead, the Paulinella chromatophore is affiliated with free-living Prochlorococcus and Synechococcus spp. (PS-clade), and thus represents the product of a second primary endosymbiosis leading to photoautotrophic eukaryotes [1,2]. It is currently debated whether the Paulinella chromatophore represents an organelle comparable to a primary plastid, or merely a stable intracellular symbiont [3-5]. However, the extent of genome reduction as well as the presence or absence of gene transfers and protein import pathways are currently unknown for Paulinella, and only three gene cluster (4.3 to 9.4 kb) on the chromatophore genome have been analyzed to date and compared to free-living cyanobacterial relatives [1,6].
Interestingly, the closest relatives of the Paulinella chromatophore (the PS-clade) possess a proteobacterial form 1A RubisCO (ribulose-1,5-bisphosphate carboxylase/oxygenase), in contrast to the remaining cyanobacteria and plastids (except rhodoplasts) with the 'typical' form 1B RubisCO [7-9]. The proteobacterial form 1A RubisCO is part of a carboxysomal operon encoding genes for both subunits of RubisCO (rbcL, rbcS = cbbL, cbbS) as well as genes for carboxysomal shell proteins and a carboanhydrase [10-12]. Cyanobacteria of the PS-clade may have acquired the complete carboxysomal operon by horizontal gene transfer (HGT) from a proteobacterial donor [10,11]. Carboxysomes containing a form 1A RubisCO are referred to as α-carboxysomes, and thus, the PS-clade has been designated as α-cyanobacteria, in contrast to β-cyanobacteria with a form 1B RubisCO integrated in β-carboxysomes [11].
Previous phylogenies based on rRNA operon sequence data resolved the Paulinella chromatophore as sister to marine Synechococcus and Prochlorococcus spp (α-cyanobacteria) [1]. The intermediate position of Paulinella, diverging between α- and β-cyanobacteria, raises the question whether the HGT of the carboxysomal operon occurred before or after the divergence of the ancestor of the Paulinella chromatophore, i.e. whether the chromatophore evolved from an α- or β-cyanobacterium. Due to missing data, affiliation to α- or β-cyanobacteria is also unknown for the Cyanobium-clade, which, besides marine Synechococcus and Prochlorococcus clades, represents the third major lineage in the PS-clade (e.g. [13-15]). Analyses of one member of the Cyanobium-clade (strain WH 5701) already indicated its individual divergence separate from Paulinella and both marine PS-subclades [6].
In this study, we have determined the sequence of the complete carboxysomal operon from the chromatophore of Paulinella and a bacterioferritin gene downstream of the carboxysomal operon. Additionally, we determined several rbcL and rDNA sequences from other cyanobacteria including the Cyanobium-clade. Our data reveal that the Paulinella chromatophore as well as the Cyanobium-clade display proteobacterial α-carboxysomes, and contain form 1A RubisCO. Furthermore, phylogenies of RubisCO and comparison of gene arrangement types of the carboxysomal operon revealed the γ-proteobacterium Nitrococcus mobilis as the closest known relative of the donor in the HGT of the carboxysomal operon. The neighbouring bacterioferritin gene was co-transferred through the same HGT event. Increased AT-content over the carboxysomal operon in Paulinella may reflect genomic adaptation to an endosymbiotic lifestyle.
Results and Discussion
Phylogenetic analysis of the ribosomal RNA operon
One goal of this study was to determine the precise phylogenetic position of the Paulinella chromatophore within the cyanobacterial radiation. Cyanobacterial phylogeny is still a challenge since the standard phylogenetic marker, 16S rDNA, is not very informative in global analyses (see below). On the other hand, previous studies using extended data sets, either the complete rDNA operon [1] or several protein coding genes [6,16] suffered from limited taxon sampling. Here, we have extended the taxon sampling of nearly complete rDNA operon sequences to include all major cyanobacterial clades [17,18] with one to several representatives. Therefore, rDNA sequences from one plastid and 12 cyanobacteria were determined (taxa in bold in Figure 1). Together with data from newly released genome projects, the resulting alignment contained sequences from 36 bacteria, 42 cyanobacteria and 23 plastids.
Figure 1.
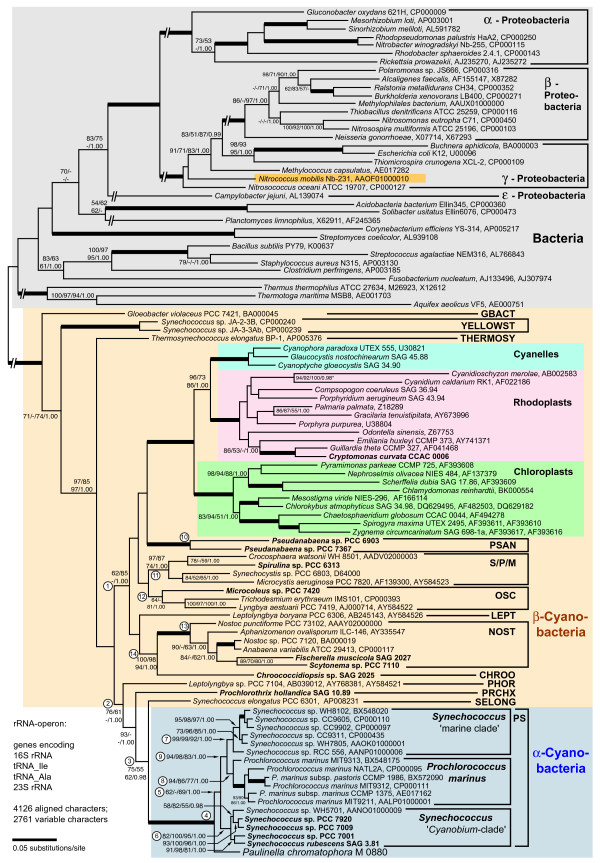
Phylogenetic position of the Paulinella chromatophore within the cyanobacteria inferred by complete rRNA operon sequence comparisons. The tree topology was generated by maximum likelihood (ML) analyses using the GTR+I+Γ model. The nodal support values are bootstrap values ≥ 50% obtained by ML (100 replicates), neighbor-joining (NJ; GTR+I+Γ model; 1000 repl.), maximum parsimony (MP; 1000 repl.), and Bayesian posterior probabilities (≥ 0.95). Branches in bold have maximal support (100%; 1.00) by all methods; interrupted branches were graphically reduced to 30% of their original length. Taxa in bold were newly determined for this study; strain designations and EMBL/GENBANK accession numbers are also given. Cyanobacterial clade abbreviations: GBACT (Gloeobacter), PSAN (Pseudanabaena), S/P/M (Synechocystis/Pleurocapsa/Microcystis), OSC (Oscillatoria), LEPT (Leptolyngbya), NOST (Nostoc), and PHOR (Phormidium): modified sequence groups after [17]; YELLOST (thermophilic "Synechococcus" from Yellowstone NP), THERMOSY (Thermosynechococcus), CHROO (Chroococcidiopsis), PRCHX (Prochlorothrix), SELONG (Synechococcus elongatus), and PS (Prochlorococcus/Synechococcus). Encircled numbers indicate clades/branches that were analyzed by single-gene analyses, and by NJ using the LogDet+I-model (see text and Additional File 1).
