Abstract
Nucleic acid double helices are stabilized by hydrogen bonding and stacking forces (a combination of hydrophobic, dispersive and electrostatic forces) of the base pairs in the helix. One would predict the hydrogen bonding contributions to increase and the stacking contributions to decrease as the water activity in the medium decreases. Study of nucleobase paired duplexes in the absence of water and ultimately in pure aprotic, non-polar organic solvents is not possible with natural phosphodiester nucleic acids due to the ionic phosphate groups and the associated cations, but could be possible with non-ionic nucleic acid analogues or mimics such as peptide nucleic acids. We now report that peptide nucleic acid (PNA) (in contrast to DNA) duplexes show almost unaffected stability in up to 70% dimethylformamide (DMF) or dioxane, and extrapolation of the data to conditions of 100% organic solvents indicates only minor (or no) destabilization of the PNA duplexes. Our data indicate that stacking forces contribute little if at all to the duplex stability under these conditions. The differences in behaviour between the PNA and the DNA duplexes are attributed to the differences in hydration and counter ion release rather than to the differences in nucleobase interaction. These results support the possibility of having stable nucleobase paired double helices in organic solvents.
INTRODUCTION
Nucleic acid double helices are stabilized by hydrogen bonding and stacking forces (a combination of hydrophobic, dispersive and electrostatic forces) of the base pairs in the helix (1–5). The most recent data suggest that stacking interactions are the more important for DNA duplex stability (6,7). Indeed these results suggest that while hydrogen bonding in GC base pairs are stabilizing the helix, hydrogen bonding in AT base pairs are slightly destabilizing (6,7). One would predict the hydrogen bonding contributions to increase and the stacking contributions to decrease as the water activity in the medium decreases. Therefore, it could be of great interest both from a structural and thermodynamic as well as from a functional point of view to study nucleobase paired duplexes in the absence of water and ultimately in pure aprotic, non-polar organic solvents.
This is not possible with natural phosphodiester nucleic acids due to the ionic phosphate groups and the associated cations, but could be possible with uncharged nucleic acid analogues or mimics such as methylphosphonates (8), phosphotriesters (9), morpholino derivatives (10) and peptide nucleic acids (PNAs) (11,12). PNAs are DNA pseudopeptide mimics capable of forming DNA-like Watson–Crick base-pair double helices with sequence complementary DNA, RNA and PNA oligomers (13–18). As the aminoethyl glycine backbone of PNA is charge neutral, no counter ions are required for stabilizing PNA·PNA duplexes, and consequently such duplexes may more conveniently be studied in organic solvents.
Results from a previous study has given preliminary indications that PNA–PNA duplexes are much less affected by the presence of organic co-solvent (50% DMF) than DNA–DNA duplexes, correlating well with the lack of measurable changes in hydration or counter ion binding upon PNA duplex formation (19). However, this observation warrants further exploration. We now report that PNA (in contrast to DNA) duplexes show almost unaffected stability in 70% dimethylformamide (DMF) or dioxane, and extrapolation of the data to conditions of 100% organic solvents indicates only a little (or no) destabilization of the PNA duplexes.
MATERIALS AND METHODS
PNAs
PNA1, PNA2, PNA3, PNA4 and PNA5 were synthesized using solid phase Boc chemistry, purified by HPLC and characterized by MALDI-TOF mass spectrometry as described previously (20). PNA concentrations were determined spectrophotometrically at 65°C using molar extinction coefficients: ε260 of adenine = 15 400 M−1 cm−1, ε260 of guanine = 11 700 M−1 cm−1, ε260 of thymine = 8800 M−1 cm−1 and ε260 of cytosine = 7400 M−1 cm−1.
Chemicals and DNAs
All chemical reagents used were of analytical grade except for dimethylformamide (DMF) and dioxane, which were spectroscopic grade from Sigma-Aldrich, Munich, Germany. The DNA oligonucleotides were purchased from DNA Technology, Aarhus, Denmark, and used without further purification.
Sample preparation
Main stock solutions of PNAs and DNAs were prepared by dissolution in deionized, double distilled water. Experimental samples were made by diluting from the corresponding main stock solutions in 10 mM phosphate buffer (pH 7.2) containing 100 mM NaCl and 0.1 mM EDTA.
Equimolar mixtures (1:1 stoichiometry in single strands) of the PNA or DNA and its complementary strand were dissolved in the buffer mentioned above with desired amount of organic co-solvents. The duplex formation was assured by heating to 90°C and then cooling slowly to room temperature to allow proper annealing. No sign of aggregation or decreased solubility of the PNAs at up to 70% of organic co-solvents was observed.
UV-melting experiments
The thermal melting experiments were performed on a Cary 300 Bio UV-visible spectrophotometer (Varian, Cary, NC, USA) connected to a temperature controller. Thermal melting profiles were obtained using heating–cooling cycles in the range of −3 to 95°C. The melting temperature (Tm) was determined from the peak of the first derivative of the heating curve. Cuvettes of 1.0 cm path length and 1.0 ml volume were used for all experiments.
Thermal melting curves at >50% of DMF or dioxane start to lose the upper baseline and show severe disturbances partly because of high absorbance of DMF at the wavelength required for the experiments. Therefore, thermodynamic data at >50% of DMF could not be obtained. However, it was possible to obtain Tm values at 60 and 70% of DMF or dioxane.
