Abstract
EBV has two lytic origins (oriLyt) of DNA replication lying at divergent sites on the viral genome within a duplicated sequence (DS). The latter contains potential hairpin loops, ‘hinge’ elements and the promoters for transcripts from viral genes BHLF1 and LF3. These genes themselves consist largely of 125 and 102 bp repetitive sequences, respectively, and encode basic proteins. We have examined these genomic regions in detail in attempts to understand why lytic replication—necessary for virus survival—is so inefficient, and to identify controlling elements. Our studies uncovered a diverse family of promoters (P) for BHLF1 and LF3, only one pair of which (P1) proved sensitive to chemical inducing agents. The others (P2–P3/4), abutting the replication ‘core’ origin elements in DS and extending into 5′-unique sequences, may play roles in the maintenance of viral latency. We further identified a family of overlapping small complementary-strand RNAs that transverse the replication ‘core’ origin elements in a manner suggesting a role for them as ‘antisense’ species and/or DNA replication primers. Our data are discussed in terms of alternative lytic replication models. We suggest our findings might prove useful in seeking better control over viral lytic replication and devising strategies for therapy.
INTRODUCTION
Epstein–Barr virus (EBV), a DNA virus and member of the herpesvirus family, has infected most humans (>90%) by the time they reach adulthood, infection having occurred orally at an early age in crowded populations, but often much later in less-populous communities. EBV is best known in Western societies for its association with the time-limited disease, infectious mononucleosis (IM) and more recently with a B-cell post-transplant lymphoproliferative disease (PTLD) that can occur in EBV-positive individuals following immunosuppressive therapy. The virus is also associated with a variety of solid tumours whose types and frequencies differ in their geographical locations (1). In sub-Saharan Africa, EBV infection contributes to the development of the B-cell malignancy, Burkitt's lymphoma (BL), the most prevalent cancer among children there (1–3). In Asia, particularly southern China, EBV is associated with an epithelial cell malignancy of adults, (undifferentiated) nasopharyngeal carcinoma (NPC) (1,2). At lesser frequencies, the virus appears as a component of a variety of other malignancies worldwide (1). Both geography and viral load may play roles in the development of specific tumours (1,4). The complete sequence of the EBV genome, a tour de force in its time (5), has stimulated work on the 100 or so viral genes associated with malignancy and/or the viral life cycle, as reviewed (6). In virions, EBV DNA exists as a double-stranded linear molecule with repetitive sequences at each terminus (TR) and unique sequences separated at roughly equal intervals by internal repeats (IR1-IR4)—mimicking as such the chromosomal organization of its human host. Recombination events at the TRs lead to generation of a circular, supercoiled plasmid species, the prevalent form of EBV found in infected cells. Repeat copy numbers dictate the genome size (from 170 to >200 kb) in different strains of the virus (7).
Lytic replication, essential for virus spread, appears to occur predominantly in epithelial cells within the oropharynx (6). For a number of oncogenic DNA viruses, notably papilloma or hepatitis B, no in vitro replication system has been found for producing progeny virus. For EBV, some viral strains undergo limited spontaneous lytic replication in B-cells in culture, others can be induced to replicate (8,9), and a third category shows no response to external reagents. No fully competent in vitro lytic system has, however, ever been identified. Of its three viral origins of replication, one (oriP) serves for EBV plasmid (supercoiled circular) replication whilst the other two (oriLyt L and R) act as sites for linear lytic replication. EBV remains mainly in its plasmid form in most infected human cells, replicating once per cell cycle (10). Its oriP—which structurally resembles replication origins of a number of other DNA viruses—has been studied in considerable detail, as have the so-called ‘latent’ viral and host genes associated with replication from this site (as reviewed 11). As yet less well-defined is the lytic mode of viral replication, although early work by Hammerschmidt and Sugden (8) suggested a complexity greater than that seen with the small DNA tumour viruses, Chavallier-Greco et al. (13) identified two EBV-encoded genes (designated EB1 and EB2) with the capacity to disrupt latency in infected cells, and Anisimova et al. (14) showed n-butyrate and the phorbol ester, TPA, to facilitate this process. Current data, including those in this manuscript, appear to support a remarkably high level of complexity, which, when understood, should increase our capacity to control this human oncogenic virus.
The two distinct genomic regions (DS) directly associated with viral lytic replication are located within duplicated sequences DL and DR (where L = left and R = right on the conventional EBV physical map; see Figure 1) adjacent to open reading frames BHLF1 (in the case of oriLytL) and LF3 (for oriLytR), whose transcripts specify proteins containing tandem repeat sequences, IR2 and IR4, respectively. BHLF1 and LF3 notably are the most abundant polyA+ viral transcripts expressed, particularly in chemically induced cells (15). Whereas some EBV genomes lack one or other of the oriLyt regions, no genome—even in strains that cannot be induced to express virus—has ever been identified which lacks both. Structural elements in the genome, within the duplicated sequence (DS) regions, appear to be essential for lytic replication. One structure involves ‘palindromic’ sequences that permit hairpin loop formations—as seen with origins of replication in other DNA viruses—and another allows for the potential existence of a ‘hinge’ (H) structure, with single- and triple-stranded elements (16). Expression of several viral transactivator functions that induce transcriptional expression of EBV lytic cycle-related early genes (17–19), the transcripts themselves (8) or a gene product that exports mRNA from the nucleus (19), also appear to be essential. The need for two lytic origins of replication has not been defined but, notably, another human herpesvirus, Kaposi sarcoma herpesvirus (KSH or HHV-8) also possesses two functional lytic origins of DNA replication (20). The topic of EBV lytic replication has been recently reviewed (12,21).
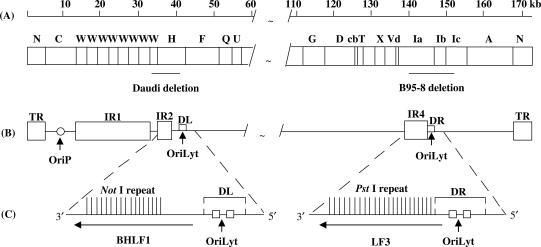
Figure 1.
EBV genome features associated with DNA lytic replication. Features shown are: (A) Truncated linear representation of the viral genome with map units relating to nucleotide sequence numbers (accession number AJ507799). BamHI DNA restriction fragments are conventionally designated alphabetically, according to size and site on the genome; locations of gene deletions (solid lines) in some viral isolates, as noted, are shown. In Daudi cells, a deletion removes the EBV BHLF1 gene and its promoter elements, as indicated; a smaller deletion in P3HR-1 (not shown) removes the gene but not its promoter sequences (31). In B95-8 cells, a deletion removes EBV LF3 coding and promoter elements. (B) A simplified version of the genome, indicating locations of tandem repetitive sequences IR1 (in BamHI W), IR2 (in BamHI H), IR4 (in BamHI Ia), as well as the terminal repeats (TR), the plasmid (oriP) and two lytic (oriLyts) origins of replication—the latter two within duplicated regions, DL and DR. (C) Expanded genomic regions indicating compositions and polarity of the viral genes, BHLF1 and LF3 that contain repeats which can be cleaved with NotI and PstI restriction enzymes, respectively, also the two oriLyt ‘core’ regions (boxes; see ref. 8). 3′-5′ polarities are used here since both BHLF1 and LF3 are expressed in a leftward orientation on the conventional EBV physical map, as given in (A).
Our initial interest in the regions encoding BHLF1 and LF3, immediately adjacent to the viral oriLyts, was stimulated by the fact that they may represent potentially important therapeutic targets in EBV-associated tumours. Their expression can be identified in subpopulations of cells in many such tumours (22,23), in a manner partly dependent upon the nature of the host cell. The basic (arginine/histidine-rich) protein product of BHLF1 has been shown to bind, preferentially, single-stranded DNA, and a structurally similar protein within LF3 postulated to behave in the same manner (24,25). Studies aimed at localizing ‘core replication’ elements identify a number of viral genes that are functionally relevant to lytic replication (26,27). Unlike the situation with the small DNA viruses, however, here no dedicated origin-binding protein was identified. More recently the work of Baumann et al. (28) has implicated cellular transcription factors that act as ‘bridging’ proteins for replication complexes and play essential roles in viral lytic replication. Earlier, Metzenberg (29), carrying out ‘run on’ transcription experiments on isolated infected cell nuclei, noted that in some instances as much as 50% of the transcriptional expression of BHLF1 appeared to come from 5′ regions ‘upstream’ of an identified promoter. Interestingly, this topic was not apparently further pursued.
Early studies using M-ABA cells (15) focused on expression of BHLF1 and LF3 controlled from a pair (P1) of promoter-related sites located in DS, in regions contiguous to the genes. Most studies relating to their transcriptional expression, in that they use cell lines (B95-8, Daudi, P3HR-1) carrying deletions of one or other of the genes (e.g. refs. 16, 29-31), do not allow for comparative expression studies of the two sites to be made. The tacit assumption appears to be that their genes and expressions are interchangeable. However, control sequences for BHLF1 and LF3 lie within different genetic environments (Figure 1) and our earlier studies identified host-cell differences in expression in virus-associated tumours, indicative of non-identity (22,23). Moreover, the genes themselves express non-identical basic proteins. Because of their presumed relevance to the viral life cycle, and their potential for exploitation, we have here examined expression control for BHLF1 and LF3 in parallel, both in cell lines and fresh EBV-positive tumours of B- or epithelial-cell origins, most of which retain the genetic information for both genes. Our data reveal for them—in addition to the P1 pair of promoter-related sequences (15)—a pattern of multiple promoters that can only in part be superimposed. These promoters, unlike P1, are not susceptible to chemical induction, yet appear to be functionally active and specify transcripts relating to BHLF1 and LF3. In addition, in the present study, we present data showing the complementary strand also to be transcriptionally active and to encode a family of small (antisense) RNA transcripts initiating in DS regions, associated with the oriLyts. We have confirmed host-cell expression differences between the two genes and ‘revisit’ the topic of lytic EBV replication with the aim of using the new data for obtaining better control over it.