Phylogenetic analyses revealed basal cyanobacterial branches (e.g. Gloeobacter), and two moderately supported lineages, one combining the majority of the β-cyanobacteria including all plastids (branch 1 in Figure 1), the other containing Paulinella and the PS-clade nested within a radiation of a few β-cyanobacteria, representing the clades PHOR, PRCHX and SELONG (branch 2 in Figure 1). Paulinella is monophyletic with the PS-clade (branch 4). Our previous study [1] had already revealed the monophyly of both marine PS-subclades to the exclusion of the Paulinella chromatophore, as confirmed here (see branch 9). The present investigation includes five sequences of the third PS-subclade, the Cyanobium-clade, which is sister to the marine subclades, with Paulinella still diverging in a basal position. The monophyly of the entire PS-clade to the exclusion of Paulinella, however, receives only moderate to low bootstrap support (branch 5), but is corroborated by unique synapomorphies in the 23S rRNA (Figure 2). As previously shown, both marine PS-subclades are characterized by unique compensatory base changes (CBCs in pairs 868/909 and 869/908), whereas Paulinella is plesiomorphic in both pairs [1]. Interestingly, the Cyanobium-clade is intermediate in sharing the unique CBC in position 868/909 with marine PS-subclades, but displaying the ancestral character state in positions 869/908, in congruence with the tree topology (Figure 2).
Figure 2.
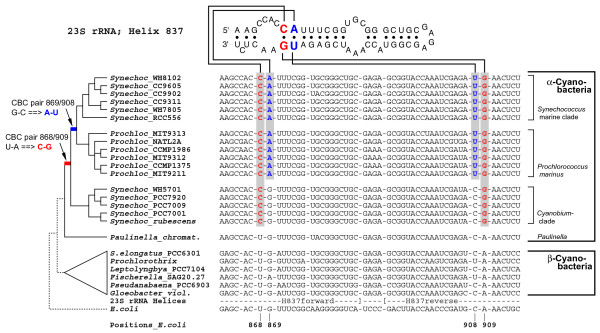
Synapomorphy support in the 23S rRNA for the sister-group relationship between Paulinella and free-living α-cyanobacteria. Shown is the alignment and secondary structure diagram of Helix 837 in the 23S rDNA, with two RNA base pairs highlighted that represent synapomorphies of α-cyanobacterial clades to the exclusion of Paulinella and other prokaryotes. Sequence data and evolutionary changes are plotted on a simplified phylogram (NJ-bootstrap consensus tree). Pair 868/909 shows a uniquely derived CBC (compensatory base change: U-A → C-G) of all free-living α-cyanobacteria; the neighbouring pair 869/908 changed in the common ancestor of the marine PS-subclades (marine Synechococcus and Prochlorococcus) whereas the Cyanobium-clade and Paulinella are plesiomorphic. Pair 869/908 shows parallel changes in a few other cyanobacteria, (e.g. in Fischerella).
Several cyanobacterial branches gained only moderate bootstrap support (e.g. branches 1, 2, 3, 5, 12 in Figure 1). One possible explanation is the base compositional bias among prokaryotic rDNAs (the Chi-square test gave p = 0.00) mainly caused by bacteria and plastids (cyanobacteria alone have no significant base compositional bias; p = 0.21). Therefore we performed distance analyses using the LogDet correction for unequal base composition (Additional File 1) that largely confirm results shown in Figure 1. Branch 5 (PS-clade without Paulinella) is even better supported (78%). The phylogenetic signal for branch 5 is confined to the 23S rRNA gene as shown by single-gene analyses (87–93% bootstrap in the 23S rRNA phylogeny), since this branch collapsed in 16S rDNA analyses, as also did the branches 9, 11, 12, and 14 (Additional File 1).
In conclusion, the rDNA data support monophyly of both marine PS-subclades (= α-cyanobacteria sensu [11] with proteobacterial form 1A RubisCO) to the exclusion of both the Cyanobium-clade and the Paulinella chromatophore, raising the question whether these taxa are affiliated with α- or with β-cyanobacteria (form 1B RubisCO), i.e. on which branch of the phylogenetic tree the horizontal gene transfer of form 1A RubisCO occurred. To answer this question, sequencing and phylogenetic analysis of RubisCO of the Cyanobium-clade and the Paulinella chromatophore was required.
Horizontal gene transfer of a carboxysomal operon into the ancestor of the Paulinella chromatophore
In this study complete rbcL (RubisCO large subunit) sequences of Paulinella and four members of the Cyanobium-clade were determined and integrated into a global alignment of form 1 RubisCO large subunit amino acid sequences. The phylogenetic analysis resolves Paulinella chromatophora and the Cyanobium-clade as monophyletic with marine Synechococcus and Prochlorococcus spp., in congruence with the rDNA phylogeny (Figure 3). The Paulinella chromatophore as well as the entire PS-clade belong to the RubisCO form 1A lineage, and in conclusion, have to be considered as α-cyanobacteria. The position of the entire α-cyanobacterial clade (including Paulinella) in the rbcL tree is not congruent with the rDNA phylogeny: α-cyanobacteria are not monophyletic with the remaining cyanobacteria (β-cyanobacteria) but are nested within a radiation of α-, β- and γ-proteobacteria in the rbcL phylogeny (Figure 3). This incongruence reflects the horizontal gene transfer (HGT) of RubisCO form 1A from proteobacteria to α-cyanobacteria, and loss of the ancestral 'cyanobacterial' RubisCO form 1B [9].
Figure 3.
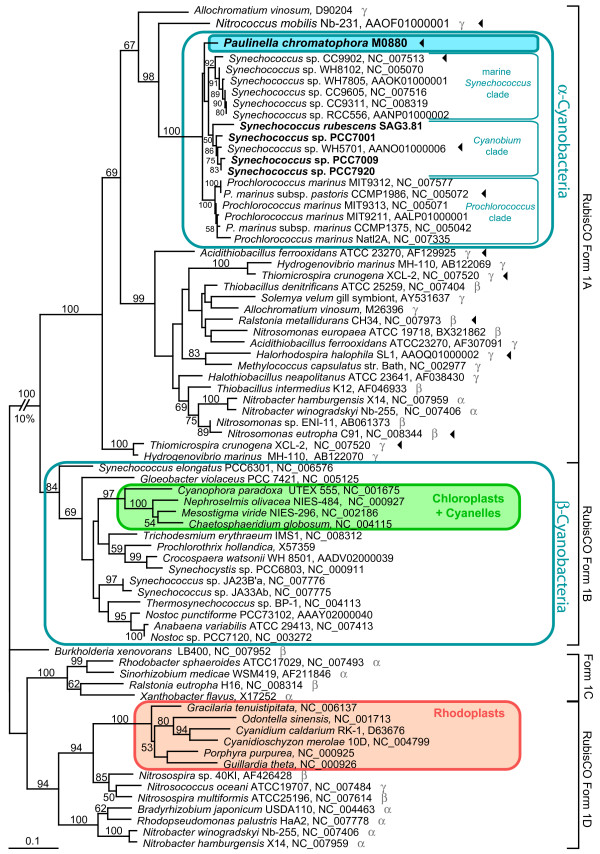
Evidence for HGT of RubisCO form 1A to the common ancestor of the Paulinella chromatophore and α-cyanobacteria. The ML tree was inferred from RubisCO large subunit (rbcL) form 1 amino acid sequences (470 aligned positions) of Paulinella chromatophora, cyanobacteria, plastids and proteobacteria under the RtREV+I+Γ model of amino acid substitution. Numbers at branches are ML bootstrap values ≥ 50%. Strain designations (when available) and NCBI accession numbers are indicated after the species name. Newly determined sequences are given in bold. Greek letters in grey indicate α-, β-, or γ- proteobacteria. Arrowheads highlight strains for which the gene arrangement of the carboxysomal operon is shown in Figure 5.