Thermodynamics
The thermodynamic parameters viz enthalpy change (ΔH0), entropy change (ΔS0), and Gibbs’ free energy change (ΔG0) were evaluated using the ‘hyperchromicity method’ (curve fitting) and/or the concentration method (21).
The hyperchromicity method
The hyperchromicity method utilizes alpha curve and van't Hoff plots (ln KT versus T −1) according to the following definitions (21): The fraction (αT) of single strands that remained hybridized in the duplex at a particular temperature T in Kelvin is represented as
| 1 |
where, Ad is the absorbance of the duplex in fully hybridized condition, As is the absorbance of the single strands in fully denatured condition and As is absorbance at a particular point on the thermal melting curve at temperature T.
For non-self-complementary sequences forming n-mer structures, the general equilibrium constant equation at a particular temperature T can be expressed as:
| 2 |
where, cts represents the total concentration of strands and n is the molecularity of the complex. Assuming a two-state model, Equation (2) reduces to
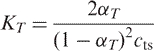 |
3 |
The van't Hoff plot ln KT versus T −1 is a straight line represented by
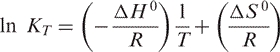 |
4 |
Hence, ΔH 0 can be obtained from the slope and ΔS 0 can be obtained from the Y-intercept of the van't Hoff plot. ΔG 0 at a particular temperature T in Kelvin can be calculated from
| 5 |
where R is the universal gas constant which is equal to 1.986 cal/mol K.
The concentration method
The concentration method utilizes a plot of 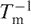 versus ln cts, where Tm is the thermal melting temperature of the duplex and cts is the total strand concentration of PNA or DNA.
versus ln cts, where Tm is the thermal melting temperature of the duplex and cts is the total strand concentration of PNA or DNA.
Since Tm is defined by the temperature where α = 0.5 for a two state transition, combining Equations (3) and (4) yields:
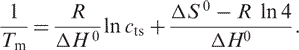 |
6 |
Thus, the thermodynamic parameters can be extracted from a linear fit to a plot of 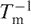 versus ln cts according to Equation (6) (21), where ΔH 0 is obtained from the slope of the linear fit and ΔS 0 from the Y-intercept.
versus ln cts according to Equation (6) (21), where ΔH 0 is obtained from the slope of the linear fit and ΔS 0 from the Y-intercept.
The values of the thermodynamic parameters calculated by this method are thus independent of strand concentration, which is not the case with the hyperchromicity method described above.
RESULTS
PNA and DNA duplexes
In order to elucidate the properties of PNA duplexes as compared to those of iso-sequential DNA duplexes at reduced water activity, we have studied the effect of organic co-solvents on the thermal stability and thermodynamics of PNA·PNA, PNA·DNA and DNA·DNA duplexes of ‘random’, mixed base-sequence (Table S1). We chose DMF (dielectric constant of DMF is 36.7) and dioxane (dielectric constant of dioxane is 2.2) as organic co-solvents, as these are aprotic and are not hydrogen bond donors but still sufficiently polar to retain solubility of the PNA·DNA complexes even above 50% organic solvent.
The thermal stability of these duplexes and the corresponding thermodynamic parameters [evaluated using both the concentration method (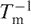 versus ln cts plot) (21) and the hyperchromicity (curve fitting) method (21)] in aqueous medium with increasing amount of DMF (extrapolated to 100% DMF) are presented in Tables 1 and S2. Representative thermal melting curves are shown in Figures S1 and S2. It is important to note that thermal denaturation curves showed essentially unperturbed monophasic behaviour up to 50% organic co-solvent. The extrapolation was performed on the basis of the linear plots of the thermal stability (Tm) and Gibbs’ free energy changes (ΔG0) as a function of increasing amount of DMF in the medium (Figure 1A and B). Because of insufficient thermal stability of the DNA1·DNA2 duplex in >30% DMF, we also designed a longer DNA·DNA duplex (DNA3·DNA4), that has thermal stability in aqueous solution similar to that of the PNA1·PNA2 duplex (data in Tables 1 and S3, Figure 1A and B, representative thermal curves in Figure S3). While the organic solvent-dependent changes in ΔG0 of all these duplexes show a very good correlation with the corresponding Tm, ΔH0 and ΔS0 values were not significantly affected within experimental error (Tables S2–S4). The extraction of thermodynamic parameters derived from thermal melting method requires that no change in heat capacity occurs in the duplex single-strand equilibrium. For the present systems we have found only minor changes. The values of specific heat capacity change (enthalpic) at constant pressure (▵Cp) were 2.0 kcal/mol.K for PNA1·PNA2, 1.4 kcal/mol.K for PNA1·DNA2, 1.9 kcal/mol.K for PNA2·DNA1 and 1.8 kcal/mol.K for DNA1·DNA2 (19).