MATERIALS AND METHODS
Tumour biopsies, cell lines and propagation
Primary NPC biopsies were obtained from Prof. M.H. Ng (Department of Microbiology) and Dr J. Nichols (Department of Pathology), Queen Mary Hospital, Hong Kong University, and NPC xenografts (C15, C17 and C18) from Dr P. Busson, Villejuif, France. The latter were propagated in athymic (nude) mice as described (32). Induction of EBV in the C15 xenograft has been described elsewhere (22). Surgically removed lymphoma biopsies, provided by Prof. E. Borgstein, the Medical College, Blantyre, Malawi, were identified as Burkitt's (non-Hodgkin's) lymphoma (BLs) by histopathology. CC is an EBV negative African lymphoma, from the same source. All tumours were suspended in maintenance media (foetal calf serum/DMSO) and stored in liquid nitrogen prior to use. BL-derived EBV-positive cell lines studied for expression of lytic-related genes, as described (23), were of marmoset (B95-8, M-ABA—generated with human EBV) or human (Raji, Daudi, P3HR-1, BL74—tumour-derived) origins; Ramos is an EBV-negative B-cell line. Cells were grown in suspension culture under standard conditions, with or without chemical induction, as described (22).
Isolation of RNA, cDNA synthesis and RT-PCR amplification
Total RNAs from fresh biopsies, xenografts or cell lines were isolated by the guanidinium–caesium chloride method, and polyadenylated (polyA+) RNAs selected on oligo(dT) mRNA purification columns (Pharmacia), as described (22). RNAs were quantified by uv-spectrophotometry (1.0 uv-absorbance reading for RNA at 260 nm corresponding to 40 µg/ml for single-stranded RNA). To remove any residual DNA, total (5 µg) or (polyA+) (1 µg) RNAs were treated with 10 U DNaseI (Boehringer) in 10 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, briefly mixed, incubated at 37°C (30 min) and the solution extracted (2×) with phenol-chloroform. The aqueous layer was made 0.3 M with NaOAc and RNAs precipitated with ethanol. To control for the DNAse digestion, pure genomic DNAs (0.1 µg) from tumour cells or cell lines were similarly treated with DNaseI and the products, DNA (T), used in PCR experiments as controls to monitor for the amplification of possible contaminating DNA. To detect BHLF1 and LF3 transcripts, expressed from different promoters by reverse transcriptase polymerase chain reaction (RT-PCR) techniques on the respective RNAs, first strand cDNAs were generated using gene-specific primers, that is: for BHLF1, 5′-GGTCCCCATGGCACAGGCCTAG (40261–40280; H-1T) and for LF3, 5′-CTGCAGCCGGGTCCGGGGTT (143189–143208; Ia-IT).
The resulting cDNA was diluted (100 μl) in l0 mM Tris–HCl (pH 8.0), 0.1 mM EDTA and amplified using a variety of primers taken either from the top (designated T) or bottom (B) strands of the conventional map of EBV.
For BHLF1, these (given in 5′–3′ orientations) are as follows:
GCCCAGCGCGCCCCGTTCA (40303–40321; H-2T);
GGGAGGACCGCGGCCGAGCCA (40325–40345; H3T);
GAGCAAGAATAAGGACGGCTC (41448–41468; H-4T);
GGTTAGTGATGAAACAGGCAAC (41514–41493; H-5B);
CGCTCGGGGGGTGCACACCT (40485–40466; Ia-3B);
GCTCCGCCTACCCCAAATCTC (40728–40708; Ia-4B);
CATTATCCTGGAGGTATCCTAAG (40833–40855; Ia-5T);
GGGGCGGAGCTTAGGATACCTC (40862–40841; Ia-5B);
GAAGGGTGGCGCACCTTAAGG (40952–40932; Ia-6B);
CACGCGGGGTGCCACGTCACC (41164–41144; Ia-7B);
TTGCCTGCCTCACCATGACAC (41345–41365; Ia-6T).
For LF3, they are as follows:
GGTCCGGGGTTCCGGCCCTG (143198–143217; Ia-2T);
CTCCGGCGGGGATGGGGGTGC (143271–143291; Ia-3T);
CTCCGGCGGGGGGTGGGGATG (143271–143291; C15-3T);
CGCTCGGGGGGTGCACACCT (143403–143384; Ia-3B);
GCTCCGCCTACCCCAAATCTC (143647–143627; Ia-4B);
CATTATCCTGGAGGTATCCTAAG (143752–143774; Ia-5T);
GGGGCGGAGCTTAGGATACCTC (143783–143762; Ia-5B);
GAAGGGTGGCGCACCTTAAGG (143871–143851; Ia-6B);
CACGCGGGGTGCCACGTCACC (144082–144062; Ia-7B);
TTGCCTGCCTCACCATGACAC (144246–144266; Ia-6T);
ACCCTCACCCCATTGCCAACT (144400–144380; Ia-8B);
GCGCCCATTAGAATCTGCTCG (144669–144649; Ia-9B);
GATTCTATTAATAAAACAAGAGAG (144704–144681; Ia-10B);
AACAGGTGTGCAGGTGTGCAT (144791–144771; Ia-11B).
Sequences were chosen by their proximity to potential ‘TATA’ box-like sequences in the DNA and genome numbers taken from a composite of B95-8 (5) and Raji (33) sequence that accommodates the EBV genomic deletion in the former, accession number AJ507799. For RT-PCR experiments, products were separated by electrophoresis and identities in most cases verified by Southern blot hybridization using appropriate radioactive probes (from the above list; see Figure legends) as previously described (23).
RNase protection assays
Construction of plasmids H-390, Ia-382, and Ia-330 DNAs
DNA templates for the synthesis of RNA probes, prepared from DNA fragments H-390 (positions 40266–40657 in BamHI H), Ia-382 (positions 143193–143575 in BamHI Ia) and Ia-330 (positions 143752–144082 in BamHI Ia) were introduced into multicloning sites of the Bluescript SK vector. To facilitate RNA synthesis, plasmid DNAs were linearized by appropriate enzymes, depending on the insert orientation, in the multicloning site between the insert and the T3 or T7 promoters. Riboprobes, oriented from left to right on the conventional map of the EBV genome were generated for identifying RNA initiation sites off different promoters.
Construction of plasmid Ia-406 DNA
To explore transcriptional expression from the strand complementary to the LF3 gene, a cDNA clone, 14A, containing the IR4 repeat sequences from a C15 xenograft cDNA library (34), cloned into the pBIIKS vector, was cleaved by XbaI and transcribed by T3 RNA polymerase to generate a riboprobe oriented from ‘right to left’ on the EBV genome. Hybridization of this probe with total RNAs from BL-derived cell lines, or from C15, produced negative results in all cases, confirming that there is no transcript on the opposite (top) strand within the IR4 repeat region itself, in the EBV genome. To search for transcripts expressed on the complementary (top) strand upstream of this repeat, within the promoter (P1) region (15), the above experiments were repeated with a DNA fragment Ia-406 (positions 143781–143375 in BamHI Ia) using the same vector, restriction enzyme and T3 RNA polymerase, to generate a riboprobe also orientated from ‘right to left’ on the genome, within the DR region containing EBV oriLyt sequence. This probe was used in RNAse protection assays to hybridize with total RNAs from various cell lines.
Radiolabelling of riboprobes by in vitro transcription of plasmid DNA (adapted from ref. 35)
Single-stranded RNA probes were generated from linearized plasmid DNAs with DNA-dependent RNA polymerase: Linearized template DNA (0.6 μg) was dissolved in DEPC-H2O (10 µl) in a 1.5 ml RNase free Eppendorf tube, together with 10× transcription buffer (2 µl) (Promega), 0.1 M DTT (2 µl), rNTP mix (A, U, G = 5 mM, C = 0.5 mM) (2 µl) and RNase inhibitor (1 µl; 10 u/µl). [α-32P]-CTP (2 µl; 40 µCi/µl) and RNA polymerase (1 µl; 20 u/µl) were added with mixing, and the reaction incubated at 37°C for 1 h. DNase I (1 µl of 1 μg/μl) was then added and the reaction further incubated for 15 min at 37°C to digest template DNA. Proteins were removed by extraction with an equal volume of phenol-chloroform. To purify residual synthesized RNA, the aqueous layer was transferred into a new tube. 3 M NaOAc (0.1 vol.) and ice-cold ethanol (2 vols.) were added to precipitate the RNA and, after 30 min at −20°C, the tube was centrifuged at 12 000g (10 min) at 4°C and the ethanol supernatant removed. The opened tube was left at room temperature (several minutes) to allow residual ethanol to evaporate, the RNA pellet was dissolved in TE buffer (200 µl, pH 7.5), ice-cold ethanol (2 vol.) was added, and the sample stored at −70°C prior to use. For hybridization, a sample aliquot was withdrawn, 3 M NaOAc (0.1 vol.) was added, and the tube left −20°C (30 min), then centrifuged. After ethanol removal, the radiolabelled RNA probe was dissolved in a desired volume of hybridization buffer from a RPAIII kit (Ambion Biotechnology).
Hybridization and RNase digestion
Total RNAs (100 μg from M-ABA and C15, and 30 μg from other cells) were hybridized with radiolabelled RNA probes (see earlier). The products were digested with RNase and purified with an RPAIII kit (Ambion Biotechnology), according to the manufacturer's instructions.
Autoradiography of RNase-protected products
Protected fragments were suspended in electrophoresis loading buffer, boiled (5 min), chilled on ice, centrifuged briefly, then separated by electrophoresis on a 6% polyacrylamide gel at 1 kV (28 mA) for 2–3 h. Simultaneously, comparable amounts of unhybridized and undigested RNA probe were diluted 500–1000 times in loading buffer, and electrophoresed to allow the full length RNA probe size to be identified. After electrophoresis, the gel was fixed in 10% acetic acid (20 min), dried under vacuum and autoradiographed.
RESULTS
Transcriptional expression of BHLF1 (IR2-containing) and LF3 (IR4-containing) genes—general findings
Published work (16, reviewed 12, 21) has focused mainly on expression of the BHLF1 gene—deleted in toto in the BL-derived cell line Daudi (31) as shown in Figure 1. Its translation product has been identified primarily as a single-stranded DNA binding protein (25); a similar property was proposed for the LF3 gene product (24). In our earlier work (22), transcriptional expression of BHLF1 was identified in primary endemic Burkitt's lymphoma (BL) biopsies and in most BL-derived cell lines, but not in several Asian nasopharyngeal carcinoma (NPC) biopsies, nor in two North African NPC xenografts, C17 and C18 (32). By comparison, LF3, except for B95-8 (gene deleted), appeared to be transcriptionally expressed in both epithelial and B-cell malignancies.