In cyanobacteria, carboxysomes are essential for the carbon concentration mechanism (CCM; [19]). Physiological differences between form 1A and form 1B RubisCO and corresponding carboxysome types (α and β) are still not understood [19,20]. It has been suspected that the occurrence of α-carboxysomes is correlated with ecological restriction to marine open ocean habitats [19]. Clearly, this view is untenable, since the Cyanobium-clade, which predominantly contains freshwater species, as well as the Paulinella chromatophore also display α-carboxysomes. Since α-cyanobacteria occur in a broad range of habitats, it is even more difficult to speculate about advantages of α-carboxysomes for survival in special ecological niches, in particular an endosymbiotic habitat (Paulinella). As the microenvironment of the chromatophore of Paulinella can presumably be characterized as CO2-rich due to host respiration, it may even be assumed that an efficient CCM may not be essential for the Paulinella chromatophore. Regrettably, experimental data on the photosynthetic properties of the Paulinella chromatophore, especially the existence and effectiveness of a CCM, are not yet available.
Previous rbcL phylogenies did not contain the γ-proteobacterium Nitrococcus mobilis (genome sequence available since Feb-2006) [10,11,20,21]. Interestingly, our rbcL-phylogeny identified this taxon as closest relative to the α-cyanobacteria: the common branch of Nitrococcus and the α-cyanobacteria gained 98% bootstrap support (Figure 3). A search for unique synapomorphies in rbcL amino acid sequences revealed 3 positions (AA 36, 59, 64 in the Paulinella sequence), which in Nitrococcus and all α-cyanobacteria share uniquely derived character states to the exclusion of all remaining proteobacterial and β-cyanobacterial sequences (Figure 4). In addition, we found two synapomorphies (positions 399, 405), which characterize the α-cyanobacterial form 1A RubisCO (Figure 4). These results highlight Nitrococcus as a key taxon for the HGT of RubisCO form 1A, being the closest known relative of the proteobacterial donor of rbcL.
Figure 4.
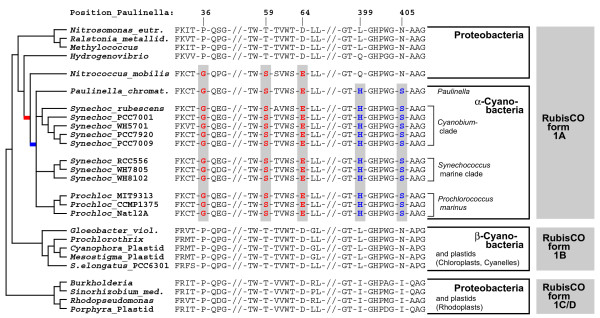
Unique synapomorphies highlighting the HGT of RubisCO from a Nitrococcus-like γ-proteobacterium to the common ancestor of the Paulinella chromatophore and free-living α-cyanobacteria. Selected regions of the RubisCO large subunit amino acid sequence were plotted against a simplified phylogram (NJ bootstrap consensus tree). Three synapomorphies shared by Nitrococcus and α-cyanobacteria are shown in red colour (positions 36, 59, 64); Histidine 399 and Serine 405 (blue) are unique for Paulinella and free-living α-cyanobacteria to the exclusion of all proteobacterial ancestors and β-cyanobacteria.
Another HGT event is responsible for the well-known polyphyly of plastids in rbcL phylogenies [9,21,22]. Chloroplasts and cyanelles are rooted in the β-cyanobacteria in congruence with ribosomal phylogenies (Figure 1). In contrast, rhodoplasts are nested within the RubisCO form 1D clade of α-, β- and γ-proteobacteria. With high significance, the analysis reveals Nitrosospira sp., Nitrosococcus oceani and Nitrosospira multiformis as a sister branch to rhodoplasts (the latter two already described in [21].)
In the α-cyanobacteria, the genes encoding RubisCO form 1A belong to an operon that further contains genes for carboxysomal proteins, and previous studies revealed that the complete carboxysomal operon was acquired by HGT [10,11]. Among proteobacteria gene content as well as gene order in rbcL-containing operons differs considerably [23]. Several proteobacteria have two or even three unrelated rbcL genes (see for example Nitrobacter, Hydrogenovibrio, Thiomicrospira in Figure 3; [23,24]). In contrast, cyanobacteria generally have only one rbcL gene. In the present study, we determined the sequence of the complete carboxysomal operon of Paulinella chromatophora (7.6 kb), and compared the gene arrangement among members of the RubisCO form 1A clade (Figure 5).
Figure 5.
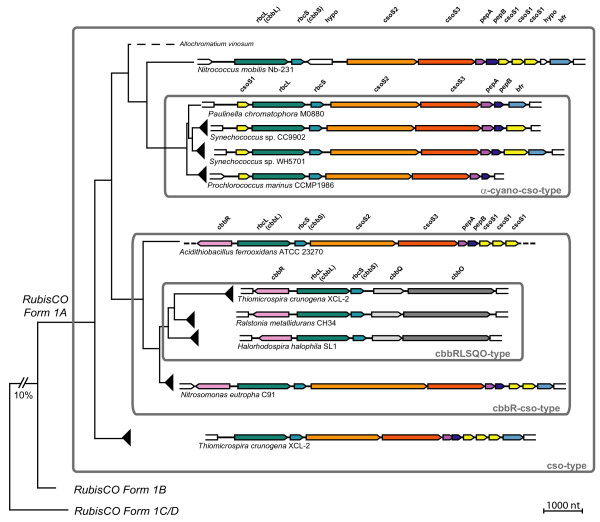
Architecture and evolution of operons containing form 1A RubisCO from proteobacteria and α-cyanobacteria including the Paulinella chromatophore. Gene arrangements from selected taxa (see arrowheads in Figure 3) are plotted against a simplified phylogram based on RubisCO amino acid sequences. Four major types of gene arrangements can be distinguished (for details, see text). The operon of Paulinella is member of the α-cyano-cso-type, which is derived from the ancestral cso-type present in proteobacteria, providing evidence for a HGT of the complete operon. Homologous genes share the same colour. Abbreviations: Carboxysomal shell proteins 1, 2, 3 (csoS1, 2, 3); RubisCO large and small subunit (rbcL, rbcS = cbbL, cbbS); carboxysomal peptides A, B (pepA, pepB); bacterioferritin (bfr); LysR-type transcriptional activator (cbbR); putative RubisCO activation proteins (cbbQ, cbbO); hypothetical proteins (hypo). Dotted lines indicate that no data are available.