versus ln cts plot) (21) and the hyperchromicity (curve fitting) method (21)] in aqueous medium with increasing amount of DMF (extrapolated to 100% DMF) are presented in Tables 1 and S2. Representative thermal melting curves are shown in Figures S1 and S2. It is important to note that thermal denaturation curves showed essentially unperturbed monophasic behaviour up to 50% organic co-solvent. The extrapolation was performed on the basis of the linear plots of the thermal stability (Tm) and Gibbs’ free energy changes (ΔG0) as a function of increasing amount of DMF in the medium (Figure 1A and B). Because of insufficient thermal stability of the DNA1·DNA2 duplex in >30% DMF, we also designed a longer DNA·DNA duplex (DNA3·DNA4), that has thermal stability in aqueous solution similar to that of the PNA1·PNA2 duplex (data in Tables 1 and S3, Figure 1A and B, representative thermal curves in Figure S3). While the organic solvent-dependent changes in ΔG0 of all these duplexes show a very good correlation with the corresponding Tm, ΔH0 and ΔS0 values were not significantly affected within experimental error (Tables S2–S4). The extraction of thermodynamic parameters derived from thermal melting method requires that no change in heat capacity occurs in the duplex single-strand equilibrium. For the present systems we have found only minor changes. The values of specific heat capacity change (enthalpic) at constant pressure (▵Cp) were 2.0 kcal/mol.K for PNA1·PNA2, 1.4 kcal/mol.K for PNA1·DNA2, 1.9 kcal/mol.K for PNA2·DNA1 and 1.8 kcal/mol.K for DNA1·DNA2 (19).
Table 1.
Thermal stability and thermodynamic parameters of PNA and DNA duplexesa
| Duplex | DMFb (%) | Tm (°C)c,d |
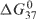 (kcal/mol)d,e (kcal/mol)d,e
|
ΔΔG0 f |
|---|---|---|---|---|
| PNA1·PNA2 | 0 | 70.2 ± 0.3 | −16.6 ± 0.5 (−17.2) | 5.6 |
| 10 | 68.3 ± 0.2 | −15.4 ± 0.5 (−16.2) | ||
| 20 | 66.2 ± 0.2 | −14.3 ± 0.4 (−15.6) | ||
| 30 | 64.3 ± 0.5 | −14.5 ± 0.8 (−15.7) | ||
| 40 | 63.4 ± 0.3 | −14.4 ± 0.5 (−15.9) | ||
| 50 | 61.3 ± 0.4 | −13.6 ± 0.7 (−15.5) | ||
| 60 | 60.0 ± 0.6g | h | ||
| 70 | 56.1 ± 0.5g | h | ||
| 100i | 52.6 | −11.0 | ||
| DNA1·DNA2 | 0 | 35.8 ± 0.5 | −8.2 ± 0.7 (−7.9) | 15.0 |
| 10 | 29.2 ± 0.4 | −6.2 ± 1.2 (−6.1) | ||
| 20 | 23.7 ± 0.5 | −4.9 ± 0.8 (−4.9) | ||
| 30j | 18.2 ± 0.3 | −3.7 ± 0.5 (−3.9) | ||
| 100i | −22.9 | 6.8 | ||
| DNA3·DNA4 | 0 | 70.0 ± 0.4 | −16.4 ± 0.4 | 19.2 |
| 10 | 62.1 ± 0.5 | −14.3 ± 0.7 | ||
| 20 | 56.2 ± 0.6 | −12.8 ± 0.8 | ||
| 30 | 49.2 ± 0.3 | −11.3 ± 1.0 | ||
| 40 | 41.3 ± 0.2 | −9.3 ± 0.6 | ||
| 50 | 30.1 ± 0.5 | −6.1 ± 0.5 | ||
| 100i | −6.2 | 2.8 | ||
| PNA1·DNA2 | 0 | 51.3 ± 0.4 | −10.3 ± 1.0 | 6.8 |
| 10 | 47.0 ± 0.5 | −9.5 ± 0.7 | ||
| 20 | 43.1 ± 0.3 | −9.0 ± 0.6 | ||
| 30 | 40.2 ± 0.4 | −8.3 ± 0.6 | ||
| 40 | 36.2 ± 0.5 | −7.7 ± 0.8 | ||
| 50 | 31.1 ± 0.2 | −6.7 ± 0.5 | ||
| 100i | 12.3 | −3.5 |
aPNA and DNA sequences: H-GTA GAT CAC T-Lys-NH2 (PNA1); H-AGT GAT CTA C-Lys-NH2 (PNA2); 5′-GTA GAT CAC T-3′ (DNA1); 5′-AGT GAT CTA C-3′ (DNA2); 5′-AGT GAT CTA CGG TGG ACG GTC C-3′ (DNA3); 5′-GGA CCG TCC ACC GTA GAT CAC T-3′ (DNA4).
bVol% in 10 mM phosphate buffer containing 100 mM NaCl and 0.1 mM EDTA, pH 7.2 ± 0.01.
cDuplex concentrations of 5.0 μM in strands were used (Tm plots are in Figures 1A, S4A and S5A).
dStandard deviations are based on five independent measurements.
eEvaluated from the hyperchromicity (curve fitting) method (21) at 37°C (ΔG037 plots are in Figures 1B, S4B and S5B). Values obtained from the concentration method (21) (Equation 6) are shown in paratheses (full details of these data are in Supplementary Data).
fΔΔG0 = ΔG037 (DMF) − ΔG037 (aqueous). Calculated only with the values obtained from the hyperchromicity method.
gThese data have poorer accuracy due to upper baseline irregularities.
hData could not be evaluated because of bad thermal curves.
iData were obtained from manual extrapolation of the linear plots in Figure 1A and B.
jData obtained at higher than 30% of DMF were not reliable because of too low values of Tm.