To confirm these data, we looked at transcriptional expression in a greater variety of materials, using RT-PCR assays that allowed us to make comparative analyses of gene expression of BHLF1 and LF3 in EBV-associated fresh tumour biopsies, as well as in B-cell lines. In the case of BHLF1, again—with the exception of a late passaged North African NPC xenograft, C15 (32)—no expression was observed in epithelial (carcinoma) cells from similar xenografts, C17 and C18, nor seven new Chinese NPC biopsies. Interestingly, this transcript was identified in the M-ABA B-cell line, generated with virus from an Asian NPC, but was not identified in a comprehensive cDNA library from an early passaged C15 tumour (34), indicative of alterations in EBV gene expression in different cellular environments, or when subjected to pressures during tumour passage in vivo. In comparable studies, 11 out of 12 new African BL samples gave bands consistent with BHLF1 gene expression; identities could be confirmed in 10 cases by hybridization. The data (not given, but summarized in Tables 1 and 2) also showed expression levels among these BLs to vary considerably, from very high to barely detectable, using comparable quantities of products. All BL-derived cell lines, except for Ramos (EBV-negative) and Daudi which served as controls, were positive. Recent sequence data on EBV virus from an NPC patient (36) or from an A-strain virus (37) show sequences in this region of the viral genome to be highly conserved and to resemble strongly those reported earlier (5,33). Since our protocols also allowed for minor sequence variations, the data must have reflected either varying viral loads in different tumours, or a marked difference in levels of cells escaping latency.
Table 1.
RT-PCR results on EBV BHLF1 transcripts in tumour biopsies and cell linesa
| Promoter-related | (P1) | (P2) | (P3) |
| primers | H-1T + Ia-3B | H-2T + Ia-4B | Ia-6T + H-5B |
| Tumours and | Products | ||
| cell lines | 40261–40485b | 40303–40728 | 41345–41514 |
| C15 | ++c | + | + |
| C17 | − | − | − |
| C18 | − | − | − |
| NPC1 | − | − | − |
| NPC2 | − | − | − |
| NPC3 | − | ND | ND |
| NPC4 | − | ND | ND |
| NPC5 | − | ND | ND |
| NPC6 | − | ND | ND |
| NPC7 | − | ND | ND |
| C15 DNA | ++ | ++ | ++ |
| CC | − | − | − |
| BL-1 | ++ | − | − |
| BL-2 | ++ | − | − |
| BL-3 | ++ | + | − |
| BL-4 | + | − | − |
| BL-5 | + | − | − |
| BL-6 | +++ | ND | ND |
| BL-7 | − | ND | ND |
| BL-8 | + | ND | ND |
| BL-9 | + | ND | ND |
| BL-10 | + | ND | ND |
| BL-11 | ++ | ND | ND |
| BL-12 | ++ | ND | ND |
| B95-8 DNA | ++ | ++ | ++ |
| Daudi(−) | − | − | ++ |
| Daudi(+) | − | − | ++ |
| M-ABA(−) | ++ | ++ | ++ |
| M-ABA(+) | ++ | ++ | ++ |
| Raji(−) | ++ | ++ | ++ |
| Raji(+) | ++ | ++ | ++ |
| P3HR-1(−) | ++ | ++ | ++ |
| P3HR-1(+) | ++ | ++ | ++ |
| B95-8(−) | ++ | ++ | ++ |
| B95-8(+) | ++ | ++ | ++ |
aRT-PCR products confirmed by Southern blot hybridizations, and in some cases by RNase protection experiments.
bFrom accession number AJ507799.
c+++, very strongly positive; ++, strongly positive; +, weakly positive, −, negative, as determined by hybridization under identical conditions and ND, not done, respectively.
Table 2.
RT-PCR results on EBV LF3 transcripts in tumour biopsies and cell linesa
| Promoter-related | (P1) | (P2) | (P2) | (P3) | (P4) | (P4) | (P4) | (P4) |
|---|---|---|---|---|---|---|---|---|
| Primers | Ia-2T + Ia-3B | Ia-2T + Ia-4B | Ia-2T + Ia-5B | Ia-2T + Ia-6B | Ia-6T + Ia-8B | Ia-6T + Ia-9B | Ia-6T + Ia-10B | Ia-6T + Ia-11B |
| Product | ||||||||
| Tumours and cell lines | 143198– 143403b | 143198– 143647 | 143198– 143783 | 143198– 143881 | 144246– 144400 | 144246– 144669 | 144246– 144704 | 144246– 144791 |
| C15 | ++c | + | + | + | + | ND | + | − |
| C17 | + | + | + | + | − | ND | − | − |
| C18 | + | + | + | + | − | ND | − | − |
| NPC1 | + | + | + | + | + | ND | + | − |
| NPC2 | + | + | + | + | + | ND | + | − |
| NPC3 | + | ND | ND | ND | ND | ND | ND | ND |
| NPC4 | + | ND | ND | ND | ND | ND | ND | ND |
| NPC5 | + | ND | ND | ND | ND | ND | ND | ND |
| NPC6 | + | ND | ND | ND | ND | ND | ND | ND |
| NPC7 | + | ND | ND | ND | ND | ND | ND | ND |
| C15 DNA | ++ | ++ | ++ | ++ | ++ | ND | ++ | ++ |
| CC | − | − | − | − | − | ND | − | − |
| BL-1 | + | + | + | + | + | ND | − | − |
| BL-2 | ++ | + | + | + | + | ND | − | − |
| BL-3 | + | + | + | + | + | ND | − | − |
| BL-4 | + | + | + | + | + | ND | − | − |
| BL-5 | + | + | + | + | + | ND | − | − |
| BL-6 | + | ND | ND | ND | ND | ND | ND | ND |
| BL-7 | + | ND | ND | ND | ND | ND | ND | ND |
| BL-8 | + | ND | ND | ND | ND | ND | ND | ND |
| BL-9 | + | ND | ND | ND | ND | ND | ND | ND |
| BL-10 | + | ND | ND | ND | ND | ND | ND | ND |
| BL-11 | + | ND | ND | ND | ND | ND | ND | ND |
| BL-12 | + | ND | ND | ND | ND | ND | ND | ND |
| Daudi DNA | ++ | ++ | ++ | ++ | ++ | ND | ++ | ++ |
| B95-8 | − | − | − | − | − | − | ND | − |
| M-ABA (−) | + | + | + | + | + | (+) | ND | − |
| M-ABA (+) | ++ | − | − | − | − | − | ND | − |
| Raji (−) | ++ | + | + | + | − | − | ND | − |
| Raji (+) | ++ | + | + | + | − | − | ND | − |
| Daudi (−) | +++ | ++ | ++ | ++ | + | + | ND | − |
| Daudi (+) | +++ | ++ | ++ | ++ | + | + | ND | − |
| P3HR-1(−) | +++ | ++ | ++ | ++ | + | + | ND | − |
| P3HR-1(+) | +++ | ++ | ++ | ++ | + | + | ND | − |
| Daudi DNA | ++ | ++ | ++ | ++ | ++ | ++ | ND | ++ |
aData with Ia-6T + Ia-8B based on ethidium bromide stained gels, all other products confirmed by Southern blot hybridisation of RT-PCR products.
bfrom accession number AJ507799.
c+++, very strongly positive; ++, strongly positive; +, weakly positive; −, negative, as determined by hybridisation data under identical conditions, and ND (not done), respectively. Some data taken from (22,23).
LF3 transcriptional expression, by contrast, using the same tumour populations and cell lines, was observed in all NPC and BL biopsy materials, as well as in the B-cell lines—except Ramos and B95-8—at levels that were generally more homogeneous than seen in the case of BHLF1.
These data confirm the cell-type bias in gene expression for BHLF1 suggested earlier (22,23) and the apparent non-cellular restricted transcription of LF3. In the M-ABA cell line, transcriptional expression from a pair of contiguous promoters (P1) was reported as being dependent on the presence of chemical inducing agents (15). With the sensitive RT-PCR assays, we identified transcripts for BHLF1 and LF3, even in the absence of induction. Our Northern blot data (22,30) on B-cell lines showed, however, that chemical induction, although not absolutely essential, generally increased transcription levels, in some cases to a very considerable degree. The RT-PCR data reported earlier (22) and confirmed here hinted, in addition, at usage of transcriptional promoters ‘upstream’ (that is, 5′) of P1, a topic followed up in subsequent studies reported here.
5′ ends of BHLF1 and LF3 mRNAs, as identified by RNase protection assays (RPA)
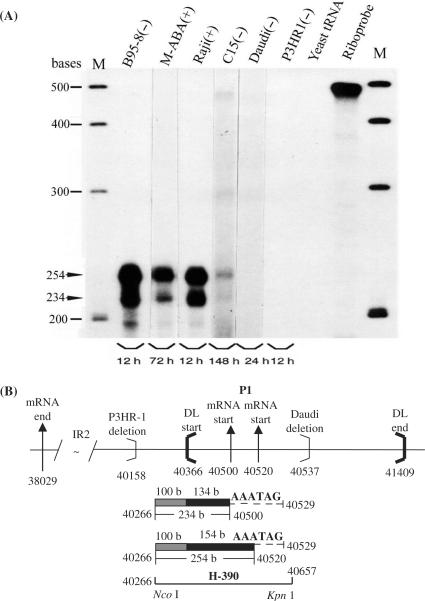
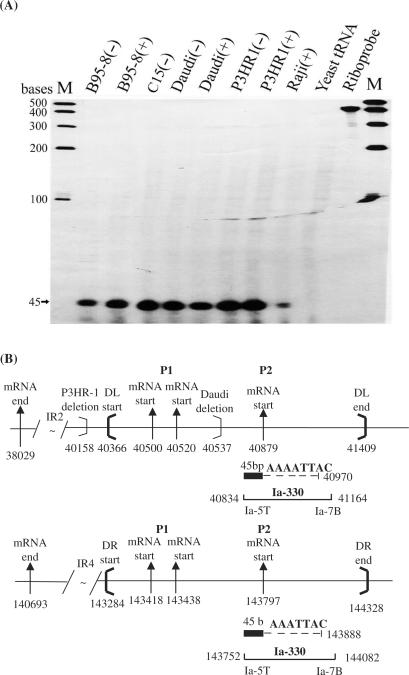
To explore transcriptional initiation sites for BHLF1 and LF3 genes, RNase protection experiments were carried out using B-cells grown in the absence (−) or presence (+) of chemical inducing agents. For BHLF1, the RPA (Figure 2A), when hybridized with a 32P-radiolabelled 490 base riboprobe, H-390 (positions 40266–40657) (Figure 2B) from the B95-8 genome, identified two closely spaced (20 nt apart) RNase-protected bands, arising from P1 and previously described in M-ABA cells (15), in all except Daudi and P3HR-1 cells. The protected fragments mapped mRNA initiation sites from this P1 pair to genome positions 40520 and 40500, respectively, within the DL region of the viral genome. The levels of protected RNA in reproducible experiments were of unequal intensities in different lines, as indicated by autoradiograph times given on the gels (Figure 2A), the larger transcript (upper band) being the more abundant of the two RNAs. In uninduced C15 NPC, the smaller band was only evident on very long exposures.