We found four major arrangement types. To analyse the evolution of these types, we plotted operon structures against a simplified rbcL tree as shown in Figure 5 (for more details, see Additional File 2). Basal branches of the RubisCO form 1A radiation (e.g. Thiomicrospira, Nitrococcus) show an almost identical operon architecture, which likely represents the plesiomorphic state. In this type, (cso-type) the following genes occur downstream of rbcL: rbcS, csoS2, csoS3, pepA, pepB, csoS1, csoS1, csoS1, and the iron storage protein bacterioferritin (bfr) [25]. The operon in the α-cyanobacteria (α-cyano-cso-type) was derived from the ancestral state by transfer of a single csoS1 gene to the 5' end of the operon, accompanied by a reduction of the number of 3' located csoS1 copies to one or zero. Interestingly, two members of the α-cyanobacteria still contain the 3' bacterioferritin gene (bfr): the Paulinella chromatophore and Synechococcus WH5701, the latter representing the Cyanobium-clade (data for the remaining Cyanobium-clade are currently missing). In the marine PS-subclades, bfr is absent (Figure 5; Additional File 2). Comparison with the ribosomal phylogeny (Figure 1) suggests that bacterioferritin was acquired by the same HGT event as the carboxysomal operon, and was secondarily lost in marine Synechococcus and Prochlorococcus species. We addressed the bfr HGT hypothesis by performing a phylogenetic analysis with α- and β-cyanobacterial ferritins and their proteobacterial relatives (Additional File 3). As in the rbcL-phylogeny, the bacterioferritins of Paulinella and WH5701 are monophyletic with Nitrococcus mobilis as their closest relative, and were nested within proteobacteria with carboxysomal operons, clearly proving co-transfer of bacterioferritin with the carboxysomal operon. Neither nonheme-ferritins of marine Synechococcus/Prochlorococcus-species nor the ferritin genes of β-cyanobacteria show any relationship to bfr of Paulinella and WH5701 (for details, see Additional File 3). In γ-proteobacteria incl. Nitrococcus, and in Prochlorococcus, the next gene downstream to the carboxysomal operon is a putative pterine-4alpha-carbinolamine dehydratase (Additional File 2) that in the remaining α-cyanobacteria is also present, but in those taxa is not linked to the carboxysomal operon. Blast searches [26] reveal their homology, suggesting that besides the carboxysomal operon and bfr even more genes may have been acquired by the same HGT event.
In parallel to the α-cyano-cso-type, another proteobacterial gene arrangement type is derived from the cso-type by the acquisition of a cbbR gene upstream to rbcL, coded by the opposite strand (Figure 5). This type is therefore here named cbbR-cso-type (e.g. Nitrosomonas eutropha, Figure 5). Though not co-transcribed with the carboxysomal genes, CbbR is linked to this operon by its specific function as a transcriptional activator [27]. Finally, the most derived gene arrangement type is nested within the cbbR-cso-type, and is named cbbRLSQO-type. The first three genes, cbbR, rbcL, and rbcS, remained unchanged whereas all carboxysomal shell proteins and bacterioferritin were lost and replaced by the genes cbbQ and cbbO, which are absent in the remaining three types of operon structures (Figure 4). cbbQ and cbbO have been shown to enhance RubisCO activity and stability [28,29].
Notably, each synapomorphic change leading to the three evolutionary derived gene arrangement types corresponds to a single branch/clade in the phylogenetic tree based on rbcL sequence data (Figure 3). This congruence provides additional credibility for the rbcL tree, including one branch without any bootstrap support that combines all taxa characterized by the cbbRLSQO-type (Figures 4, 5). However, the loss of bacterioferritin in the marine Synechococcus and Prochlorococcus clades, which can also be assumed to be a synapomorphic change, cannot be traced to a single branch in the rbcL tree due to low resolution among α-cyanobacteria. Although the four major lineages (Paulinella, Cyanobium-clade, marine Synechococcus- and Prochlorococcus-clades) are recovered, relationships between these lineages remain unresolved (Figure 3). Based on the rDNA phylogeny, it appears likely that bacterioferritin was uniquely lost in the common ancestor of marine α-cyanobacterial clades as a single synapomorphic change (branch 9 in Figure 1). In general, the branching order within the α-cyanobacteria shows no significantly supported conflict between rDNA and rbcL phylogenies, and thus, both data sets were used separately and in combination to resolve phylogenetic relationships within the α-cyanobacteria.
Phylogenetic resolution of the Synechococcus/Prochlorococcus-clade with the concatenated dataset
In Figure 6, three phylogenetic analyses of P. chromatophora and 17 taxa of Synechococcus and Prochlorococcus using three different datasets are compared: (A) rbcL nucleotide sequences, (B) complete rDNA operon sequences, and (C) concatenated rbcL and rDNA sequences. Prior to phylogenetic analyses, nucleotide frequencies of rbcL and rDNA sequence data were determined to prevent artefacts caused by base compositional bias. Whereas the base composition of rbcL codon positions one and two was homogenous across taxa (Figure 7A), the third codon position revealed strong differences between AT-rich (Paulinella, and Prochlorococcus strains: ca. 50–80%) and AT-poor taxa (Synechococcus strains ca. 10–30%) (Figure 7B). Therefore, only first and second codon positions were used for phylogenetic analyses of the rbcL gene.
Figure 6.
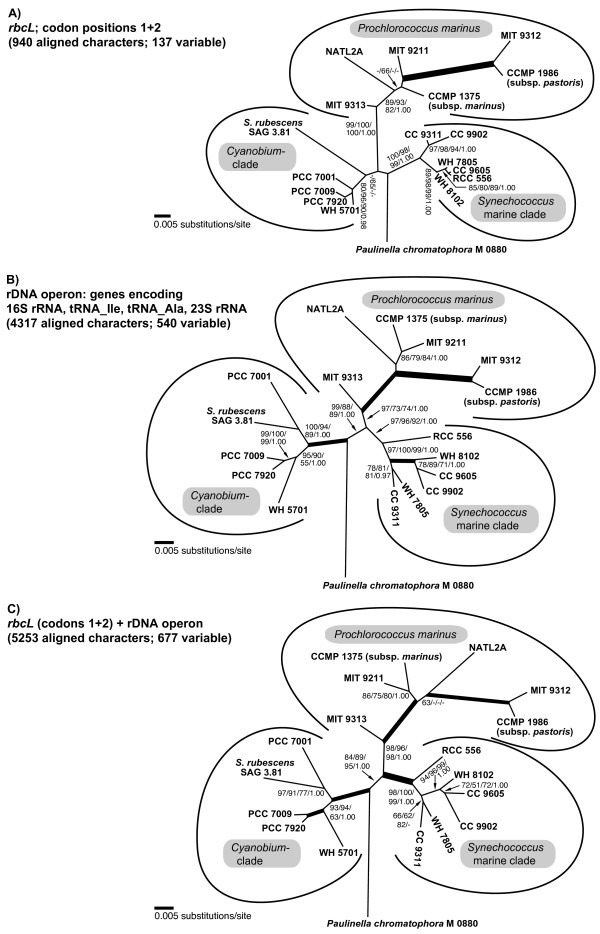
Comparison of phylogenetic relationships among Paulinella and free-living α-cyanobacteria inferred by rbcL and/or rDNA nucleotide sequence data. A. Unrooted analysis of codon positions 1+2 of the rbcL gene. B. Phylogeny of the rDNA operon, using more aligned positions as in Figure 1 (4317 vs. 4126 characters). C. Phylogeny inferred from concatenated rbcL and rDNA sequences. Tree topologies resulted from ML analyses using a GTR+I+Γ model; significance values shown as in Figure 1.
Figure 7.