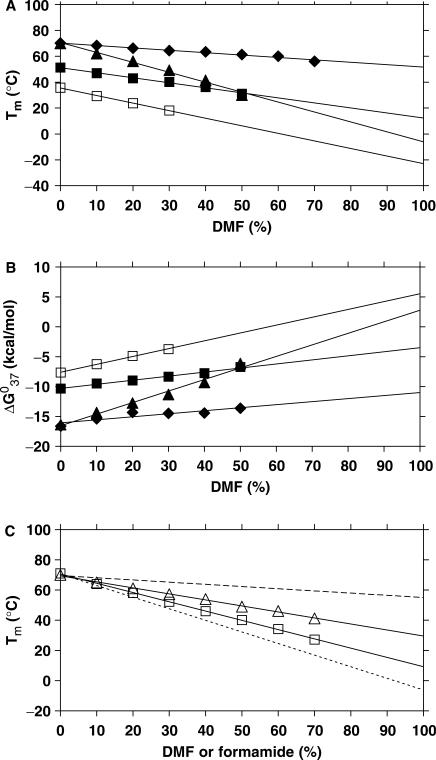
Figure 1.
Plots of (A) Tm and (B) ΔG0 of PNA1·PNA2 (solid diamond), PNA1·DNA2 (solid square), DNA1·DNA2 (open square) and DNA3·DNA4 (solid triangle) as a function of the amount of DMF in the medium. (C) Plots of Tm of PNA1·PNA2 (open triangle) and DNA3·DNA4 (open square), as a function of the amount of formamide in the medium, compared to that of PNA1·PNA2 (dashed line) and DNA3·DNA4 (dotted line), as a function of the amount of DMF in the medium (DMF data taken from Figure 1A). The aqueous buffer was 10 mM phosphate buffer containing 100 mM NaCl and 0.1 mM EDTA, pH 7.2 ± 0.01 (data in Tables 1, S2–S5).
It is clearly evident that the presence of DMF has a much smaller effect on the thermal stability and free energy of the PNA·PNA duplex than on the DNA·DNA duplex, whereas the effect on the PNA·DNA duplex is intermediate. The plots in Figure 1A and B show a linear dependence of both Tm and ΔG0 on the DMF concentration. The change in thermal stabilities as a function of the amount of DMF in the medium did not deviate within experimental error from linearity up to 70% of DMF, thereby supporting a hypothetical approximation beyond 70% of DMF (and an extrapolation of the data to 100% DMF) (Figure 1A and B, Table 1). These results strongly indicate that PNA·PNA duplexes in contrast to DNA·DNA (and PNA·DNA) duplexes will have appreciable stability even in the absence of water. Naturally, we cannot exclude that non-linearity might occur at very low water contents.
Analogous studies of PNA1·PNA2, PNA1·DNA2 and DNA3·DNA4 in dioxane show that, despite the significant difference between the dielectric constants of these solvents, there is no significant difference between the effects of DMF and dioxane on Tm and ▵G0 (Tables S3 and S4, representative thermal curves in Figures S2 and S3, plots in Figures S4 and S5).
PNA and DNA duplexes in formamide
In order to compare the effect of aprotic solvent (DMF and dioxane, where the destabilization of the duplex is assumed to be predominantly caused by dehydration (and perhaps change in dielectrics) with that of a hydrogen-bond donor (and breaking) solvent, we studied the thermal and thermodynamic properties of PNA1·PNA2 and DNA3·DNA4 in formamide (dielectric constant of formamide is 109), which is a well-established nucleic acid denaturant (Table S5, representative thermal curves in Figure S6, plots in Figures 1C, S7 and S8). In formamide, the destabilization is caused by a combined effect of H-bond disruption and dehydration. Notably, the destabilizing effect of formamide is almost as pronounced for the PNA duplex as for the DNA duplex. The slopes of the linear plots of Tm of PNA1·PNA2 and DNA3·DNA4 as a function of increasing amount of DMF in the medium are −0.17 (without taking the values at 60 and 70% into account) and −0.77, respectively, whereas, those values in formamide are −0.40 and −0.61, respectively (Figure 1C). On the other hand, the decrease in Tm is paralleled by the increases in both ΔG0 and ΔH0. Thus in stark contrast to the effects of DMF and dioxane, formamide has comparable effects on PNA and DNA duplex stabilities.
Self-complementary PNA and DNA hairpins
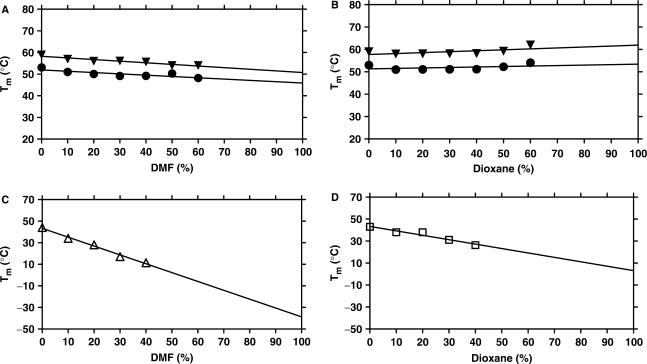
Additionally, we examined the effect of organic co-solvent on the stability of self-complementary (foldback) PNA hairpins (Figure 2). Unfolding of hairpin PNAs, H-AGAG-(eg1)3-CTCT-Lys-NH2 (PNA3) and H-ACAG-(eg1)3-CTGT-Lys-NH2 (PNA4) also showed little change in Tm and ΔG0 with increasing amount of DMF or dioxane in the medium (Tables S6 and S7, Figures 2A, B and S9). In fact, the hairpins were slightly stabilized in dioxane (Table S7 and Figure 2B), and in this case the stabilization appears to be enthalpic (Table S7). Similar studies with an analogous DNA hairpin (DNA5: 5′-AGAGTTTTCTCT-3′) in DMF and dioxane as a control showed a dramatic thermal destabilization by both co-solvents (Tables S8 and S9, Figures 2C, D and S10).