Figure 2.
P1 promoter usage for BHLF1 transcription, mapped by RPA. RNase protection experiments carried out on total RNAs from EBV-positive B-cell lines and the C15 NPC xenograft, as indicated, to re-examine P1 promoter usage for BHLF1 in a semi-quantitative manner, and in relation to LF3 (Figure 3). (A) Protected products from chemically uninduced (−) or induced (+) cells, as indicated, run on gels and identified using the 32P-radiolabelled cloned riboprobe, H-390. Bands at 254 and 234 bases (arrows) were obtained from all except substrates from Daudi, P3HR-1 and the yeast negative control. Intensities of the larger protected fragment are ca. 4× that of the smaller product suggesting that, of the P1 pair, the site further from the gene itself is the stronger. Gels shown were developed at times indicated to illustrate comparable cellular product levels, in all except C15, where bands were consistently weak. (Not shown are smaller cross-hybridizing protected bands relating to LF3, observed with all but B95-8 substrates). M = molecular weight size markers. (B) Diagram (with nucleotide numbers) showing features of the BHLF1 gene, including its mRNA 5′-ends as initiated from P1 (doublet, vertical arrows), 3′-ends of deletions found in P3HR-1 and Daudi cells (light brackets) and site locations of the DL termini (heavy brackets). The location and nucleotide numbers of the RNA probe, H-390, and the protected 234 and 254 base products (in 2A) lying within DL (134 and 154 bases, solid bars) or within BamHI unique sequence (100 bases, hatched bars), are indicated. Location of the promoter signal probably controlling transcriptional expression of BHLF1 (5′GATAAA, position 40529) is indicated by dashed lines.
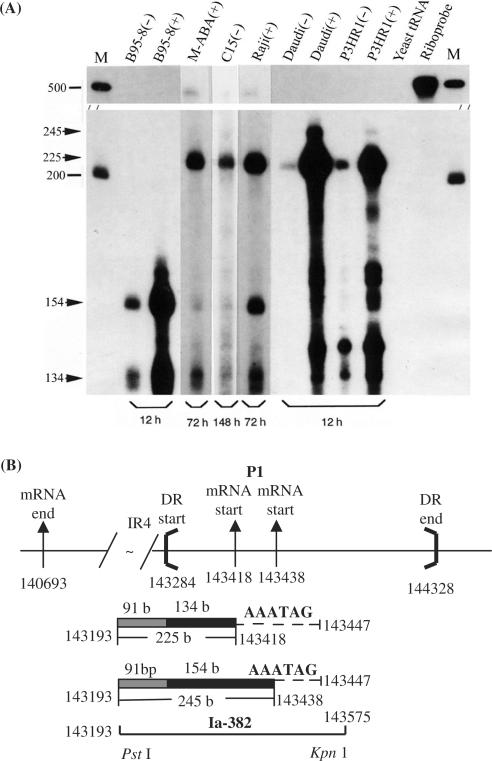
Similar RPA experiments were carried out with respect to LF3 transcripts (Figure 3A), using a radiolabelled 500 base riboprobe, Ia-382 (encompassing genome positions 143193–143575, partly within DR, Figure 3B). They identified one major protected fragment (225 nt) in all samples except that from B95-8 cells. In RNAs from induced cells, a minor, larger, band (245 nt) was also observed in Daudi and P3HR-1 cells. A negative result with B95-8 is consistent with this product coming from LF3, rather than the BHLF1 region of the genome (Figure 1). In the lower portion of this gel, bands (in B95-9, M-ABA and Raji cells) are attributable to cross-hybridization with transcripts expressed from DL whilst bands observed in Daudi and P3HR-1, with aberrant sizes, are likely to be cross-reacting artefacts. The data for LF3 transcripts are consistent with Northern blot hybridization data (22) and suggest that the first (P1 pair) promoter site for LF3, although sensitive to chemical induction, is also active to a lesser extent in uninduced B-cells and in the C15 tumour. Also for LF3, in contrast to BHLF1, the smaller RNA (lower band) of the P1 pair is found to be the more abundant in all cells examined.
Figure 3.
P1 promoter usage for LF3 transcription, mapped by RPA. (A) RNase protection data, using cell lines, as indicated, here exploring further the role of chemical induction on transcription by including pairs of corresponding RNAs from uninduced (−) and induced (+) cells, as indicated. With the 32P-radiolabelled Ia-382 riboprobe, a major protected LF3 band of 225 bases, highly sensitive to chemical induction, was observed (in all but B95-8 cells), and very minor bands (at 245 bases) are seen in induced Daudi and P3HR-1 cells. Notably, for LF3, as opposed to BHLF1 (Figure 2A), the smaller product predominates, suggesting the major use of the P1 pair nearest the gene. Bands at 154 and 134 bases, also present in B95-8, reflect cross-hybridization of the probe with the DL BHLF1 transcript. (Random-sized bands in Daudi and P3HR-1 cells are artefacts of the protocol). (B) Diagram (with nucleotide numbers) showing features, as noted in the legend to Figure 2, using sizes and numbers that correspond to the LF3 gene. The location and nucleotide numbers of the RNA probe, Ia-382, and the protected products of 225 and 245 bases (in 3A) lying within DR (134 and 154 bases, solid bars) or within BamHI Ia IR4 repeat (91 bases, hatched bars), are indicated. Location of the promoter signal possibly controlling the transcriptional expression of LF3 (5′GATAAA, position 143447) is indicated by dashed lines. The deletion in B95-8 (not shown) covers this entire region.
Identification of unique multiple ‘upstream’ transcriptional initiation sites for BHLF1 and LF3 genes
In RT-PCR confirmatory experiments, in addition to P1 some minor higher molecular weight band products, lying in the ‘core’ lytic origin sequences, were identified, both in ethidium bromide stained gels and blots hybridized with 32P-radiolabelled probes, suggesting that RNAs were also being expressed from promoters 5′ to P1. To explore ‘upstream’ transcriptional expression (that is, 5′ to P1 in the leftward transcripts) at what might prove low levels, since it has not been previously reported, we used RT-PCR initially as a diagnostic protocol because of its sensitivity. Data covering these regions of BHLF1 and LF3 are shown in Figure 4, panels A and B, respectively.
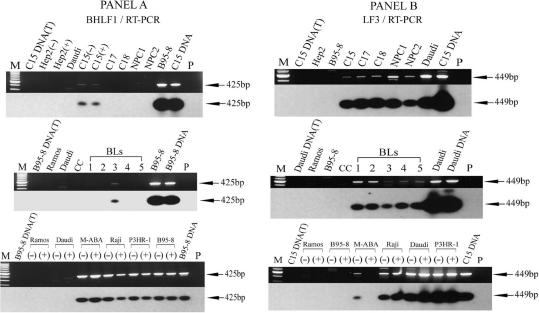
Figure 4.
RT-PCR detection of upstream promoters, P2, for expression of BHLF1 and LF3. An RT-PCR technique was adapted to identify potential transcriptional expression of BHLF1 and LF3 from regions upstream of P1. Combinations of H-2T and Ia-4B (for BHLF1), and Ia-2T and Ia-4B (for LF3) were used for the PCR amplifications; products were hybridized with 32P-radiolabelled H-3T (for BHLF1), and a combination of Ia-3T and C15-3T (for LF3) to accommodate minor sequence alterations observed with C15 (our data). Panel A. Promoter ‘upstream’ usage for BHLF1, identified as 425 bp products from uninduced (−) or induced (+) cells, as indicated. There is no evidence that chemical induction alters transcription from this site, designated P2. Except for the C15 xenograft and BL-3, P2 was not apparently active in other tumour samples (Table 1) as opposed to cell lines. Panel B. Similar protocols used for studying ‘upstream’ promoter usage for LF3. A product of 449 bp was obtained from all substrates [except those from B95-8, induced (+) M-ABA cells or EBV-negative controls]. Transcripts in these panels correspond to usage of P2, a promoter-related site that is not responsive to, and may even be down-regulated (see M-ABA data) by, chemical agents that induce transcriptional expression from P1.
For BHLF1, among the NPCs examined (panel A), bands coming from a promoter we designate P2 were—as with P1—only identified in C15 xenografts. Using tumours propagated in the absence (−) or presence (+) of chemical inducing agents, the data showed no evidence that this RNA product was subject to chemical induction. Similar findings were obtained in all the B-cell lines examined (B95-8, M-ABA, Raji and P3HR-1) where this gene is present. It was not observed in Daudi cells, but one of the primers used, H2T, lies within the deletion in these cells (31, see Figure 1). Whilst active in established B-cell lines, P2 usage was only observed in 1/5 of the tumour cells—BL3 being the single exception. We have at present no explanation for this finding, although heterogeneity among primary BLs is not uncommon (3).
Similar experiments on P2 usage for LF3, showed little difference among the samples populations, whether from NPC or BL primary tumours, or established B-cell lines (Figure 4, panel B). In established lines using both uninduced (−) and induced (+) cells, no differences in usage were apparent, suggesting promoter insensitivity to the action of these agents. The exception to this is M-ABA, where the promoter appears to be negatively regulated. Throughout, RNAs from B95-8 were negative, confirming the nature of the transcript as being derived from LF3.
