Abstract
The origin of DNA replication (oriC) of the hyperthermophilic archaeon Pyrococcus abyssi contains multiple ORB and mini-ORB repeats that show sequence similarities to other archaeal ORB (origin recognition box). We report here that the binding of Cdc6/Orc1 to a 5 kb region containing oriC in vivo was highly specific both in exponential and stationary phases, by means of chromatin immunoprecipitation coupled with hybridization on a whole genome microarray (ChIP-chip). The oriC region is practically the sole binding site for the Cdc6/Orc1, thereby distinguishing oriC in the 1.8 M bp genome. We found that the 5 kb region contains a previously unnoticed cluster of ORB and mini-ORB repeats in the gene encoding the small subunit (dp1) for DNA polymerase II (PolD). ChIP and the gel retardation analyses further revealed that Cdc6/Orc1 specifically binds both of the ORB clusters in oriC and dp1. The organization of the ORB clusters in the dp1 and oriC is conserved during evolution in the order Thermococcales, suggesting a role in the initiation of DNA replication. Our ChIP-chip analysis also revealed that Mcm alters the binding specificity to the oriC region according to the growth phase, consistent with its role as a licensing factor.
INTRODUCTION
The duplication and transmission of genetic information without loss, is a very important issue for life. Cell division must be accompanied by DNA replication with an appropriate timing and frequency. One mechanism to achieve this goal is the regulated initiation of DNA replication. In bacteria and eukaryotic organisms, specific initiator protein(s) initiate DNA replication by activating specific region(s), namely the replication origin(s).
Archaea, the third domain of life, are unicellular organisms whose genome size is comparable to that of bacteria. Most of the putative DNA replication proteins encoded in the archaeal genome, however, are related to eukaryotic genes involved in DNA replication (reviewed in 1–4). The hyperthermophilic anaerobe Pyrococcus abyssi belongs to the order Thermococcales in Euryarchaeota. We have used P. abyssi as a model organism to study the molecular physiology of archaeal DNA replication. P. abyssi has a single gene similar to cdc6 or orc1 (cdc6/orc1) and a single mcm gene in the 1.8 M bp chromosome. We have discovered a single replication origin in the circular chromosome (5,6). Although the minimal requirements for designating the replication origin (oriC) have not yet been determined, DNA replication initiates from the region containing an 800 bp intergenic region upstream of the cdc6/orc1 gene. Interestingly, the single origin per chromosome rule is not applicable to all archaeal genomes, as three replication origins were experimentally identified in Sulfolobus solfataricus and S. acidocaldarius (7–9). Another potential origin has been reported in S. solfataricus (10). Two replication origins were predicted in Halobacterium sp. Strain NRC-1 (11,12), and one of them was isolated as an ARS element (13). Marker frequency analyses detected a single origin in Archaeoglobus fulgidus and no single replication origin in Methanocaldococcus jannaschii (14). Recently, Aeropyrum pernix was also shown to have a replication origin (15). In S. solfataricus, DNaseI footprint experiments identified 36 bp sequences (ORB, origin recognition box) in oriC1 and 17 bp sequences (mini-ORB) in oriC2 that are recognized by purified Cdc6-1 protein (7). The putative replication origin of Methanothermobacter thermautotrophicus has multiple 13 bp mini-ORBs (16), which are minimized versions of Sulfolobus ORBs (17). A single mini-ORB is capable of binding M. thermautotrophicus Cdc6-1 specifically, while this protein binds cooperatively to a DNA fragment containing multiple mini-ORB repeats (17). The replication origin in A. pernix also contains ORB repeats, and ORC1 dimer binds to these repeats (15). Furthermore, the Halobacterium ARS element lost its replication activity when a half of the inverted repeat containing ORB-related sequences was deleted (13). ORB-related sequences are found in many archaeal origins, and specific binding of ORB by Cdc6/Orc1 is likely to be a common mechanism for origin recognition in Archaea (7,17).