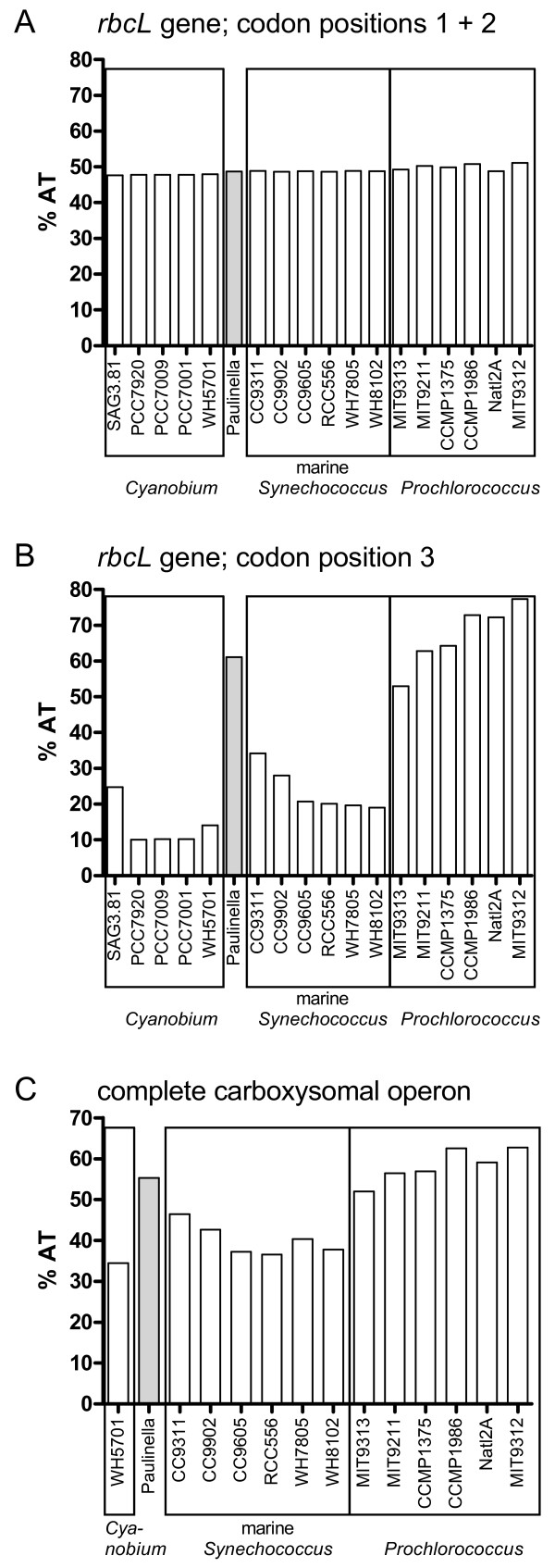
AT content bias in rbcL and complete carboxysomal operon sequences in Paulinella and Prochlorococcus. Whereas the AT-content in codon positions 1 + 2 in the rbcL gene is balanced across Paulinella and 17 free-living α-cyanobacteria (A), the third codon position displays a sharply elevated AT-content in Paulinella as well as Prochlorococcus strains (B). The AT-content integrated over the complete carboxysomal operon (from csoS1 to pepB) including intergenic spacer regions shows a similar bias, although less pronounced (C).
Both rbcL and rDNA phylogenies recover the monophyly of three major clades: Cyanobium-clade, marine Synechococcus-clade and Prochlorococcus-clade. However, rbcL data fail to recover the Cyanobium-clade with significance, probably due to the long-branched Synechococcus rubescens (see also the amino acid analysis; Figure 3). On the other hand, Prochlorococcus displays a much longer basal branch in the rbcL analysis compared to rDNA, due to a higher number of synapomorphies, accompanied by a considerably higher bootstrap support (99–100% in rbcL compared to 73–97% in rDNA). The higher-level order of the major clades remains unresolved in the rbcL tree, whereas the rDNA analysis recovers the sister-group relationship of marine Synechococcus and Prochlorococcus clades (88–99% bootstrap; see also Figure 1). Within Prochlorococcus, relationships among strains are resolved almost congruently by rbcL and rDNA data: MIT9313 appears as the basal divergence, followed by strains NATL2A, CCMP1375 and MIT9213 (relationships among these strains are not resolved), and a derived, long-branched clade consisting of MIT9312 and CCMP1986. In the remaining clades (Cyanobium-clade and marine Synechococcus-clade), the branching pattern of most strains remains unresolved by rbcL analyses, due to their almost redundant rbcL sequences. In contrast, the higher rDNA sequence diversity allows a better resolution in these lineages. For instance, Synechococcus rubescens, a single long branch in the rbcL tree, is confidently identified as sister of PCC 7001 by rDNA data. Similarly, Synechococcus strain RCC 556, which has an almost identical rbcL sequence as three other strains (CC9605, WH7805, WH8102), is resolved as the basal divergence of the marine Synechococcus clade by rDNA analyses. The concatenated data set (5253 nucleotide characters) is largely congruent with the rDNA tree, albeit with even better bootstrap support for the monophyly of the major clades (especially Prochlorococcus and marine Synechococcus). However, in a few cases (e.g. position of Synechococcus rubescens, position of Synechococcus CC9311) the combined analysis showed slightly reduced support in comparison with the rDNA phylogeny (Figure 6B, 6C).
In the phylogenetic analyses performed in this study, we found no specific and robustly supported relationship between Paulinella and any of the three major clades of free-living α-cyanobacteria. RbcL data fail to resolve the monophyly of the Cyanobium-clade to the exclusion of Paulinella with significance, irrespective of whether amino acid or nucleotide data are analyzed (Figures 3, 6A), in contrast to the remaining phylogenies that receive high significance for this clade (Figures 1, 6B, 6C). Although the prokaryote-wide rDNA phylogeny (Figure 1) uses fewer positions than the unrooted rDNA phylogeny of the α-cyanobacteria (Figure 6B; 4126 vs. 4317 characters), we recognize almost complete congruence between the two phylogenies among α-cyanobacteria. In addition, our tree topologies corresponded almost completely to phylogenetic analyses of the 16S-23S rDNA spacer ('ITS'; 233 aligned positions) from 57 free-living α-cyanobacteria (Figure 4A in [30]). Probably due to the much higher number of aligned/variable (4317/540) positions in the rRNA and tRNA genes, our analyses gained high significance values for most basal and internal branches that were largely non-significant in ITS-phylogenies [30], e.g. the branch uniting all Prochlorococcus strains including the basal divergence MIT9313. Other phylogenies that included only a few α-cyanobacteria had often failed to position strain MIT9313 as monophyletic with other Prochlorococcus strains (e.g. [1,31,32]), even in multigene analyses of complete cyanobacterial genomes [31,32]. Superiority of a 233-character-analysis over whole-genome phylogenies may highlight the importance of a sufficient taxon sampling. Moreover, phylogenetic information content of protein gene data may be impaired by unequal base composition and amino acid frequencies across taxa: whereas derived Prochlorococcus strains have a high AT-content and show a preference for amino acids encoded by AT-rich codons, base composition and amino acid usage of Prochlorococcus MIT9313 are more similar to Synechococcus strains with lower AT-content [33], explaining artificial tree topologies. In the more conserved rbcL gene, the AT bias is confined to the third codon positions (Figure 7A, 7B), and thus, phylogenies using only the first and second positions are not affected by base compositional bias. In fact, our rbcL tree does not contradict analyses using rDNA data, which are known to have a more balanced base composition across taxa ([33], and our own results). Interestingly, Paulinella shows the same tendency towards high AT-content as known for Prochlorococcus, as evident from third rbcL codon positions (Figure 7B) as well as the complete carboxysome operon (Figure 7C). An elevated AT content, accompanied by genome size reduction, is a highly unusual phenomenon for free-living organisms, and in the case of Prochlorococcus, was interpreted as adaptation to oligotrophic marine environments with low nitrogen and phosphorus availability [33]. In contrast, endosymbionts or intracellular pathogens are known to tend to evolve towards AT rich genomes (e.g. [34-37]). We suspect that the complete genome of the Paulinella chromatophore is characterized by high AT content since not only the carboxysomal operon (this study) but also two DNA fragments of 9.4 kb and 4.3 kb (see Figure 1 in [6]) support this view. Together with its isolated position in molecular phylogenies ([1,6], and this study), the elevated AT content further indicates that the chromatophore of Paulinella has significantly diverged from its free-living ancestor, and undergone typical steps in the evolution of an intracellular symbiont such as genome reduction.