Confirmation of the second (P2) promoter by RPA
RNase protection assays were carried out to confirm the use of a P2 promoter-related sequence for expression (in part) of BHLF1 and for LF3, to determine the respective transcript 5′-end initiation sites—using a 32P-radiolabelled RNA probe, plasmid Ia-330 (position 143752–144082), constructed for this purpose from Raji DNA (33)-and confirm its apparent insensitivity to inducing agents. Hybridization of total RNAs following RNase digestion using this 440 base riboprobe from the DS region revealed a single fragment of 45 bases in all of the cell lines employed, as shown (Figure 5A), mapping the transcript initiation site for BHLF1 to DL, at position 40879; for LF3, this site in DR maps to position 143797 (Figure 5B). The data show transcripts from P2 in different cells to be expressed from non-inducible promoters, suggesting that they may be regulated in a manner distinguishable from transcripts off P1 promoters which are generally sensitive to chemical induction.
Figure 5.
RPA confirming ‘upstream’ P2 promoter usage for BHLF1 and LF3. (A) Total RNAs from chemically uninduced (−) or induced (+) cell lines (as noted), hybridized with a 440 base riboprobe transcribed from the DNA clone Ia-330. Bands at 45 bases were protected from RNase degradation in transcripts from all samples, except the yeast tRNA control. The data identify usage of the ‘upstream’ promoter, P2, in all EBV-positive cells examined, and are consistent with P2, unlike P1, being non-responsive to chemical inducing agents. (B) Schematic diagram showing the gene structure of BHLF1 and LF3, locations and nucleotide numbers of the riboprobe Ia-330, and the RNase protected 45 base product within DS. For BHLF1 and LF3, P2 transcription is initiated at positions 40 879 and 143 797, respectively. The transcriptional controlling signals for P2 is probably from ‘CATTAAA’ boxes at positions 40 970 (for BHLF1) or 143 888 (for LF3).
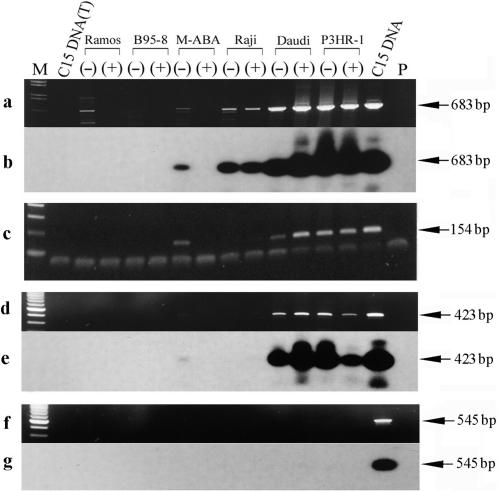
Mapping LF3 promoters P3 and P4 by RT-PCR
To ask whether there are transcripts being expressed off promoters beyond P2, RT-PCR experiments were further carried out using several different sets of primers and 32P-labelled probes, as found in ‘Materials and Methods’, for examining LF3 expression. As shown (Figure 6, panels a and b), when a combination of primers, Ia-2T and Ia-6B, was used, the same expression profile was observed as seen in Figure 4 (panel B, bottom tracts, using primers Ia-2T and Ia-4B). Since primer Ia-6B is located ‘upstream’ of the 5′-end of the RNA transcribed from the P2 promoter, these results demonstrate that transcripts are still being expressed from a third promoter, P3, in these cell lines. When the combination of Ia-6T and Ia-8B was used (panel c) Raji cells gave negative results, suggesting the existence of a P3 promoter between nucleotide positions 143 851 (Ia-6B) and 144 400 (Ia-8B). Positive bands with Daudi, P3HR-1 and uninduced M-ABA cells suggested that there should be a fourth promoter, P4, for LF3, used by these cells, but not by Raji. When the combination of primers Ia-6T and Ia-9B was used, and product probed with 32P-radiolabelled Ia-8B, positive bands were obtained in comparable amounts in Daudi and P3HR-1 lines (uninduced or induced) but only at low levels in uninduced and not at all in induced M-ABA cells (Figure 6, panels d and e). As there is a TATA box at position 144 720, this signal probably directs use of the fourth promoter for the leftwardly expressed LF3 RNAs, from unique sequence in the genome. Similar experiments carried out with a combination of Ia-6T and Ia-11B (panel f), hybridized with radiolabelled Ia-8B, were negative (panel g), setting an upper limit for transcriptional expression of LF3 at genome position 144 704.
Figure 6.
Mapping of the LF3 promoters P3 and P4 by RT-PCR. cDNAs were synthesized by reverse transcription of total RNAs from cell lines, as indicated, using a gene-specific primer Ia-1T. Primers used for PCR amplifications to detect RNA species ‘upstream’ of the P2 promoter are indicated (below) and their locations on the EBV genome map given in ‘Materials and Methods’. Panel a shows ethidium bromide stained gel results using primers Ia-2T and Ia-6B in PCR amplifications to detect a possible third promoter, P3, and panel b the same gel hybridized with 32P-labelled Ia-3T. Panel c confirms the data in a using a different set of primers, a combination of Ia-6T and Ia-8B. Notably, this P3 promoter was not identified in the case of BHLF1 (see ‘Results’ section). To detect a possible fourth promoter, P4, for the LF3 transcript, the same cDNA samples were used in further experiments given in panels d–g. In panel d, primers Ia-6T and Ia-9B were used and the products probed with 32P-labelled Ia-8B (panel e). In panel f, primers Ia-6T and Ia-11B were used, with probe 32P-labelled Ia-8B (panel g). Notably, whereas products were observed in panels d and e, none were observed in f and g. M-tracks contain size markers, P-tracks, primers only, and (T)-tracks, DNase I-treated DNA from the cells.
When primers and probes were adapted to assess BHLF1 expression from possible P3 and P4 promoters, all cell-line samples used, and the C15 xenograft (but not other tumours) gave positive results, even when primers came from outside DL in unique BamHI H sequence. Thus we could identify no specific P4 promoter usage for transcription of this gene. But a P3 promoter not comparable to that of LF3 P3 (described earlier) was identified. We designated this promoter P3′. Although we did not definitely map it, P3′ lies beyond positive 41 514 in the unique region. In experiments using primers Ia-6T and H-5B, with a radiolabelled H-4T probe, positive results were obtained with Daudi, M-ABA, Raji, P3HR-1 and B95-8 cells, and the C15 tumour. This promoter proved insensitive to inducing agents, and for BHLF1 gene expression, comparable results were obtained with M-ABA cells, whether induced or not (data not shown).
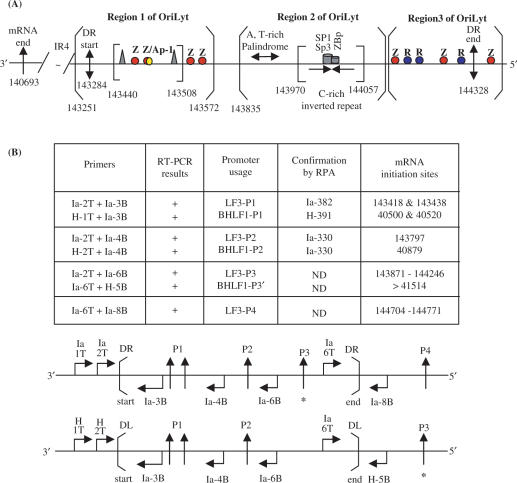
These data are summarized in Figure 7A, together with locations of some known transcriptional transregulation functions (viral and cellular). Of note is a ‘zeta’ transcriptional activation motif just 5′ to the DR regions.
Figure 7.
Summary of transcriptional responsive elements (TREs) and promoters identified for BHLF1 and LF3, within oriLyt, as given here for LF3 and elsewhere for BHLF1 (21). (A) The IR4 repeat region and the 3′-mRNA end (single arrow) are indicated. The duplicated sequence (DS) regions of EBV DNA contain potential structural elements that divide it into three distinct regions, as noted (open brackets). Region 1 contains the DR start (in LF3 at position 143 284; double-headed arrow), as well as the LF3 mRNA start site from the inducible promoter pair, P1 (not shown), a number of zta (Z or EB1)-responsive elements (in red) and an Ap-1 binding sequence (yellow). The core OriLyt sequence in region 1 is indicated (closed brackets) and contains ‘TATA’ and ‘CAAT’ boxes (triangles) and TREs, as shown. The P2 non-inducible promoter maps to position 143 797, between regions 1 and 2. Region 2 contains a DNA sequence that permits formation of three short A,T-rich hairpin loops in BHLF1 (between positions 40 974 and 41 044) and in LF3 (between 143 893 and 143 964), and a putative ‘hinge’ (H) G,C-rich region with triple and single-stranded components, in BHLF1 (between 41 077 and 41 140) and in LF3 (predicted, between 143 995 and 144 023), as well as SP1/SP3 and ZBp TRE elements (21), as noted. Region 3, further ‘upstream’, contains the 5′-end of DR (in LF3 at position 144 328; double-headed arrow) as well as both Z (red) and R (rta or EB2, blue) responsive elements, interspersed. These may compete with each other, or alternatively, act cooperatively in transcription events. Immediately outside the DR region is a further Z responsive element. Sp1, Sp3 and ZBP are cellular transcription factors (21). (B) Summary of promoter mapping data. To determine the transcriptional expression from different promoters, each transcript was first detected by RT-PCR (shown in part in Figures 4 and 6), then fine-mapped and confirmed by RNase protection assays (Figures 2, 3 and 5). RT-PCR data are summarized in Tables 1 and 2. Another promoter, designated P3′, for BHLF1 and P3 for LF3 transcripts, was identified and partly, but not precisely, mapped in these studies. For BHLF1, it lay outside DL, whereas for LF3 it was within DR. A fourth promoter, P4, was found for the LF3 transcript and localized by RT-PCR, outside DR, as shown. It may functionally correspond to P3′ for BHLF1, but this is not confirmed. The laboratory data are compiled here in tabular and linear forms and the precise, or approximate (*), locations of the various promoter sites within or outside DS, are identified for LF3 (P1–P4) and for BHLF1 (P1–P3′). PCR primers used in each case (see ‘Materials and Methods’) are given for each gene, and their locations noted.