Pyrococcus abyssi oriC contains multiple 13 bp mini-ORB repeats, two AT rich regions, and two 34 bp ORB repeats that form an inverted repeat (6,18). Both the mini-ORB and ORB show significant sequence similarities to known ORBs (7,17). Furthermore, ORB sequences of P. furiosus can bind S. solfataricus Cdc6-1 in vitro (7). In order to demonstrate the role of a putative archaeal initiator protein in vivo, we have shown, using a chromatin immunoprecipitation (ChIP) assay, that P. abyssi oriC is recognized by Cdc6/Orc1 protein referred to as Cdc6 hereafter for simplicity; (6). The Cdc6 protein binds with much higher affinity to the oriC region in exponentially growing cells as opposed to three other unrelated regions used as controls. This interaction between oriC and Cdc6 is also observed in non-replicating cells. The ChIP technique was also employed in S. solfataricus to demonstrate preferential binding of Cdc6 proteins to replication origins by comparison with a control locus in S. solfataricus (7). However, the localization pattern of the Cdc6/Orc1 protein on a genomewide scale remains to be elucidated in Archaea. Although the Mcm protein of P. abyssi was shown to bind the oriC in exponentially growing cells (6), the genomewide distribution of archaeal Mcm remains to be examined, too.
In eukaryotes, genomewide localization analyses of ORC and/or MCM have identified multiple binding sites (19–21). We developed an experimental method combining ChIP and microarray analysis to investigate the distribution of the Cdc6/Orc1 and Mcm proteins in the entire P. abyssi genome. Sequence analysis of the region identified as the binding site for Cdc6 and Mcm revealed a novel cluster of ORB repeats ∼ 1.5 kb apart from the known ORB repeats in oriC. Furthermore, ChIP at a higher resolution and biochemical studies indicated that both of these ORB clusters were bound specifically by Cdc6. The organization of these ORB repeats is conserved during evolution, suggesting a role for the initiation of DNA replication. The ChIP-chip analysis also showed the regulated loading of Mcm to oriC.
MATERIALS AND METHODS
Microarray preparation
The P. abyssi genome was divided into 353 regions, each of which is 5 kb in length. Each of the 353 DNA regions has a specific ID number. For example, the microarray region 25 corresponds to the genome coordinates 120 000–125 000. For the microarrays, each of the 353 regions was amplified with the BD Advantage 2 PCR kit (BD Biosciences, Palo Alto, CA, USA, Clontech). The primers were designed using the P. abyssi genome sequence so that they have very low possibility of miss-hybridization. The quality (appropriate size and purity) of each PCR product was confirmed by agarose gel electrophoresis. The purified PCR products were quantitated and distributed into 384-well source plates at final concentrations of 100 ng/μl in final volumes of 8 μl of water. PCR products were dried using centrifugation under vacuum and then resuspended in 8 μl of 1% CHAPS and 50% formamide (22).
The DNAs were arrayed onto activated GAPSII slides (Corning, NY, USA) using a GeneTAC G3 robot (Genomic Solutions, Ann Arbor, MI, USA) operating under a controlled environment (20°C and 35% humidity) using thirty-two 150 μm diameter pins. Each fragment was spotted six times (three spots in two different blocks originating from two different pins). P. abyssi plasmid pGT5 and negative controls (lambda DNA, pBluescript, pESP1, 1 kb ladder) were also added after linearization by appropriate restriction enzymes. After spotting, the slides were dried overnight, protected from light, at room temperature. The DNA was then UV-crosslinked to the slide using 250 mJ/cm2 (Hoeffer UVC500; Amersham Biosciences, Freiburg, Germany). For quality control, one slide from every print batch was stained with SYBR Green II (Invitrogen, Carlsbad, CA, USA) diluted 1:10 000. The slides were pre-processed by washing for 5 min each with 1) 2× SSC/0.1% SDS, 2) 0.2× SSC and 3) water and then dried using centrifugation under vacuum. Identical pre-processing was used for slides destined for hybridization.
Labeling of chromatin immunoprecipitated DNA
The basic design of the ChIP-chip experiment followed the protocol by Wyrick et al. (19). ChIP was performed as described previously (6). The specificity of the antisera used in this study was confirmed by ChIP using preimmune antiserum and by western blotting of P. abyssi cell extracts. The DNA fragments obtained by the ChIP method, using a 500 μl cell extract, were treated with RNaseA (0.3 mg/ml) and purified with the QIAquick PCR purification kit (Qiagen, Valencia, CA, USA). The purified DNA fragments were blunted with T4 DNA polymerase (0.03 U/μl, New England Biolabs, Beverly, MA, USA) and purified with phenol/chloroform treatment. The blunted DNA fragments were then ligated to the unidirectional linker (19) by incubating with 8 U/μl ligase (New England Biolabs) at 16°C overnight. The resulting DNA fragments were purified again by ethanol precipitation before being subjected to PCR based DNA labeling. The experimental conditions for DNA-labeling were the same as described (19); Cy5-dUTP and Cy3-dUTP, respectively (Amersham Biosciences), were used for labeling ChIP co-immunoprecipitated DNA and DNA from mock-treated cell extract. Labeled samples were purified by using the StrataPrep PCR purification kit (Stratagen, La Jolla, CA, USA), and concentrated with a vacuum evaporator just prior to hybridization to the microarray.