Conclusion
The basal divergence of the Paulinella chromatophore as sister to free-living α-cyanobacteria was revealed by phylogenetic analysis of the complete rDNA operon with an extended taxon sampling, especially by addition of the Cyanobium-clade. Paulinella and free living α-cyanobacteria share a proteobacterial carboxysomal operon with a form 1A RubisCO, indicating that the HGT of the carboxysomal operon predated the divergence of the Paulinella chromatophore. The γ-proteobacterium Nitrococcus mobilis was identified as the closest known relative to the proteobacterial donor of the carboxysomal operon. The isolated position of Paulinella among α-cyanobacteria in molecular phylogenies as well as the elevated AT content of several of its genes indicates that Paulinella has already undergone typical steps in reductive genome evolution associated with an intracellular lifestyle.
Methods
Algal cultures
Strains used in this study were obtained from the following sources: Spirulina sp. PCC 6313, Microcoleus sp. PCC 7420, Scytonema sp. PCC7110, Pseudoanabaena sp. PCC 7367, Pseudoanabaena sp. PCC 6903, Synechococcus sp. PCC 7001, Synechococcus sp. PCC 7920, Synechococcus sp. PCC 7009: Pasteur Culture Collection of Cyanobacteria, Institute Pasteur, Paris, France [38]; Fischerella muscicola SAG 2027, Prochlorothrix hollandica SAG 10.89, Chroococcidiopsis sp. SAG 2025, Synechococcus rubescens SAG 3.81: Sammlung für Algenkulturen, University of Göttingen, Germany [39]. Paulinella chromatophora M0880: Culture collection Melkonian, University of Cologne, Germany;Cryptomonas curvata CCAC 0006: Culture Collection of Algae at the University of Cologne, Germany [40].
DNA extraction, PCR and sequencing
Complete genomic DNA was extracted using a CTAB protocol. PCR primers for amplification of the rDNA operon, and sequencing methods were described previously [1]. For amplification of the full length rbcL gene from Paulinella chromatophora and Synechococcus strains, α-cyanobacteria-specific primers were designed using an alignment of cyanobacterial rbcL and surrounding genes. PCR primers bind in the neighbouring genes of rbcL: csoS1 and rbcS. Primer sequences: Al_csoS1_F2: (GARGCWGCWGAYGCHATGACCAAGG) and Al_rbcS_R1: (TGRTCGTADATYTCKTCCTGGGTCATMGG). If primary products were too weak a reamplification was done with Al_csos1_F3 (GCHGAAGTKCGYCTKATYGGTCGTG) or Al_csos1_F4 (CGYCCYCAYMGNGAAGTKGAGCCWGC) and Al_rbcS_R1. Using the same alignment new sequencing primers were designed: Al_csoS1_F4 (sequence see above), Al_rbcL_F1 (TTYGARTTYGTHGCBGAAGC), Al_rbcL_R1 (GGCATRTGCCANACRTGRATRCC), Al_rbcL_R2 (ARYTTHGGYTTRATRGTRCARCC). RbcL PCR products were purified using the QIAquick PCR Purification Kit (Qiagen) and sequenced by the cycle sequencing method using an ABI 3730 sequencer.
Alignments and phylogenetic analyses
Newly obtained sequences were combined with database sequences to construct amino acid and nucleotide sequence alignments. Accession numbers of new sequences are AM709625 – AM709637 (rRNA operon), AM701774 – AM701778 (rbcL), and EF589049 (carboxysomal operon of Paulinella); accession numbers of database sequences are given in Figures 1 and 3. RbcL and ferritin protein alignments were obtained using clustalW, and refined manually; rDNA operon alignments were constructed manually, guided by rRNA and tRNA secondary structure. Unalignable positions were excluded from datasets prior to phylogenetic analyses. RbcL nucleotide sequences were aligned according to the RubisCO amino acid alignment. The rDNA-operon analyses contained 4126 aligned characters for prokaryotes (Figure 1), and 4317 for α-cyanobacteria (Figure 6B, 6C); the RubisCO large subunit amino acid dataset contained 470 aligned positions (Figure 3). The α-cyanobacterial rbcL nucleotide dataset was reduced to first and second codon position resulting in 940 aligned nucleotides (Figure 6A, 6C). Nucleotide sequence analyses used PAUP 4.0b10, MrBayes_3.1.1, and MODELTEST, as previously described [1]. The homogeneity of base frequencies across taxa was investigated by Chi-square tests (PAUP 4.0b10). When distance analyses used the LogDet+I model, the I-value was copied from the GTR+I+Γ model parameters that were estimated by MODELTEST for the same dataset. ML bootstrap analyses in Figure 1 were constrained towards 3000 rearrangements per replicate. For Bayesian analyses, two MCMC chains with 500000 generations were performed, and the 'burnin' determined by the convergence criterion (see [1]). The search procedure to find molecular characters, which represent unique synapomorphies for a clade of interest, has been described previously [1], and was here applied to nucleotide as well as protein alignments. Protein datasets were subjected to maximum likelihood bootstrap (n = 1000) analyses with Phyml V2.4.5 [41]. The evolutionary model fitting best (RtREV+I+Γ) was determined using ProtTest 1.3 [42] according to the Akaike Information Criterion. Proportion of invariable sites and Γ-shape parameter were calculated in Phyml.
Carboxysomal operon data
Sequence data of the carboxysomal operon of the Paulinella chromatophores were taken from the ongoing Paulinella chromatophore genome project. DNA was extracted from crude chromatophore fractions obtained by density centrifugation on a percoll gradient. This DNA was used to generate a small insert library. Random sequencing of 400 clones yielded raw reads overlapping the previously obtained rbcL sequence. Using a combination of primer walks and PCR all gaps in the rbcL region were closed.