Identification of overlapping, non-coding ‘rightward’ (antisense) transcripts
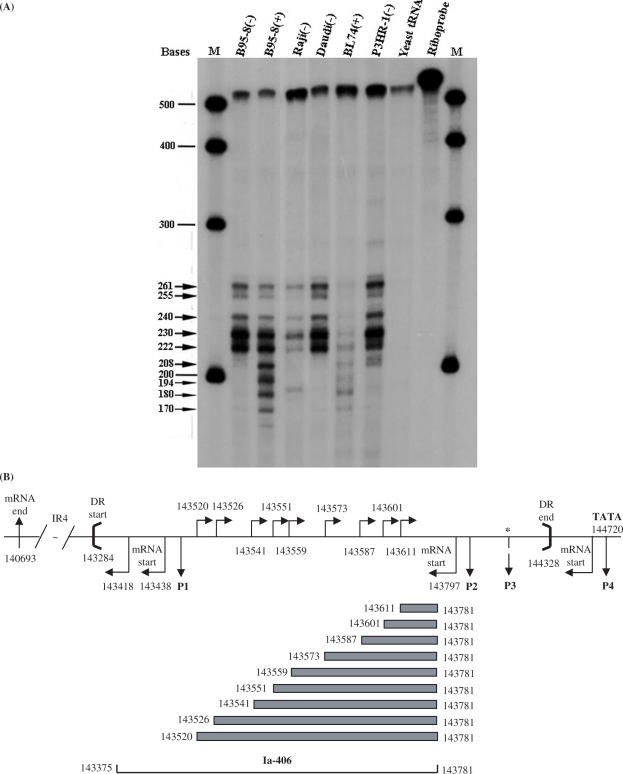
To search for complementary strand RNAs, suggested by examining the primary structure on the strand complementary to BHLF1 and LF3 genes, and potential promoters, we initially generated a cDNA riboprobe, 14A, transcribed from ‘right to left’ on the EBV genome, containing the IR4 repeat sequences from the C15 tumour (34). When this 32P-radiolabelled probe was hybridized with total RNAs from a variety of cell lines or the C15 tumour itself, and products subjected to RNase digestion, only negative results were obtained (data not shown), suggesting no transcription on the opposite strand, within the main body of the coding region of LF3 (38).
In a second set of experiments, similar RNase protection assays were carried out with another 32P-radiolabelled complementary strand riboprobe, Ia-406 (nucleotides 143781–143375) that incorporated the first promoter (P1) region of the LF3 gene and part of the ‘core’ oriLyt sequences. When Ia-406 was hybridized with total RNAs from B95-8 [uninduced (−) or induced (+)], Raji (−), Daudi (−), BL74 (+) and P3HR-1 (−) cells, after RNase digestion a series of 9–10 related protected bands were observed (Figure 8A). Although the bands varied in intensity among transcripts derived from different cells, the product patterns were similar or identical. Multiple bands were observed in all samples (except control, yeast tRNAs).
Figure 8.
Identification of complementary (rightward) strand transcripts by RNase protection (RPA) assays. (A) A 32P-radiolabelled 520 base riboprobe (Ia-406, see track, Riboprobe) from the DR region of the viral genome was used in RPA assays to explore ‘rightward’ transcription on the strand complementary to LF3, that is, in a ‘rightward’ direction on the EBV genome (Figure 1). In cells, as noted, a series of small, overlapping products were observed as seen here. Due to deletions in their respective genomes, bands in B95-8 cells must arise from the DL region and in Daudi cells from DR. Raji, BL74 or P3HR-1 bands may represent mixtures of products from both DL and DR. Notably, the patterns are identical, although band intensities vary. For BL74, bands were only observed in induced cells, as shown, which was not the case with other lines. Data given here use a low-specific activity probe, in great excess. Larger bands appear more prominent in uninduced cells, with induction appearing to favour in general the smaller bands. M = molecular weight markers; size calculations of protected bands are given (arrows). (B) An illustrative diagram showing location and nucleotide numbers of the riboprobe (Ia-406) and the protected products from the rightward ‘complementary strand’ transcripts, relative to the BamHI Ia fragment in the EBV genome (33) and DR (between heavy brackets). The locations of the leftward promoter-related sequences, P1–P4 (single-headed arrows) and LF3 mRNA start sites (bent arrows, below horizontal line), both below the horizontal line, are shown. The 5′-end initiation sites of ‘rightward’ transcripts over this same region are indicated (bent arrows) above the horizon line. If expressed simultaneously in cells, the rightward strand transcripts would overlap leftward transcripts, possibly to form double-stranded RNAs which, as such, might be targeted for degradation by cellular enzymes. (*) = mapped only approximately.
This family of RNA molecules—transcribed with the opposite polarity to the BHLF1/LF3 transcripts—was initiated within the so-called ‘upstream region’ of EBV oriLyt (8) and extended into the ‘downstream’ region (Figure 8B). Transcripts were identified in both B95-8 and Daudi cells indicating that both DL and DR support the expression of ‘antisense’ transcription. The promoter region, as mapped for the largest of the transcripts, notably overlaps that of the inducible P1 regions for BHLF1 and LF3.
DISCUSSION
EBV lytic replication has been a subject of intense, but sporadic, attention in a number of laboratories for at least 20 years (8, reviewed 6,12,21,39). Two nearly identical viral repetitive sequences (DS), with oriLyt activity, fulfil this role for EBV. These lie about 40 000 bp from each end of the linear viral genome and are designated DL and DR—that is, left and right duplications of DNA. In addition to containing the ‘core’ sites, or origins, of lytic replication, the duplications also contain strong promoters for the major mRNAs found in many EBV-related cell lines and tumours. These control expression of two viral ‘early’ genes, BHLF1 (associated with DL) and LF3 (with DR), each of which specifies a protein. These genes are transcribed from ‘leftward’ transcripts on the conventional map of EBV DNA (Figure 1). Most viral strains contain both genes and their associated DS regions, but some have lost one or other of them—not both—yet remain viable. The two repetitive elements themselves do not reside within identical genetic environments and data reported elsewhere (22,23), and confirmed in the present study (summarized, Tables 1 and 2), show that they may also be differently regulated. For example, whereas LF3 expression can be identified in tumours of both lymphoid (Burkitt's lymphoma, BL) and epithelial cell (nasopharyngeal carcinoma, NPC) origins, in general BHLF1 expression seems largely confined to the former.
Our interest in the two viral components, DL and DR, their usage and in turn the genes they control, stems initially from the assumption that studying them in concert might identify sequences that could be used to regulate EBV lytic replication in a cell-type specific manner. And further—since in the case of ‘endemic BL’ (3,40,41) genes associated with EBV lytic replication appear to mark tumours most likely to respond to simple chemotherapy—in the longer term such studies might prove of relevance to tumour control. The data we present here concern transcriptional control of BHLF1 and LF3, and suggest roles these genes might have in controlling viral lytic replication.
From earlier studies, EBV lytic replication has been shown to be dependent upon transcription (8), and to be tightly controlled by, or dependent on, expression of two viral genes originally designated EB1 and EB2 (13), and later also BZLF1 (Zebra, zta) and BRLF1 (Rta), respectively (18). Lytic replication may be enhanced by transcription inducing agents such as the tumour promoter, phorbol 12-myristate 13-acetate (TPA), histone deacetylase inhibitors such as n-butyrate or surface immunoglobulin cross-linking agents (discussed 6,9,14). Our studies here reveal the apparent complexities of promoters used for transcription of these two genes, with at least four distinct promoters used for expression of LF3, three of which are also probably involved in expression of BHLF1. Most of these promoter-related sequences lie within DL and DR, but only one of them (designated here, the P1 pair; ref. 15), but not the others, appear responsive to chemical inducing agents. Moreover, at least one promoter identified for each gene lies in unique DNA regions, outside DS, allowing possibly for differential responsiveness to transcriptional activators or repressors, and interactions with host-cell functions. Since the promoter elements identified mostly (or all) contain one or more typical ‘TATA’ and ‘CCAAT’ boxes, and their transcripts contain ‘AATAAA’ polyadenylation signals (22), we assume, but have not proved, that they are transcribed by RNAse Pol II. Our data on these transcripts are summarized in Figure 7, and evidence leading to this summary figure is presented in Figures 2–6 and Tables 1 and 2. Whether transcripts expressed from each of these promoters are translated is yet to be determined. This need not be the case. Interestingly, the role of the proteins associated with BHLF1 (25) and LF3 (24) also remains unknown, but both are highly basic and known to associate with DNA. The structure of the LF3 translation products, as a consequence of slippage events inherent in its small tandem 102 bp repetitive sequences, may be highly variable within EBV from different individuals (38). We proposed that such data are consistent with a molecular mechanism that permits their escape from immune surveillance.
In the present article, our transcription data show that chemical induction, known to increase lytic replication, generally increases expression from P1 at the expense of the alternative, non-inducible, promoters identified. Our data raise important questions concerning the role(s) of the latter promoters. We address this topic from several points of view. We question whether they, one or all, might act as replication primers or whether one or more of their products might play roles in the maintenance of latency. We argue that such transcripts may all be part of a complex lytic replication pathway used by EBV.
With regard to these questions, by virtue of their orientation, the viral ‘leftwardly’ transcribed RNAs must, in crossing the lytic origin ‘core’ sequences, pass sequentially first through the postulated G,C-rich ‘hinge’ structure (16) before encountering less-stable short A,T-rich palindromic loops (Figure 7A). Without ‘help’, it seems energetically unlikely that transcripts from P3/P4 for LF3, or P3′ from BHLF1, would thus function as the required transcriptional adjuncts (8) to EBV lytic replication. Were specified translation products (from P1 and P2) to act as replication-specific DNA binding proteins, on the other hand, this apparently energetic unfavourable event might receive ‘support’, i.e. allow for strand separation. Or, if their transcripts specifically bound to DNA sites that contain the G,C-rich inverted structure, or a hinge variant of it, then, again, they might contribute directly to lytic replication. In preliminary work, however, we did not detect these transcripts in association with DNA, arguing against the latter suggestion. Notably, the promoters that we designate P3′ for BHLF1 (Table 1) and P4 for LF3 (Table 2) are located 5′ to the ‘core’ lytic origin of replication, outside DS (Figure 6). Transcribed products from such promoters must cross this entire origin sequence, potentially destabilizing it and assisting in the generation of a hinge (H)-sequence that would release a single-stranded loop in the genome for lytic replication, as projected (16). It seems energetically unlikely, however, that such a structure would persist in the absence of a lytic origin-binding protein.