Microarray hybridization and image acquisition
Experiments were done using at least three (Cdc6 in exponential and stationary phases, MCM in stationary phase) or four (MCM in exponential phase) independently prepared samples. Fluorescently labeled samples were resuspended in 110 μl of Micromax buffer (Perkin Elmer Life Sciences, Foster City, CA, USA). All arrays were hybridized and washed using an automated GeneTAC hybridization station (Genomic Solutions). The hybridization protocol consisted of incubations at (i) 65°C for 3 h; (ii) 55°C for 3 h and (iii) 50°C for 12 h. Slides were then washed twice with medium stringency buffer for 40 s at 50°C; and then twice with high stringency buffer for 40 s at 42°C and then twice with postwash buffer for 40 s at 42°C. All the buffers were from Genomic Solutions. The slides were scanned using a GenePix 4000B Microarray Scanner. The resulting 16 bit TIFF images were analyzed using the GenePix Pro 3 software (Axon Instruments, Aberdeenshire, Scotland). All slides were scanned using 100% laser power and the same PMT voltage (580 V for Cy5 and 520 V for Cy3).
Data analysis
For each spot, the log base 2 of the ratio of the median intensity of Cy5 to Cy3 (M) was calculated. Normalization by the lowess method across an array was done using the Bioconductor LIMMA package (23). Scale normalization across slides was also performed (24). Peak detection was performed as follows: first, the average value of M for each of the fragments, i, on a slide, Mi , was calculated. Then, for each slide, the average and SD of the Mi , Mall and σMi , respectively, were calculated. The Mi were centered around zero, with a constant SD, σ, by means of the following relationship:
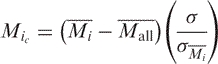 |
where σ is the geometric mean of the 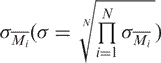 .
.
Finally, for each series of experiments (Cdc6 exponential phase, Cdc6 stationary phase, Mcm exponential phase, Mcm stationary phase), the average Mic and its SD, Mc and σMc , respectively, were determined.
Student's t-tests comparing the Mi value of each fragment to the mean of all the spots were performed independently for each slide. Associated P-values were calculated and the minimum P-value is used to indicate the degree of significance of each fragment. Selected peaks (1 − P > 0.95) and the associated values of Mc and σMc are shown in Table 1.
Table 1.
Comparison of DNA-binding specificities of Cdc6/Orc1 and Mcm
| Cdc6/Orc1 | No. 24 | No. 25 | No. 42 | No. 43 | No. 228 | No. 229 |
|---|---|---|---|---|---|---|
| Expo Mc ± σMc | 0.72 ± 0.29 | 3.06 ± 0.10 | 0.14 ± 0.10 | 0.04 ± 0.08 | 0.36 ± 0.08 | 0.34 ± 0.11 |
| 1 − P | 0.999 | 1 | 0.218 | 0.504 | 0.999 | 0.998 |
| Stat Mc ± σMc | 0.61 ± 0.07 | 2.91 ± 0.16 | 0.23 ± 0.32 | 0.06 ± 0.04 | 0.25 ± 0.20 | 0.27 ± 0.27 |
| 1 − P | 1 | 1 | 0.166 | 0.332 | 0.264 | 0.268 |
| Mcm | No. 24 | No. 25 | No. 42 | No. 43 | No. 228 | No. 229 |
| Expo Mc ± σMc | 0.08 ± 0.05 | 0.72 ± 0.18 | 0.47 ± 0.19 | 0.26 ± 0.06 | −0.01 ± 0.08 | −0.06 ± 0.13 |
| 1 − P | 0.109 | 1 | 0.999 | 0.999 | 0.350 | 0.874 |
| Stat Mc ± σMc | 0.12 ± 0.04 | 0.37 ± 0.13 | 0.76 ± 0.31 | 0.28 ± 0.05 | −0.08 ± 0.02 | −0.06 ± 0.01 |
| 1 − P | 0.855 | 0.999 | 1 | 0.999 | 0.807 | 0.645 |
Data are the average of at least three independent replicates with the corresponding standard deviation. 1 − P expresses the significance of the peak height (see Materials and Methods section for details). Values should not be compared directly among different growth conditions, as ‘input DNA’ is specific for each condition. Expo, exponentially growing cells; Stat, cells in stationary phase.