Figure 2.
Plots of Tm of self-complementary (hairpin) PNAs PNA3 (solid inverted triangle) and PNA4 (solid circle) as a function of the amount of (A) DMF and (B) dioxane in the medium. Plots of Tm of hairpin DNA control DNA5 as a function of the amount of (C) DMF (open triangle) and (D) dioxane (open square) in the medium. The aqueous buffer was 10 mM phosphate buffer containing 100 mM NaCl and 0.1 mM EDTA, pH 7.2 ± 0.01 (PNA3: H-AGAG-(eg1)3-CTCT-Lys-NH2, PNA4: H-ACAG-(eg1)3-CTGT-Lys-NH2, DNA5: 5′-AGA GTT TTC TCT-3′) (data in Tables S6–S9).
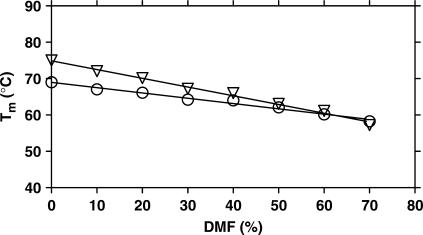
PNA duplex with an endstacked tricyclic thymine
In order to evaluate the effect of removal of water from the medium (by increasing DMF content) on base stacking forces more directly, we studied the effect of DMF on the behaviour of a PNA duplex (PNA5·PNA6) containing a tricyclic thymine analogue (benzo[b]-1,8-naphthyridin-2(1H)-one, designated as tT) (22) attached at the end of one strand, thereby stabilizing the duplex predominantly (or entirely) by end-stacking (22). It is evident that, DMF preferentially destabilizes the PNA5·PNA6 duplex (Tm slope is −0.24) compared to the control PNA6·PNA7 duplex (Tm slope is −0.15) (Table S10, Figures 3 and S11). Most interestingly, the stabilization of the duplex contributed by the tT base is completely lost at 70% DMF (Figure 3), clearly indicating a dramatic reduction of the contribution of stacking interactions to duplex stability under these conditions (decrease in thermal stability of PNA5·PNA6 at 70% DMF with respect to 0% DMF, ΔTm = 18.8, whereas, that of the control duplex PNA6·PNA7 is only 10.7).
Figure 3.
Plots of Tm of PNA5 · PNA6 (open inverted triangle) containing tricyclic thymine (tT) and its control PNA6 · PNA7 (open circle), as a function of the amount of DMF in the medium (data in Table S10). The aqueous buffer was 10 mM phosphate buffer containing 100 mM NaCl and 0.1 mM EDTA, pH 7.2 ± 0.01 (PNA5: H-tT-GTA GAT CAC T-NH2, PNA6: H-AGT GAT CTA C-NH2, PNA7: H-GTA GAT CAC T-NH2).
Mismatched duplexes
Sequence discrimination is critically dependent on hydrogen bonding recognition, but base-pair mismatches may also change the geometry of the helix and thus influence stacking interactions. For instance, X-ray crystallography and NMR studies have revealed that G·T and T·T mismatches in DNA likely form wobble base pairs (23–30). The homopurine A·A mismatch in DNA involves one hydrogen bond between the amino group of one adenine and the nitrogen N1 of the opposite adenine, whereas, the mispaired T·T bases in DNA involve two imino-carbonyl hydrogen bonds (23–30). A C·T mismatch is the most unstable in DNA and makes one regular H-bond involving carbonyl groups along with one weak H-bond bridged by a water molecule at neutral pH (23–30). Effects of mispairs are rather localized and the duplex retains a global B-form conformation (23–30). As hydration and stacking forces could be more important factors for mismatch stabilization, removal of water may preferentially destabilize these.