In proposing that the very basic (arginine/histidine-rich) LF3 and BHLF1 translation products might themselves fulfil a role in lytic EBV replication—by specifically binding components of the origin sequences in the viral DNA, and stabilizing them—we are confounded by the problem that a dedicated EBV oriLyt-binding protein was not identified in early in vitro studies aimed at defining viral proteins associated with lytic replication (26,27). From these published data, the conclusion allowed was either that a dedicated EBV origin-binding function is not required for lytic replication or, alternatively, a cellular function may fulfil this role. There is another explanation, however. That is, the Vero cell line in which these in vitro replication experiments were carried out might have supplied this function, not from its host genes but from the virus it harbours. Vero is derived from a New World primate and is now known to harbour its own endogenous herpesvirus, whose DNA sequence bears many homologies with that of EBV (42). These include genes structurally related to the EBV IR2- and IR4-containing genes, BHLF1 and LF3. Thus, proteins in the VERO cells could have provided in-trans the origin-binding functions for EBV lytic replication, in the earlier studies. If so, then arguably, the leftward transcripts might be essential components of viral lytic replication, helping with ‘opening up’ the energetically tight structures in the OriLyts to allow replication to proceed.
Evenso, it is obvious that a more energetically favourable sequence of events would result if, in transcription events, the small, less stable, A,T-rich hairpin loop structures in the oriLyts were encountered initially, followed then by the putative ‘hinge regions’ (15, see Figure 7A). We thus examined the possibility of transcription events occurring on the complementary DNA strands of DS. Using RNase protection assays and total cellular RNAs from a variety of cell lines, small protected bands, of varying lengths, were indeed identified (Figure 8A and B). These ‘rightwardly transcribed’ overlapping RNAs transverse oriLyt sequences with a polarity that should be energetically preferred, as argued earlier, permitting them to encounter sequentially first the A,T-rich palindromes, then the ‘hinge’ sequences. As such, they could provide potential primers for DNA polymerase and serve direct roles in lytic replication. Their most 5′-end initiation site is adjacent to, and possibly overlaps, the P1 promoters identified earlier for the ‘leftwardly’ expressed transcripts (15). (They terminate beyond DS at a site we have yet precisely to identify.) In the tumour cell line, BL74, chemical induction was apparently necessary for their transcription, but in other cells a role for inducing agents was less clear (Figure 8A). We found such ‘rightward’ transcripts in both B95-8, Daudi/P3HR-1 and other cells, suggesting transcription from both DL and DR. Our data, which were reproducible, do not show whether the variable 5′-ends of these small ‘rightwardly expressed’ RNAs represent primary transcription events or, alternatively, cleavage products of an RNA/DNA hybrid using host enzymes (43). In preliminary studies, again we did not identify RNA/DNA hybrids. Notably the sequence of the ‘rightward’ strand—complementary to that containing the promoters mapped for BHLF1 and LF3 and the respective genes themselves—contain ‘TATA’-box initiation sites, in sufficient numbers to account for the nine major products observed (Figure 8)—suggesting these may be primary transcription products. In the absence of data to the contrary, we propose that the polarity and properties of the ‘rightward’ transcripts suggest they also may represent events directly associated with the control of EBV lytic replication. As ‘antisense’ RNAs, they may block transcription on the other strand from some of the promoters, or vice versa.
Basically, we show here that BHLF1 and LF3 genes reside in highly complex regions of the EBV genome, and that this complexity is reflected in respective transcriptional events, most likely associated with viral lytic replication and/or blocking this event. One possible pair of promoters, but probably not the others, respond to chemical inducing agents. Like their positions in the genome, these viral genes possess both numerous similarities and differences that may be exploited in different cell types. The identification of three viral products surrounding the oriLyt regions—that is, RNAs that are transcribed from both strands of the viral DNA, and possible DNA-binding proteins (24,25) specified by transcripts from one of the two strands—is reminiscent of the model deduced for ColE1 plasmid replication in the elegant studies of Tomizawa (reviewed, 44) and colleagues. Their findings may thus provide a useful model for further studies relating to EBV lytic replication.
We entertained the possibility that these small, ‘complementary’ RNAs (Figure 8A) act as ‘classical’ micro (mi) RNAs. However, although miRNAs have been identified elsewhere in the EBV genome (45–47), they have not been found within DS. We cannot at this stage exclude the possibility that the transcripts may function as small interfering (si) RNAs, although these species appear to arise from double-stranded precursors, which is not obviously the case here.
EBV BHLF1 and LF3 genes are so-called ‘early antigens’, and cells in which the virus can be induced to synthesize early antigens are targeted for cell death. Moreover, lytic cycle repression is important for tumour maintenance. One of the prime reasons in studying EBV lytic replication must lie with the potential for exploiting knowledge derived for ‘translational research’ programmes aimed at tumour control (3,48). Field studies on BL patients suggest that stimulating the B-cells to undergo lytic replication increases their responsiveness to ‘classical’ chemotherapy protocols (40,41). Immunohistochemistry studies show that the LF3-specified early antigen co-localizes, in the cell nucleus and cytoplasm, with both the major viral capsid proteins, VCAs (our unpublished data), and the viral transcription transactivator (BZLF1 or EB1) zta protein (38). What the multiple promoters and antisense transcripts for BHLF1 and LF3 offer us thus is an opportunity, for example, of using miRNA technologies to study the ablation of individual promoters in the viral genome, and analysing subsequent responses by immunocytochemistry or corresponding changes in cellular phenotypes and genotypes in in vitro studies. Moreover, whereas the ‘leftward’ gene RNAs have all the hallmarks of RNA polymerase Pol II transcripts in human cells, the small complementary ‘rightwardly expressed’ RNAs, on the other hand, more resemble TATA+ Pol III transcripts (49): there are numerous ‘TATA’ boxes in the sequence, but no data to support these RNAs as exons of larger messenger species, and no obvious polyadenylation (AATAAA) signal in adjacent downstream viral sequences. The likelihood is that these ‘rightward’ transcripts may directly interfere with transcriptional expression of BHLF1 and LF3. In practical terms, if it should prove that Pol II and Pol III polymerases invoke ‘leftward’ and ‘rightward’ transcripts, respectively, they may provide alternative easily defined approaches for further studies on the control and regulation of lytic replication of this medically important virus. A third topic to be taken into account is that of epigenetic events associated with transcription of BHLF1 and LF3. Our preliminary studies, both in cell lines and tumours (not shown) identify a considerable degree of methylation over corresponding regions of the genome with, for example, many CCGG sequences lacking sensitivity to cleavage with the methylation-sensitive HpaII restriction enzyme. Thus epigenetic events will undoubtedly prove important in unravelling events relevant to the transcriptional control of these genes, as well as the ‘rightwardly expressed’ transcripts. Dissecting this important topic will be complicated by the very large numbers of supposedly ‘rare’ CpG sequences found in these regions (30), on both strands of the DNA.
We also note the striking question of two apparently genetic and functionally related sequences in EBV, since viruses on the whole, by virtue of their size and dependency on their hosts, have little scope for redundant information. Gene duplication has been argued to be a main source of evolutionary innovation in that where one copy of the gene becomes repositioned—as here—the creation of a new function may occur (50). For EBV, the existing data argue against functional redundancy but rather suggest that duplication may have occurred in the past to create an extended host range for this virus. For EBV—and possibly HHV-8 (20)—gene duplication in the past may represent an expansionist event, a quest for survival. Notably among DNA viruses, EBV has a remarkably wide host range (1). In our study, we have examined transcription in only two (B-lymphocytes and epithelium) of these cell types, encountering cell specificity with regard to expression of the two genes studied, BHLF1 and LF3. Such specificities may also exist in other cell types (T-cells, fibroblasts) associated with this virus.
Since EBV infection can be associated with serious pathological consequences for some individuals, understanding how to control its proliferation is more than an academic exercise and warrants continued study. Further examining the rationale and control of the numerous promoters identified in our work will, we trust, offer possibilities for new, future molecular studies relating to the regulation of EBV lytic replication, a topic that has proved problemative to date.
ACKNOWLEDGEMENTS
We thank numerous individuals for generous gifts of materials, as noted in the text, and Dr Q-L Lu for his assistance with immunochemical staining experiments. We thank colleagues for discussions on ColE1 replication. Funding to pay the Open Access publication charges for this article was provided by the BL Research Unit within Imperial College.
Conflict of interest statement. None declared.