Purification of the Cdc6/Orc1 protein of P. furiosus
We used the EasySelect Pichia expression system (Invitrogen) to express histidine-tagged P. furiosus Cdc6/Orc1 protein in recombinant yeast cells. As the majority of the Cdc6/Orc1 was found in the insoluble fraction, the histidine-tagged Cdc6/Orc1 was purified under denaturing conditions using a Ni+–NTA agarose column (Qiagen), followed by renaturation in the buffer containing 50 mM Tris–HCl (pH = 8.0), 250 mM NaCl, 5 mM β-mercaptoethanol and 10% glycerol.
Gel retardation assay
The purified, histidine-tagged Cdc6/Orc1 and 0.1 pico mole of 32P-labeled DNA were incubated in 20 μl of reaction buffer containing 20 mM triethanolamine (pH = 7.5), 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, 2% glycerol, 10 μg/ml poly (dI-dC)-poly(dI-dC) and 0.1 mg/ml BSA, for 20 min at 55°C. The reaction mixture was terminated and the protein–DNA complex was chemically fixed by 0.1% glutaraldehyde. The samples were electrophoresed on a 4% polyacrylamide gel (37.5:1) in 1× TAE. The gel was dried and analyzed using an FLA-5000 image analyzer (Fujifilm, Allendale, Tokyo, Japan). When substrate DNA was bound by Cdc6/Orc1, we observed two major ‘shifted’ DNA bands and they were included in the quantification. Sequences of the 49 bp DNA fragments used in the assay are as follows: ORB1, AAAACCCCCCAGAGTTTCATTTCCACTGGAACCAGGTTTTGAAAGGTAA; ORB1-mut, AAAACCCCCCAGAGTTTCATGCTAACTTACGCCAGGTTTTGAAAGGTAA; ORB2, GAAGTCCCCCAGAGTTTCATTTCCACTGGAGCCGGGCGGCAAACACCAG; ORB2-mut1, GAAGTCCCGATCAGTTTCATTTCCACTGGAGCCGGGCGGCAAACACCAG; ORB2-mut2, GAAGTCCCCCAGAGTTTCATTTCCACTGGAGCCGGATCGCAAACACCAG; ORB3, AAAGAAGGAGAGAGTTTTATTTCCACTGGAAGTGAACCACTTGAAGAGG; mini-ORB, AGCTACCATGCCTGCACGATTTCCACTGGAGGTAATCATGGTCATAGCT; control, AGCTACCATGCCTGCACGAATTAAGCAATTCGTAATCATGGTCATAGCT.
RESULTS
Cdc6/Orc1 binds to oriC in vivo with high specificity
We previously reported that P. abyssi Cdc6 binds preferentially to P. abyssi oriC in vivo as compared to three other regions of the P. abyssi genome by using the ChIP method (6). However, it was not known whether this specificity persists at the level of the whole genome. To address this issue, we devised a method to analyze the distribution of Cdc6 over the entire genome by combining ChIP and microarray based hybridization techniques (ChIP-chip analysis). DNA fragments obtained by ChIP using Cdc6 specific antiserum and those from mock treated cell extracts were labeled by incorporating fluorescent-labeled nucleotides, Cy5-dUTP and Cy3-dUTP, respectively. Both labeled DNA samples were hybridized simultaneously on the microarray, the spots of which represent the entire genome of P. abyssi. The specificity of Cdc6 for each region was evaluated by comparing the hybridization efficiency between co-immunoprecipitated and mock-treated DNA on the microarray (see Materials and methods).
When exponentially growing cells were analyzed using this method, we found that the Cdc6 protein bound specifically to region 25 containing oriC (genome coordinate 120 000–125 000) in a statistically significant manner (P << 0.0001). The Mc -value for region 25 was extremely high compared to the averages of all the other regions (Figure 1A and Table 1). Binding to region 24 was also significant (P = 0.001), but the value of Mc was much lower. It should be noted that under the conditions used in the ChIP-chip analyses, a portion of co-immunoprecipitated DNA fragments was relatively long (up to 1.5 kb), and thus can hybridize to two flanking regions such as regions 24 and 25. Cdc6 binding to region 25 was also statistically significant (P << 0.0001) with similar Mc -values in stationary phase cells, consistent with our previous results implying constitutive binding of Cdc6 to oriC (Figure 1B and Table 1).
Figure 1.
The binding of the Cdc6 and Mcm proteins to the P. abyssi genome in exponentially growing (A and C) and stationary cells (B and D) is represented by the value of Mc for each of the 353 fragments of the genome. Cdc6 binds to the region 24–25 containing oriC with an extremely high specificity in exponentially growing and stationary cells. Mcm binds to two regions 25 and 42–43. The data are the results of 3 or 4 independent experiments in each case.