Authors' contributions
BM determined 13 rDNA operon sequences, and was responsible for rDNA alignments and analyses, constructed the ferritin alignment, and performed synapomorphy searches, leading to Figures 1, 2, 4, 6B, 6C, Additional Files 1, and 3. ECMN isolated chromatophores of P. chromatophora for the ongoing genome project, determined 5 rbcL sequences, was responsible for RubisCO alignments and analyses, and investigated gene arrangement types and base composition of carboxysomal operons, leading to Figures 3, 5, 6A, 6C, 7, and Additional File 2. BM and ECMN wrote the manuscript. GG determined the sequence of the carboxysomal operon of P. chromatophora. MM conceived the study, contributed to its design, and critically revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Significance measures for 14 selected clades (encircled numbers in Figure 1) in single-gene and combined analyses of rRNA genes. Analyses used 1000 bootstrap replicates of NJ (LogDet+I model), NJ (GTR+I+Γ model), and MP, and Bayesian posterior probabilities. Numbers of aligned (ch) and variable characters (var) are given for each partition.
Gene arrangement of operons containing form 1A RubisCO for all strains included in Figure 3. The table describes the typical gene arrangements in the 4 different arrangement types, defined in Figure 5, and indicates presence (x) or absence () of the gene in a specific species. Abbreviations as given in Figure 5; further abbreviations: n.d.: no data available; ham1: Ham1 like protein; ndhF3: NADH dehydrogenase subunit L; PCD_DCoH: possible pterin-4alpha-carbinolamine dehydratase; GlnK: nitrogen regulatory protein P-II; GGPS: Glucosylglycerol-phosphate synthase; Transp: transposase, mutator type; REC: response regulator receiver protein; Rpe: Ribulose-phosphate 3-epimerase; TktA: Transketolase; DedA: Uncharacterized membrane-associated protein; CBD_II: Cellulose binding domain protein. chlN: light-independent protochlorophyllide reductase subunit N.
Evidence that bacterioferritin in Paulinella and Synechococcus WH5701 was acquired by HGT from a Nitrococcus -like γ-proteobacterium. Phylogeny of three ferritin-families occuring in cyanobacteria (α-cyanobacteria in blue; β-cyanobacteria in orange colour) together with their proteobacterial relatives: Bacterioferritin, Nonheme-Ferritin, and Ferritin and Dps ("DNA-binding protein from starved cells"). The ML analysis was performed as in Figure 3, and used 159 aligned amino acid positions. The chromatophore of Paulinella and Synechococcus WH5701 (representing the Cyanobium-clade) are the only cyanobacteria, which possess bacterioferritin linked to the carboxysomal operon (see Figure 5). Similar to the RubisCO phylogeny, Nitrococcus mobilis is sister to these taxa in the bacterioferritin clade. Note that many taxa contain more than one ferritin (up to five in Synechococcus CC9311), e.g. Nitrococcus displays two unrelated bacterioferritin genes, and Synechococcus WH5701 has 1 bacterioferritin, one nonheme-ferritin, and one member of the Ferritin and Dps family (indicated by numbers in curly braces). Cyanobacterial ferritins are dispersed into several separate branches, usually nested within bacterial divergences, suggesting many independent HGT events. One to five (in CC9311) nonheme-ferritin genes are characteristic for members of all PS-subclades (presence in the Paulinella chromatophore is currently unknown), and their tree topology indicates an early gene duplication followed by later duplication/gene loss events.
Acknowledgments
Acknowledgements
This study was supported by the University of Cologne (BM), and by a grant from the International Graduate School in Genetics and Functional Genomics at the University of Cologne (ECMN).
Contributor Information
Birger Marin, Email: birger.marin@uni-koeln.de.
Eva CM Nowack, Email: eva.nowack@uni-koeln.de.
Gernot Glöckner, Email: gernot@fli-leibniz.de.
Michael Melkonian, Email: michael.melkonian@uni-koeln.de.
References
- Marin B, Nowack ECM, Melkonian M. A plastid in the making: Evidence for a second primary endosymbiosis. Protist. 2005;156:425–432. doi: 10.1016/j.protis.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Philippe H. Plastid origin: Replaying the tape. Curr Biol. 2006;16:R53–R56. doi: 10.1016/j.cub.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Theissen U, Martin W. The difference between organelles and endosymbionts. Curr Biol. 2006;16:R1016–R1017. doi: 10.1016/j.cub.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Archibald JM. The difference between organelles and endosymbionts – Response to Theissen and Martin. Curr Biol. 2006;16:R1017–R1018. doi: 10.1016/j.cub.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Archibald JM. Endosymbiosis: Double-take on plastid origins. Curr Biol. 2006;16:R690–R692. doi: 10.1016/j.cub.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Reyes-Prieto A, Melkonian M, Bhattacharya D. Minimal plastid genome evolution in the Paulinella endosymbiont. Curr Biol. 2006;16:R670–R672. doi: 10.1016/j.cub.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Shimada A, Kanai S, Maruyama T. Partial sequence of ribulose-1,5-bisphosphate carboxylase/oxygenase and the phylogeny of Prochloron and Prochlorococcus (Prochlorales) J Mol Evol. 1995;40:671–677. doi: 10.1007/BF00160516. [DOI] [PubMed] [Google Scholar]
- Watson GMF, Tabita FR. Regulation, unique gene organization, and unusual primary structure of carbon fixation genes from a marine phycoerythrin-containing cyanobacterium. Plant MolBiol. 1996;32:1103–1115. doi: 10.1007/BF00041394. [DOI] [PubMed] [Google Scholar]
- Delwiche CF, Palmer JD. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol Biol Evol. 1996;13:873–882. doi: 10.1093/oxfordjournals.molbev.a025647. [DOI] [PubMed] [Google Scholar]
- Cannon GC, Heinhorst S, Bradburne CE, Shively JM. Carboxysome genomics: a status report. Funct Plant Biol. 2002;29:175–182. doi: 10.1071/PP01200. [DOI] [PubMed] [Google Scholar]
- Badger MR, Hanson D, Price GD. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct Plant Biol. 2002;29:161–173. doi: 10.1071/PP01213. [DOI] [PubMed] [Google Scholar]
- So AKC, Espie GS, Williams EB, Shively JM, Heinhorst S, Cannon GC. A novel evolutionary lineage of carbonic anhydrase (ε class) is a component of the carboxysome shell. J Bacteriol. 2004;186:623–630. doi: 10.1128/JB.186.3.623-630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A, Becker S, Wollenzien UIA, Postius C. Ecosystem-dependent adaptive radiations of picocyanobacteria inferred from 16S rRNA and ITS-1 sequence analysis. Microbiology. 2003;149:217–228. doi: 10.1099/mic.0.25475-0. [DOI] [PubMed] [Google Scholar]
- Crosbie ND, Pockl M, Weisse T. Dispersal and phylogenetic diversity of nonmarine picocyanobacteria, inferred from 16S rRNA gene and cpcBA-intergenic spacer sequence analyses. Appl Environ Microbiol. 2003;69:5716–5721. doi: 10.1128/AEM.69.9.5716-5721.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller NJ, Marie D, Partensky F, Vaulot D, Post AF, Scanlan DJ. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl Environ Microbiol. 2003;69:2430–2443. doi: 10.1128/AEM.69.5.2430-2443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Löffelhardt W, Bohnert HJ, Philippe H, Lang BF. Monophyly of primary photosynthetic eukaryotes: Green plants, red algae, and glaucophytes. Curr Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Turner S, Pryer KM, Miao VPW, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small submit rRNA sequence analysis. J Eukaryot Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- Fewer D, Friedl T, Büdel B. Chroococcidiopsis and heterocyst-differentiating cyanobacteria are each other's closest living relatives. Mol Phylogenet Evol. 2002;23:82–90. doi: 10.1006/mpev.2001.1075. [DOI] [PubMed] [Google Scholar]