REFERENCES
- 1.IARC. Lyon: IARC Press; 1997. Monographs on the evaluation of carcinogenic risks to humans. Epstein-Barr Virus and Kaposi's Sarcoma Herpesvirus/Human Herpesvirus 8; pp. 47–373. [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson MP, Kurzrock R. Epstein-Barr virus and cancer. Clin. Cancer Res. 2004;10:803–821. doi: 10.1158/1078-0432.ccr-0670-3. [DOI] [PubMed] [Google Scholar]
- 3.Griffin BE, Rochford R. ‘Endemic Burkitt's lymphoma’. In: Robertson ES, editor. Epstein-Barr Virus. Horizon Scientific Press, Norwich, UK; 2005. pp. 113–137. [Google Scholar]
- 4.Griffin BE. Epstein-Barr virus (EBV) and human disease: facts, opinions and problems. Mutat. Res. 2000;462:395–405. doi: 10.1016/s1383-5742(00)00028-4. [DOI] [PubMed] [Google Scholar]
- 5.Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC, et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 6.Kieff E, Rickinson AG. ‘Epstein-Barr virus and its replication’. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology 4. Vol. 2. Philadelphia, PA: Lippincott-Raven; 2001. pp. 2572–2629. [Google Scholar]
- 7.Kinchington D, Griffin BE. Size heterogeneity of EBV and mitochondrial DNAs in Burkitt's lymphoma lines. Nucleic Acids Res. 1987;15:10345–10354. doi: 10.1093/nar/15.24.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 9.Inman GJ, Binne UK, Parker GA, Farrell PJ, Allday MJ. Activators of the Epstein-Barr virus lytic program concomitantly induce apoptosis, but lytic gene expression protects from cell death. J. Virol. 2001;75:2400–2410. doi: 10.1128/JVI.75.5.2400-2410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yates JL, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugden B, Leight ER. EBV's plasmid replicon: an enigma in cis and trans. Curr. Topics Microbiol. Immunol. 2001;258:3–11. doi: 10.1007/978-3-642-56515-1_1. [DOI] [PubMed] [Google Scholar]
- 12.Israel BF, Kenney SC. ‘EBV lytic replication’. In: Robertson ES, editor. Epstein-Barr Virus. Norwich, UK: Horizon Scientific Press; 2005. pp. 139–155. [Google Scholar]
- 13.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anisimova E, Prachova K, Roubal J, Vonka V. Effects of n-butyrate and phorbol ester (TPA) on induction of Epstein-Barr virus antigens and cell differentiation. Arch. Virol. 1984;81:223–237. doi: 10.1007/BF01309995. [DOI] [PubMed] [Google Scholar]
- 15.Laux G, Freese UK, Bornkamm GR. Structure and evolution of two related transcription units of Epstein-Barr virus carrying small tandem repeats. J. Virol. 1985;56:987–995. doi: 10.1128/jvi.56.3.987-995.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portes-Sentis S, Sergeant A, Gruffat H. A particular DNA structure is required for the function of a cis-acting component of the Epstein-Barr virus OriLyt origin of replication. Nucleic Acids Res. 1997;25:1347–1354. doi: 10.1093/nar/25.7.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feederle R, Kost M, Baumann M, Janz A, Drouet E, Hammerschmidt W, Delecluse H-J. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 2000;19:3080–3089. doi: 10.1093/emboj/19.12.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruffat H, Batisse J, Pich D, Neuhierl B, Manet E, Hammerschmidt W, Sergeant A. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J. Virol. 2002;76:9635–9644. doi: 10.1128/JVI.76.19.9635-9644.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiriart E, Bardouillet L, Manet E, Gruffat H, Penin F, Montserret R, Farjot G, Sergeant A. A region of the Epstein-Barr virus (EBV), RNA export factor EB2 containing an arginine-rich motif, mediates direct binding to RNA. J. Biol. Chem. 2003;278:37790–37798. doi: 10.1074/jbc.M305925200. [DOI] [PubMed] [Google Scholar]
- 20.AuCoin DP, Colletti KS, Xu Y, Cei SA, Pari GS. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 2002;76:7890–7896. doi: 10.1128/JVI.76.15.7890-7896.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsurumi T, Fujita M, Kudoh A. Latent and lytic Epstein-Barr virus replication strategies. Rev. Med. Virol. 2005;15:3–15. doi: 10.1002/rmv.441. [DOI] [PubMed] [Google Scholar]
- 22.Xue S-A, Lu Q-L, Poulsom R, Karran L, Jones MD, Griffin BE. Expression of two related viral early genes in Epstein-Barr virus-associated tumours. J. Virol. 2000;74:2793–2803. doi: 10.1128/jvi.74.6.2793-2803.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue S-A, Labrecque LG, Lu QL, Ong SK, Lampert IA, Kazembe P, Molyneux P, Broadhead RL, Borgstein E, et al. Promiscuous expression of Epstein-Barr virus genes in Burkitt's lymphoma from the central African country, Malawi. Int. J. Cancer. 2002;99:635–643. doi: 10.1002/ijc.10372. [DOI] [PubMed] [Google Scholar]
- 24.Nuebling CM, Mueller-Lantzsch N. Identification of the gene product encoded by PstI repeats (IR4) of the Epstein-Barr virus genome. Virology. 1991;185:519–523. doi: 10.1016/0042-6822(91)90812-p. [DOI] [PubMed] [Google Scholar]
- 25.Nuebling CM, Mueller-Lantzsch N. Identification and characterization of an Epstein-Barr virus early antigen that is encoded by the NotI repeats. J. Virol. 1989;63:4609–4615. doi: 10.1128/jvi.63.11.4609-4615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fixman ED, Hayward GS, Hayward SD. trans-Acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 1992;66:5053–5059. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fixman ED, Hayward GS, Hayward SD. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumann M, Feederle R, Kremmer E, Hammerschmidt W. Cellular transcription factors recruit viral replication proteins to activate the Epstein-Barr virus origin of lytic DNA replication, oriLyt. EMBO J. 1999;18:6095–6105. doi: 10.1093/emboj/18.21.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metzenberg S. Relative rates of RNA synthesis across the genome of Epstein-Barr virus are highest near oriP and oriLyt. J. Virol. 1989;63:4938–4944. doi: 10.1128/jvi.63.11.4938-4944.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Smith PR, Karran L, Lu QL, Griffin BE. Induction of an exceptionally high level transcript of double-stranded, non-translated, EBV-encoded RNA in the interferon sensitive Burkitt's lymphoma line, Daudi. J. Virol. 1997;71:84–94. doi: 10.1128/jvi.71.1.84-94.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones MD, Foster L, Sheedy T, Griffin BE. The EB virus genome in Daudi Burkitt's lymphoma cells has a deletion similar to that observed in a non-transforming strain (P3HR-1) of the virus. EMBO J. 1984;3:813–821. doi: 10.1002/j.1460-2075.1984.tb01890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Busson P, Ganem G, Flores P, Mugneret F, Clausse B, Caillous B, Brahan K. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinoma. Int. J. Cancer. 1988;42:599–606. doi: 10.1002/ijc.2910420422. [DOI] [PubMed] [Google Scholar]
- 33.Parker BD, Bankier A, Satchwell S, Barrell B, Farrell PJ. Sequence and transcription of the Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology. 1990;179:339–346. doi: 10.1016/0042-6822(90)90302-8. [DOI] [PubMed] [Google Scholar]
- 34.Hitt MM, Allday MJ, Hara T, Karran L, Jones MD, Busson P, Tursz T, Ernberg I, Griffin BE. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Little PF, Jackson IJ. Application of plasmids containing promoters specific for phage-encoded RNA polymerases. In: Glover DM, editor. DNA Cloning III, A Practical Approach. Oxford: IRL Press; 1987. pp. 1–18. [Google Scholar]
- 36.Zeng M-S, Li D-J, Liu Q-L, Song L-B, Li M-Z, Zhang R-H, Yu X-J, Wang H-M, Ernberg I, et al. Genomic sequence analysis of Epstein-Barr virus strain GD1 from a nasopharyngeal carcinoma patient. J. Virol. 2005;79:15323–15330. doi: 10.1128/JVI.79.24.15323-15330.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolan A, Addison C, Gatherer D, Davison AJ, McGeoch DJ. The genome of Epstein–Barr virus type 2 strain AG876. Virology. 2006;350:164–170. doi: 10.1016/j.virol.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 38.Xue S-A, Jones MD, Lu Q-L, Middeldorp JM, Griffin BE. Genetic diversity: frameshift mechanisms alter coding of a gene (Epstein-Barr virus LF3 gene) that contains multiple 102-base-pair direct sequence repeats. Mol. Cell. Biol. 2003;23:2192–2201. doi: 10.1128/MCB.23.6.2192-2201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujii K, Yokoyama N, Kiyono T, Kuzushima K, Homma M, Nishiyama Y, Fujita M, Tsurumi T. The Epstein-Barr virus Pol catalytic subunit physically interacts with the BBLF4-BSLF1-BBLF2/3 complex. J. Virol. 2005;74:2550–2557. doi: 10.1128/jvi.74.6.2550-2557.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labrecque LG, Xue S-A, Kazembe P, Phillips J, Lampert I, Wedderburn N, Griffin BE. Expression of Epstein-Barr lytically related genes in African Burkitt's lymphoma: correlation with patient response to therapy. Int. J. Cancer. 1999;81:6–11. doi: 10.1002/(sici)1097-0215(19990331)81:1<6::aid-ijc2>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Ong SK, Xue SA, Molyneux R, Broadhead RL, Borgstein E, Ng MH, Griffin BE. African Burkitt's lymphoma: a new perspective. Trans. Roy. Soc. Trop. Med. Hyg. 2001;95:93–96. doi: 10.1016/s0035-9203(01)90348-7. [DOI] [PubMed] [Google Scholar]
- 42.Rivailler P, Jiang H, Cho Y-G, Quink C, Wang F. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J. Virol. 2002;76:421–426. doi: 10.1128/JVI.76.1.421-426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frank P, Braunshofer-Reiter C, Poltl A, Holzmann K. Cloning, subcellular localization and function expression of human RNAse HII. Biol. Chem. 1998;379:1407–1412. doi: 10.1515/bchm.1998.379.12.1407. [DOI] [PubMed] [Google Scholar]
- 44.Tomizawa J. Control of ColE1 plasmid replication. Intermediates in the binding of RNA1 and RNA11. J. Mol. Biol. 1990;212:683–694. doi: 10.1016/0022-2836(90)90230-j. [DOI] [PubMed] [Google Scholar]
- 45.Pfeffer S, Zavolan M, Grasser FA, Cien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 46.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by gamma-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. Epub. Mar 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng WH, Hong G, Delecluse HJ, Kenney SC. Lytic induction therapy for Epstein-Barr virus-positive B-cell lymphomas. J. Virol. 2004;78:1893–1902. doi: 10.1128/JVI.78.4.1893-1902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao X, Schramm L, Hernandez N, Herr W. A shared surface of TBP directs RNA polymerase II and III transcription via association with different TFIIB family members. Mol. Cell. 2003;11:151–161. doi: 10.1016/s1097-2765(02)00797-9. [DOI] [PubMed] [Google Scholar]
- 50.Rodin SN, Parkhomchuk DV, Riggs AD. Epigenetic changes and respositioning determine the evolutionary fate of duplicated genes. Biochem. (Moscow) 2005;70:559–567. doi: 10.1007/s10541-005-0149-5. [DOI] [PubMed] [Google Scholar]