In exponentially growing cells, we found that two other regions, 228 (genome coordinate 1 135 000–1 140 000) and 229 (genome coordinate 1 140 000–1 145 000) bound Cdc6 significantly (P < 0.002), albeit with markedly lower Mc -values, despite the lack of ORB-related sequence (Figure 1A and Table 1). Although a peak corresponding to these regions was also apparent with Cdc6 and stationary phase cells, this binding was not statistically significant (P > 0.7; Figure 1B and Table 1). Among the 11 putative ORFs present in these regions, only one ORF (PAB7298), whose function is unknown, has homologs in the two other Pyrococcus sp. Currently, in the absence of any other information, the possible role of Cdc6 binding to this region remains unclear.
Two clusters of ORB repeats are present in the region bound by Cdc6
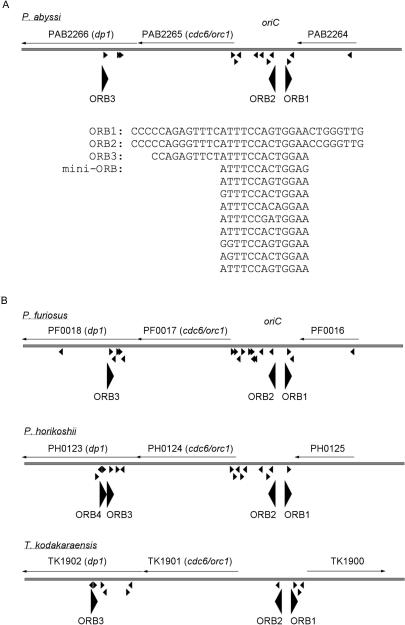
There are multiple 13 bp mini-ORB and two 34 bp ORB in oriC (ORB1 and ORB2). In addition to the cluster of these repeats in oriC, we found another cluster of 13 bp mini-ORB and 23 bp ORB (ORB3) repeats in the dp1 gene encoding the small subunit of family D DNA polymerase II (Figure 2A). This ORB cluster in dp1 resides in the region where Cdc6 binds specifically (Figure 1A and B). Interestingly, the organization of these repeated sequences is conserved in all three Pyrococcus genomes and in the recently sequenced genome of Thermococcus kodakaraensis that also belongs to the order Thermococcales (Figure 2B). We also prepared a consensus sequence based on the eight variations of the mini-ORB repeats observed in the P. abyssi oriC and used it to map all possible 13 bp repeats in the P. abyssi genome. We found 30 repeats widely distributed over the entire genome (Supplementary Figure 1). Although these 13 bp repeats are scattered over the entire genome, no more than two repeats appeared in clusters except in the 5 kb region spanning the oriC, cdc6/orc1 and dp1 gene.
Figure 2.
The cdc6/orc1 gene is located between two clusters of ORB repeats in Thermococcales genomes. (A) The region surrounding the cdc6/orc1 gene and oriC in P. abyssi is presented schematically. The location of the mini-ORB repeats is indicated by small arrowheads, while the ORB repeats are indicated by large arrowheads (not to scale). The 5 kb region presented here corresponds to the region 25 of the microarray. Nucleotide sequences of the ORB and mini-ORB are aligned below. (B) ORB clusters in other Pyrococcus and Thermococcus genomes are presented as in (A).
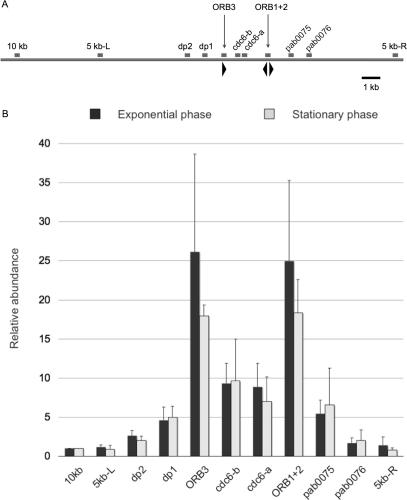
To address the question whether Cdc6 binds both of these ORB clusters, we employed the conventional ChIP technique in which specific binding is evaluated by measuring PCR efficiency (6). We analyzed 11 loci including the two ORB clusters, its neighboring regions and control regions, as indicated in Figure 3A. The results indicated that Cdc6 binds preferentially to both of the ORB clusters in vivo (Figure 3B). The binding affinity to the ORB cluster in dp1 (ORB3) was comparable to that of oriC (ORB1 + 2). Furthermore, this binding is observed in exponential and stationary phases (Figure 3B).