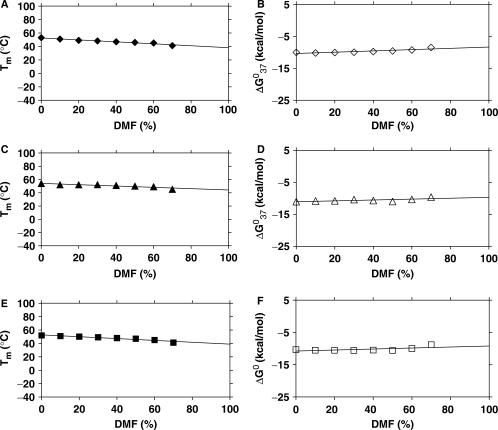
However, surprisingly we find that the relative effects of DMF on the stability of single base mismatched (T·T, A·A and C·T) duplexes of PNA8 (H-AGT GTT CTA C-Lys-NH2), PNA9 (H-GTA GAA CAC T-Lys-NH2) and PNA10 (H-GTA GCT CAC T-Lys-NH2) with PNA1 and PNA2 are not distinguishable within experimental error from that observed with the corresponding fully matched PNA duplexes (The Tm slope of the fully matched PNA1·PNA2 is −0.17, of T·T mismatched PNA1·PNA8 is −0.13, of A·A mismatched PNA2·PNA9 is −0.08, and of C·T mismatched PNA2·PNA10 is −0.12) (Tables 1 and S11, Figures 1A, 4, S12 and S13). These results suggest that, in these base pair mismatched duplexes, hydrogen bonding and stacking interactions are equally compromised or that the mismatched base pair contributes only very little to the overall duplex stability.
Figure 4.
Thermal stabilities of T·T, A·A and C·T mismatched PNA duplexes in DMF (data in Table S11). Plots of (A) Tm (solid diamond) and (B) ▵G0 (open diamond) of PNA1·PNA8 as a function of the amount of DMF in the medium. Plots of (C) Tm (solid triangle) and (D) ▵G0 (open triangle) of PNA2·PNA9 as a function of the amount of DMF in the medium. Plots of (E) Tm (solid square) and (F)▵G0 (open square) of PNA2·PNA10 as a function of the amount of DMF in the medium. The aqueous buffer was 10 mM phosphate buffer containing 100 mM NaCl and 0.1 mM EDTA, pH 7.2 ± 0.01.
DISCUSSIONS AND CONCLUSIONS
The relative insensitivity of PNA duplexes to the reduced water content is consistent with our previous observations showing that in contrast to the case of DNA·DNA (and to a lesser extent PNA·DNA duplexes), no change in hydration occurs upon duplex formation (19) and no change in counter ion binding is observed either (19).
Thus the effect of organic solvents on PNA duplex stability may be ascribed to changes in base-pair hydrogen bonding and stacking forces of the base pairs. Stacking forces have contributions from hydrophobic, dispersion and dipole electrostatic forces (31–33), and therefore both hydrogen bonding as well as stacking interactions are affected by the dielectric constant of the solvent. However, we observe no correlation between PNA duplex stability and solvent dielectric constant. Water, formamide, DMF and dioxane have dielectric constants of 78, 109, 37 and 2.2, respectively, and in particular the comparable effect of dioxane and DMF strongly argue against a simple dependence of the dielectric constant. On the other hand, it is clear that the contribution of hydrogen bonding to PNA duplex stability should significantly increase when the dielectric constant and especially the hydrogen bonding donor/acceptor activity of the solvent decreases. (It should be kept in mind that the hydrogen bond stabilization results as the difference between nucleobase hydrogen bonding to solvent versus base pairing.) Furthermore, our results very clearly show that the stabilizing effect of end stacking decreases upon addition of DMF to the solvent. Indeed, the end-stacking effect using a tricyclic thymine analogue is absent already at 70% DMF. This thymine analogue in essence contains an additional phenyl ring compared to thymine (22), and therefore the stacking interactions of the tricyclic thymine are expected to contain a relatively larger hydrophobic contribution than natural nucleobase pairs (32). Thus a similar effect on the natural nucleobases is expected to occur at higher DMF (lower water) content. Nonetheless, our results strongly support the contention that relative helix stabilizing contribution of base-pair hydrogen bonding and base stacking will be shifting towards hydrogen bonding as the water content of the medium (as well as the dielectric constant) decreases. In this context, we note that for the PNA hairpins an increased stability is indicated with increasing concentration of the low dielectric constant solvent dioxane (in contrast to DMF).
It is well established that formamide and urea destabilize DNA and RNA duplexes (34,35), and in agreement with the present data, the effect of DMF and dioxane (at 10 mol%) on RNA duplex stability was found to be comparable to that of formamide (33). However, also in case of RNA, no direct correlation between the duplex stabilizing effect and the solvent dielectric constant was apparent (33). Indeed, from our data comparing the effects of formamide and DMF on PNA duplex stability, we would argue that the effects of organic solvents on DNA and RNA duplexes is to a larger extent due to hydration (solvation) and counterion effects, whereas the effects on PNA duplexes more directly reflect changes in hydrogen bonding and stacking contributions to helix stability. Thus more systematic studies on the different behaviour of PNA versus DNA duplexes could possibly shed light on these relative contributions to nucleobase (including DNA) helix stability.
Interestingly, the data (Tables S2 and S3) could hint that the decreased stability of the DNA duplexes upon removal of water appears to be primarily entropic rather than enthalpic, though the data are not conclusive. This would be compatible with the differences in behaviour between the PNA and the DNA duplexes (as mentioned above) being caused by the differences in hydration and counter ion release rather than via nucleobase interaction. Thus, these results can be of great significance in understanding nucleobase pairing systems, and it will be of great interest to learn whether other non-ionic DNA analogues such as methylphosphonates (8), phosphotriester (9) and morpholino (10) derivatives behave similarly.