- Badger MR, Price GD, Long BM, Woodger FJ. The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J Exp Bot. 2006;57:249–265. doi: 10.1093/jxb/eri286. [DOI] [PubMed] [Google Scholar]
- Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- Rice DW, Palmer JD. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006;4:31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin K, Zetsche K. The genes of both subunits of ribulose-1,5-bisphosphate carboxylase constitute an operon on the plastome of a red alga. Curr Genet. 1989;16:203–209. doi: 10.1007/BF00391478. [DOI] [PubMed] [Google Scholar]
- Scott KM, Sievert SM, Abril FN, Ball LA, Barrett CJ, Blake RA, Boller AJ, Chain PSG, Clark JA, Davis CR, Detter C, Do KF, Dobrinski KP, Faza BI, Fitzpatrick KA, Freyermuth SK, Harmer TL, Hauser LJ, Hugler M, Kerfeld CA, Klotz MG, Kong WW, Land M, Lapidus A, Larimer FW, Longo DL, Lucas S, Malfatti SA, Massey SE, Martin DD, et al. The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol. 2006;4:e383. doi: 10.1371/journal.pbio.0040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa Y, Toyoda K, Arai H, Ishii M, Igarashi Y. CO2-responsive expression and gene organization of three ribulose-1,5-bisphosphate carboxylase/oxygenase enzymes and carboxysomes in Hydrogenovibrio marinus strain MH-110. J Bacteriol. 2004;186:5685–5691. doi: 10.1128/JB.186.17.5685-5691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Aurora R, Pakrasi HB. Critical roles of bacterioferritins in iron storage and proliferation of cyanobacteria. Plant Physiol. 2004;135:1666–1673. doi: 10.1104/pp.104.042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI BLAST http://www.ncbi.nlm.nih.gov/blast/
- van den Bergh ERE, Dijkhuizen L, Meijer WG. CbbR, a LysR-Type transcriptional activator, is required for expression of the autotrophic CO2 fixation enzymes of Xanthobacter flavus. J Bacteriol. 1993;175:6097–6104. doi: 10.1128/jb.175.19.6097-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi NR, Arai H, Kodama T, Igarashi Y. The novel genes, cbbQ and cbbO, located downstream from the RubisCO genes of Pseudomonas hydrogenothermophila, affect the conformational states and activity of RubisCO. Biochem Biophys Res Commun. 1997;241:565–569. doi: 10.1006/bbrc.1997.7853. [DOI] [PubMed] [Google Scholar]
- Hayashi NR, Arai H, Kodama T, Igarashi Y. The cbbQ genes, located downstream of the form I and form II RubisCO genes, affect the activity of both RubisCOs. Biochem Biophys Res Commun. 1999;265:177–183. doi: 10.1006/bbrc.1999.1103. [DOI] [PubMed] [Google Scholar]
- Rocap G, Distel DL, Waterbury JB, Chisholm SW. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O, Gogarten JP, Charlebois RL, Doolittle WF, Papke RT. Phylogenetic analyses of cyanobacterial genomes: Quantification of horizontal gene transfer events. Genome Res. 2006;16:1099–1108. doi: 10.1101/gr.5322306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci USA. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6 doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J, Ganatra M, Kamal I, Ware J, Makarova K, Ivanova N, Bhattacharyya A, Kapatral V, Kumar S, Posfai J, Vincze T, Ingram J, Moran L, Lapidus A, Omelchenko M, Kyrpides N, Ghedin E, Wang S, Goltsman E, Joukov V, Ostrovskaya O, Tsukerman K, Mazur M, Comb D, Koonin E, Slatko B. The Wolbachia genome of Brugia malayi : Endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, Silva FJ, Zientz E, Delmotte F, Gonzalez-Candelas F, Latorre A, Rausell C, Kamerbeek J, Gadau J, Holldobler B, van Ham RCHJ, Gross R, Moya A. The genome sequence of Blochmannia floridanus : Comparative analysis of reduced genomes. Proc Natl Acad Sci USA. 2003;100:9388–9393. doi: 10.1073/pnas.1533499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Moran NA. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteur culture collection of Cyanobacteria, Institute Pasteur, Paris, France http://www.pasteur.fr/recherche/banques/PCC/
- Sammlung für Algenkulturen, University of Göttingen, Germany http://www.epsag.uni-goettingen.de/html/sag.html
- Culture collection of algae at the University of Cologne, Germany http://www.ccac.uni-koeln.de/
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Significance measures for 14 selected clades (encircled numbers in Figure 1) in single-gene and combined analyses of rRNA genes. Analyses used 1000 bootstrap replicates of NJ (LogDet+I model), NJ (GTR+I+Γ model), and MP, and Bayesian posterior probabilities. Numbers of aligned (ch) and variable characters (var) are given for each partition.
Gene arrangement of operons containing form 1A RubisCO for all strains included in Figure 3. The table describes the typical gene arrangements in the 4 different arrangement types, defined in Figure 5, and indicates presence (x) or absence () of the gene in a specific species. Abbreviations as given in Figure 5; further abbreviations: n.d.: no data available; ham1: Ham1 like protein; ndhF3: NADH dehydrogenase subunit L; PCD_DCoH: possible pterin-4alpha-carbinolamine dehydratase; GlnK: nitrogen regulatory protein P-II; GGPS: Glucosylglycerol-phosphate synthase; Transp: transposase, mutator type; REC: response regulator receiver protein; Rpe: Ribulose-phosphate 3-epimerase; TktA: Transketolase; DedA: Uncharacterized membrane-associated protein; CBD_II: Cellulose binding domain protein. chlN: light-independent protochlorophyllide reductase subunit N.
Evidence that bacterioferritin in Paulinella and Synechococcus WH5701 was acquired by HGT from a Nitrococcus -like γ-proteobacterium. Phylogeny of three ferritin-families occuring in cyanobacteria (α-cyanobacteria in blue; β-cyanobacteria in orange colour) together with their proteobacterial relatives: Bacterioferritin, Nonheme-Ferritin, and Ferritin and Dps ("DNA-binding protein from starved cells"). The ML analysis was performed as in Figure 3, and used 159 aligned amino acid positions. The chromatophore of Paulinella and Synechococcus WH5701 (representing the Cyanobium-clade) are the only cyanobacteria, which possess bacterioferritin linked to the carboxysomal operon (see Figure 5). Similar to the RubisCO phylogeny, Nitrococcus mobilis is sister to these taxa in the bacterioferritin clade. Note that many taxa contain more than one ferritin (up to five in Synechococcus CC9311), e.g. Nitrococcus displays two unrelated bacterioferritin genes, and Synechococcus WH5701 has 1 bacterioferritin, one nonheme-ferritin, and one member of the Ferritin and Dps family (indicated by numbers in curly braces). Cyanobacterial ferritins are dispersed into several separate branches, usually nested within bacterial divergences, suggesting many independent HGT events. One to five (in CC9311) nonheme-ferritin genes are characteristic for members of all PS-subclades (presence in the Paulinella chromatophore is currently unknown), and their tree topology indicates an early gene duplication followed by later duplication/gene loss events.