Figure 3.
Cdc6 binds specifically to the two ORB clusters in vivo. (A) Location of the regions analyzed by conventional ChIP is indicated as small boxes. Arrowheads indicate the ORB repeats. (B) DNA-binding specificity of Cdc6 to the regions indicated in (A) was analyzed by relative abundance of each PCR product. ChIP analysis was performed as described previously (6). The error bar indicates the SD (n ≥ 3).
Cdc6 protein binds to DNA with the conserved ORB repeat in vitro
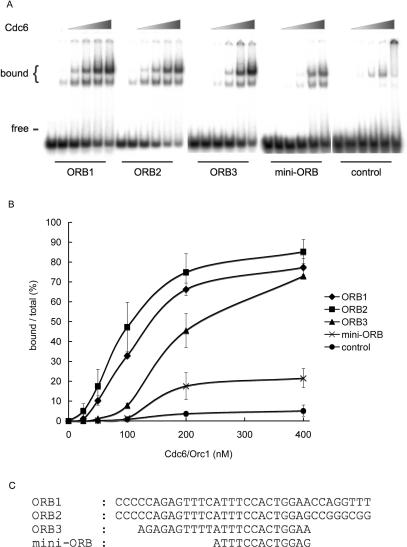
In Pyrococcus sp., there have been no studies that address the DNA-binding activity of Cdc6 in vitro. The amino acid sequence identity between P. furiosus Cdc6 and P. abyssi Cdc6 is 93%, thus confirming their functional identity. We, therefore, purified the Cdc6 protein of P. furiosus from which many DNA replication proteins have been purified in our laboratory, and examined its DNA-binding properties in vitro (Figure 4). We found, using a gel retardation assay that the recombinant Cdc6 protein binds to a DNA fragment carrying either one of the ORB repeats (ORB1, ORB2 or ORB3; see also Figure 2B). The concentration of Cdc6 to achieve 50% binding of the substrate DNA was ∼ 150, 110 and 240 nM for ORB1, ORB2 and ORB3, respectively. On the contrary, a control DNA fragment whose sequence is irrelevant to the ORB repeats bound Cdc6 with a very low affinity (Figure 4). We also evaluated the binding of Cdc6 to one of the 13 bp mini-ORB repeats in oriC in the same gel retardation assay (Figure 4). Cdc6 showed a low affinity to a DNA fragment carrying the mini-ORB.
Figure 4.
DNA-binding activity of the purified Cdc6 protein was analyzed by the gel retardation assay. (A) A representative result obtained with a 49 bp DNA fragment containing ORB, mini-ORB, or the control sequence is shown. The reaction contained 0, 25, 50, 100, 200 or 400 nM Cdc6 protein. (B) Free and bound forms of DNA were quantitated and the ratio of bound DNA was plotted against the concentration of Cdc6. The error bar indicates the SD (n ≥ 3). (C) Nucleotide sequences of P. furiosus ORB1, ORB2, ORB3 and mini-ORB used in the assay are indicated.
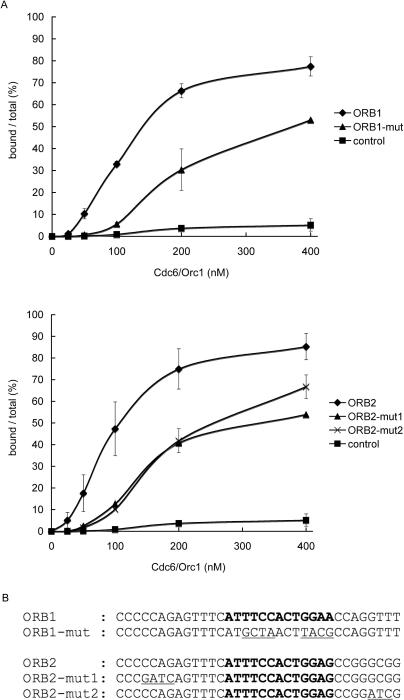
To further understand the mechanism of ORB recognition by Cdc6, we introduced nucleotide substitutions into the ORB repeat and analyzed the DNA-binding activity of Cdc6 to the mutated versions of ORB. The mini-ORB and ORB repeats share a consensus sequence with perfectly conserved motifs, ‘TTCC’ and ‘GAA.’ We introduced nucleotide substitutions into these ‘core motifs’ or into their surrounding sequences (Figure 5B). We found that both the ‘core motifs’ and their surrounding sequences are important for binding with Cdc6, as every nucleotide substitution examined results in partial reduction of the binding affinity (Figure 5A).