The possibility of having stable nucleobase paired double helices in organic solvents has several implications. First of all, we note that it should be possible to reach the goal of finding PNA duplexes that are freely soluble in non-polar organic solvents by chemically modifying the PNA backbone, for instance using hydrophobic amino acids in place of glycine (36). Studying the behaviour of these in the absence of water could provide valuable information about fundamental properties of nucleobase double helices. Furthermore, PNA duplexes spanning lipid membranes could be envisaged as a means for mediating electron transport over the membrane. Sequence-dependent electron transport (or hole migration) through DNA duplexes is now well established (37–41), and although electron transport involving PNA still require more extensive studies (42,43) this should be worthwhile pursuing. Finally, entirely new dimensions would be added to the emerging technique of ‘DNA sequence’ directed organic synthesis (44). Thus novel avenues of exploiting nucleobase recognition systems can be enabled by transferring these to non-aqueous environments.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
ACKNOWLEDGEMENT
This work is supported by the European Commission in the 6th framework PACE project, contract no. 002035. Funding to pay the Open Access publication charges for this article was provided by the European Commission.
Conflict of interest statement. None declared.
REFERENCES
- 1.Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 2.Saenger W. Principles of Nucleic acid Structure. New York: Springer; 1988. [Google Scholar]
- 3.Kool ET. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu. Rev. Biophys. Biomol. Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Oostenbrink C, van Gunsteren WF. Efficient calculation of many stacking and pairing free energies in DNA from a few molecular dynamics simulations. Chemistry. 2005;11:4340–4348. doi: 10.1002/chem.200401120. [DOI] [PubMed] [Google Scholar]
- 5.Sundaralingam M, Ponnuswamy PK. Stability of DNA duplexes with Watson–Crick base pairs: a predicted model. Biochemistry. 2004;43:16467–16476. doi: 10.1021/bi048158+. [DOI] [PubMed] [Google Scholar]
- 6.Protozanove E, Yakovchuk P, Frank-Kamenetskii MD. Stacked-unstacked equilibrium at the nick site of DNA. J. Mol. Biol. 2004;342:775–785. doi: 10.1016/j.jmb.2004.07.075. [DOI] [PubMed] [Google Scholar]
- 7.Yakovchuk P, Protozanove E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic. Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller PS, Yano J, Yano E, Carroll C, Jayaraman K, Ts’o PO. Nonionic nucleic acid analogues. Synthesis and characterization of dideoxyribonucleoside methylphosphonates. Biochemistry. 1979;18:5134–5143. doi: 10.1021/bi00590a017. [DOI] [PubMed] [Google Scholar]
- 9.Tosquellas G, Alvarez K, Dell'Aquila C, Morvan F, Vasseur J-J, Imbach J-L, Rayner B. The pro-oligonucleotide approach: solid phase synthesis and preliminary evaluation of model pro-dodecathymidylates. Nucleic Acids Res. 1998;26:2069–2074. doi: 10.1093/nar/26.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen PE. Peptide nucleic acid. A molecule with two identities. Acc. Chem. Res. 1999;32:624–630. [Google Scholar]
- 13.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, et al. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 14.Wittung P, Nielsen PE, Buchardt O, Egholm M, Nordén B. DNA-like double helix formed by peptide nucleic acid. Nature. 1994;368:561–563. doi: 10.1038/368561a0. [DOI] [PubMed] [Google Scholar]
- 15.Jensen KK, Ørum H, Nielsen PE, Nordén B. Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry. 1997;36:5072–5077. doi: 10.1021/bi9627525. [DOI] [PubMed] [Google Scholar]
- 16.Brown SC, Thomson SA, Veal JM, Davis DG. NMR solution structure of a peptide nucleic acid complexed with RNA. Science. 1994;265:777–780. doi: 10.1126/science.7519361. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson M, Nielsen PE. Solution structure of a peptide nucleic acid-DNA duplex. Nature Struct. Biol. 1996;3:410–413. doi: 10.1038/nsb0596-410. [DOI] [PubMed] [Google Scholar]
- 18.Rasmussen H, Kastrup JS, Nielsen JN, Nielsen JM, Nielsen PE. Crystal structure of a peptide nucleic acid (PNA) duplex at 1.7 A resolution. Nature Struct. Biol. 1997;4:98–101. doi: 10.1038/nsb0297-98. [DOI] [PubMed] [Google Scholar]
- 19.Sen A, Nielsen PE. Unique properties of purine/pyrimidine asymmetric PNA.DNA duplexes: differential stabilization of PNA.DNA duplexes by purines in the PNA strand. Biophys. J. 2006;90:1329–1337. doi: 10.1529/biophysj.105.073213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen L, Fitzpatrick R, Gildea B, Petersen K, Hansen HF, Koch T, Egholm M, Buchardt O, Nielsen PE, et al. Solid-phase synthesis of peptide nucleic acids. J. Pept. Sci. 1995;1:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 21.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 22.Eldrup A, Nielsen BB, Haaima G, Rasmussen H, Kastrup JS, Christensen C, Nielsen PE. 1,8-Naphthyridin-2(1H)-ones—novel bicyclic and tricyclic analogues of thymine in peptide nucleic acids (PNAs) Eur. J. Org. Chem. 2001;9:1781–1790. [Google Scholar]