Figure 5.
Mutation analysis of the ORB repeat in the gel retardation assay. (A) DNA-binding activity of Cdc6 to ORB1 or ORB2 with nucleotide substitutions was analyzed as in Figure 4. (B) Position and sequences of nucleotide substitutions are shown (underlined), together with the original sequence. The 13 bp sequences that match the consensus of mini-ORB repeats are indicated in bold letters. These are the ORB repeats of P. furiosus and not identical to those shown in Figure 2.
Eukaryotic ORC can bind single stranded DNA (25), and structure rather than sequence dependent DNA-binding activity of archaeal Cdc6 has been reported, too (26–28). We then tested if Pyrococcus Cdc6 can bind single-stranded DNA or flayed duplex in the gel retardation assay. The results, however, indicated that the Pyrococcus Cdc6 protein does not bind these substrates (data not shown).
Genomewide analysis of Mcm distribution
ChIP-chip analysis revealed that the Mcm protein in exponentially growing cells also binds specifically to region 25 (P << 0.0001) that contains oriC (Figure 1C and Table 1). In contrast to the result obtained with Cdc6, Mcm binding to this region decreased greatly in stationary phase (Figure 1D and Table 1). Surprisingly, we also observed that region 42 (genome coordinate 205 000–210 000) is specifically bound by Mcm in exponential phase (P = 0.001; Figure 1C and Table 1). Mcm remained bound specifically to region 42 in stationary phase (P < 0.001).
DISCUSSION
We developed the ChIP-chip method and explored, for the first time in Archaea and Bacteria, the genomewide distribution of the initiator of DNA replication in vivo. In contrast to eukaryotic ORC that has multiple binding sites on the genome (19–21), our ChIP-chip analyses have shown that region 25 containing oriC, cdc6/orc1 and dp1 is the only one where Cdc6 protein binds with an extremely high specificity in P. abyssi cells (Figure 1). The result is also in contrast to the observation that DnaA in Escherichia coli recognizes the promoter/operator region of certain genes that are irrelevant to the regulation of DNA replication (29). This highly specific binding of P. abyssi Cdc6 to oriC was observed both in replicating and non-replicating cells, indicating a constitutive mode of oriC-binding, as was suggested by our previous ChIP analysis (6). In S. cerevisiae, many of the sites that bind both ORC and MCM proteins colocalize well with replication origins (19,21). Our ChIP-chip analysis has shown that Mcm, as well as Cdc6, binds specifically to the fragment 25 containing oriC (Figure 1 and Table 1), and that it is the only region where both protein binds specifically. Furthermore, the binding of Mcm to oriC decreased markedly in stationary phase cells. This is consistent with our previous hypothesis that loading of Mcm to oriC is a key step for regulating DNA replication (6).
In the course of the sequence analysis of the 5 kb region bound by Cdc6 and Mcm, we found a previously unnoticed cluster of ORB-related repeats (13 or 23 bp) in the dp1 gene. The cdc6/orc1 gene, therefore, is flanked by two clusters of the ORB repeats (Figure 2). The organization of the ORB clusters is conserved in the three sequenced Pyrococcus genomes and in the T. kodakaraensis genome (Figure 2), suggesting that evolution has conserved a role for this structure in the initiation of DNA replication in the order Thermococcales. Supporting this idea, our ChIP and gel retardation assays demonstrated that Cdc6 binds to both of the two ORB clusters (Figures 3 and 4). The dnaA gene in certain gram-positive bacteria is flanked by multiple target sequences (DnaA box) on both sides (30). In Bacillus subtilis, both of the DnaA box clusters are required for the origin function, and DNA synthesis starts from within one of the DnaA-box clusters. Whether binding of Cdc6 to the two ORB clusters introduces a higher order looping structure in Pyrococcus remains to be investigated. Such a nucleo–protein complex with a higher-order structure has also been proposed to occur in Archaea (15).