- 23.Patel DJ, Shapiro L, Hare D. In: In Nucleic acid and Molecular Biology. Eckstein F, Lilley DMJ, editors. Berlin: Springer; 1987. pp. 70–84. [Google Scholar]
- 24.Germann MW, Kalisch BW, Varnum JM, Vogel HJ, van de Sande JH. NMR spectroscopic and enzymatic studies of DNA hairpins containing mismatches in the EcoRI recognition site. Biochem. Cell Biol. 1998;76:391–402. doi: 10.1139/bcb-76-2-3-391. [DOI] [PubMed] [Google Scholar]
- 25.Allawi HT, SantaLucia J., Jr Thermodynamics of internal C.T mismatches in DNA. Nucleic. Acids Res. 1998;26:2694–2701. doi: 10.1093/nar/26.11.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boulard Y, Cognet JAH, Fazakerley GV. Solution structure as a function of pH of two central mismatches, C.T and C.C, in the 29 to 39 K-ras gene sequence, by nuclear magnetic resonance and molecular dynamics. J. Mol. Biol. 1997;268:331–347. doi: 10.1006/jmbi.1997.0975. [DOI] [PubMed] [Google Scholar]
- 27.Nemecek D, Stepanek J, Turpin PY, Rosenberg I. Raman study of potential ‘antisense’ drugs: nonamer oligonucleotide duplexes with a central mismatch as a model system for the binding selectivity evaluation. Biopolymers. 2004;74:115–119. doi: 10.1002/bip.20056. [DOI] [PubMed] [Google Scholar]
- 28.Hunter WN, Brown T, Kneale G, Anand NN, Rabinovich D, Kennard O. The structure of guanosine-thymidine mismatches in B-DNA at 2.5-A resolution. J. Biol. Chem. 1987;262:9962–9970. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- 29.Ke S-H, Wartell RM. The thermal stability of DNA fragments with tandem mismatches at a d(CXYG).d(CY′X′G) site. Nucleic. Acids Res. 1996;24:707–712. doi: 10.1093/nar/24.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gervais V, Cognet JA, Le Bret ML, Sowers C, Fazakerley GV. Solution structure of two mismatches A.A and T.T in the K-ras gene context by nuclear magnetic resonance and molecular dynamics. Eur. J. Biochem. 1995;228:279–290. [PubMed] [Google Scholar]
- 31.Guckian KM, Schweitzer BA, Ren RX-F, Sheils CJ, Tahmassebi DC, Kool ET. Factors contributing to aromatic stacking in water: Evaluation in the context of DNA. J. Am. Chem. Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim TW, Kool ET. A series of nonpolar thymidine analogs of increasing size: DNA base pairing and stacking properties. J. Org. Chem. 2005;70:2048–2053. doi: 10.1021/jo048061t. [DOI] [PubMed] [Google Scholar]
- 33.Hickey DR, Turner DH. Solvent effects on the stability of A7U7p. Biochemistry. 1985;24:2086–2094. doi: 10.1021/bi00329a042. [DOI] [PubMed] [Google Scholar]
- 34.Hutton JR. Renaturation kinetics and thermal stability of DNA in aqueous solutions of formamide and urea. Nucleic. Acids Res. 1977;4:3537–3555. doi: 10.1093/nar/4.10.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blake RD, Delcourt SG. Thermodynamic effects of formamide on DNA stability. Nucleic. Acids Res. 1996;24:2095–2103. doi: 10.1093/nar/24.11.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Püschl A, Sforza S, Haaima G, Dahl O, Nielsen PE. Peptide nucleic acids (PNAs) with a functional backbone. Tetrahedron Lett. 1998;39:4707–4710. [Google Scholar]
- 37.Schuster GB. In Long-Range Charge Transfer in DNA I-II. Heidelberg: Springer; 2004. [Google Scholar]
- 38.Delaney S, Barton JK. Long-range DNA charge transport. J. Org. Chem. 2003;68:6475–6483. doi: 10.1021/jo030095y. [DOI] [PubMed] [Google Scholar]
- 39.Fink HW, Schonenberger C. Electrical conduction through DNA molecules. Nature. 1999;398:407–410. doi: 10.1038/18855. [DOI] [PubMed] [Google Scholar]
- 40.Wan C, Fiebig T, Schiemann O, Barton JK, Zewail AH. Femtosecond direct observation of charge transfer between bases in DNA. Proc. Natl Acad. Sci. USA. 2000;97:14052–14055. doi: 10.1073/pnas.250483297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao F, Augustyn K, Barton JK. Sequence dependence of charge transport through DNA domains. J. Am. Chem. Soc. 2005;127:17445–17452. doi: 10.1021/ja0563399. [DOI] [PubMed] [Google Scholar]
- 42.Armitage B, Ly D, Koch T, Frydenlund H, Orum H, Batz HG, Schuster GB. Peptide nucleic acid-DNA duplexes: long range hole migration from an internally linked anthraquinone. Proc. Natl Acad. Sci. USA. 1997;94:12320–12325. doi: 10.1073/pnas.94.23.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanabe K, Yoshida K, Dohno C, Okamoto A, Saito I. Control of electron transfer in DNA by peptide nucleic acids (PNA) Nucleic Acids Symp. Ser. 2000;44:35–36. doi: 10.1093/nass/44.1.35. [DOI] [PubMed] [Google Scholar]
- 44.Gertner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR. DNA-templated organic synthesis and selection of a library of macrocycles. Science. 2004;305:1601–1605. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.