Although the ORB repeat is a common characteristic of archaeal oriC, the length of ORB is variable. In S. solfataricus, Cdc6-1 recognizes not only the 36 bp ORB of oriC1 but also the 17 bp mini-ORB of oriC2 (7), while in A. pernix, ORC1 recognizes a 22 bp ORB (15). For M. thermoautotrophicus Cdc6-1, a 13 bp mini-ORB repeat is sufficient for specific binding (17). Interestingly, there are mini-ORB (13 bp) repeats and longer ORB repeats (23 or 34 bp) in Pyrococcus oriC. Our gel retardation assay revealed that purified Cdc6 binds to a single mini-ORB only weakly (Figure 4). The ChIP-chip analyses also indicate that a single mini-ORB repeat is not sufficient for stable binding of Cdc6 in vivo. For example, although a 13 bp sequence ‘TTCCAGTGGAACT’ in the region 348 is identical to that which overlaps ORB1 repeat in oriC, the region 348 does not show any preferential binding to Cdc6/Orc1 (Figure 1). The role of the 13 bp repeats located outside oriC and dp1 in the genome is unclear since their numbers and locations are not identical in the three sequenced Pyrococcus genomes. Multiple mini-ORB repeats in oriC and dp1, on the other hand, may contribute to the highly specific binding of Cdc6 observed in vivo. Unlike the mini-ORB, a single ORB repeat showed high affinity for purified Cdc6 protein (Figure 4). The ORB repeats share the same motifs with the mini-ORB repeats in Pyrococcus sp., and similar motifs in ORB of oriC1 in S. solfataricus are essential for the binding purified Cdc6-1 protein (7). However, our mutation analyses indicated that not only the conserved motifs but also their surrounding sequences are important for the binding between ORB and Cdc6 in Pyrococcus sp. (Figure 5). This explains why Cdc6 binds to ORB more efficiently than to mini-ORB. The binding of Cdc6 to ORB or mini-ORB did not fit a simple hyperbolic curve (Figure 4B). Furthermore, the shapes of the curves are not identical (Figure 4B and Supplementary Figure 2). The gel retardation assays also indicated that the complex of Cdc6 and ORB/mini-ORB is not uniform: we observed two major ‘shifted’ bands in the gel (Figure 4A). We, therefore, do not exclude the possibility that the binding of Cdc6 to a single ORB/mini-ORB repeat involves a complex mechanism such as cooperative binding. In M. thermoautotrophicus, Cdc6-1 binds cooperatively to a DNA fragment containing multiple mini-ORB repeats (17).
We also found that the two flanking regions, 228 and 229, showed statistically significant Cdc6 binding in the ChIP-chip assay in the exponential phase (P < 0.002), although the value of Mc was less than that for the region 25 (Figure 1 and Table 1). These regions do not have any ORB-like repeats and they show no DNA replication initiating activity (5). Furthermore, the genetic environment of these regions is not conserved in the three Pyrococcus genomes, suggesting that a possible role for Cdc6 is not specific for these genes. Unexpectedly, ChIP-chip analysis revealed that Mcm has a second binding site, the specificity of which is comparable to oriC. This region becomes the major binding site for Mcm in stationary phase, while the specificity for the region containing oriC decreased greatly (Figure 1 and Table 1). The region 42 contains two rRNA genes and a tRNA gene and is extremely rich in GC nucleotides (Supplementary Figure 3), raising the possibility that this GC-rich region is an obstacle to the replication fork. Although it is possible, this does not sufficiently explain the binding between Mcm and this region in stationary phase. Also, it should be noted that the three genes in region 42 are transcribed in the same direction as the replication fork movement, thus eliminating the possibility of collision between transcription machinery and DNA replication fork. Eukaryotic MCM protein is known to interact with the C-terminal domain of RNA polymerase II, HBO1 histone acetyltransferase and histone H3 (reviewed in 31). Furthermore, MCM is identified in a Yph1 complex that is involved in the biogenesis of the 60S ribosome (32). MCM is also suggested to move along with the RNA polymerase II during transcription activated by Stat1 (33). However, with the limited information available, understanding of the involvement of archaeal Mcm in such biological contexts remains elusive.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by Human Frontier Science Program, the CNRS (Programme Puces-à-ADN), the Programme InterEPST Bioinformatique, the Curie Institute, the Association de Recherche contre le Cancer Research (ARC) and the Ile-de-France Region. F.M. thanks Japan Society for the Promotion of Science for financial support. Funding to pay the Open Access publication charges for this article was provided by Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cann IK, Ishino Y. Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics. 1999;152:1249–1267. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacNeill SA. Understanding the enzymology of archaeal DNA replication: progress in form and function. Mol. Microbiol. 2001;40:520–529. doi: 10.1046/j.1365-2958.2001.02390.x. [DOI] [PubMed] [Google Scholar]
- 4.Grabowski B, Kelman Z. Archeal DNA replication: eukaryal proteins in a bacterial context. Annu. Rev. Microbiol. 2003;57:487–516. doi: 10.1146/annurev.micro.57.030502.090709. [DOI] [PubMed] [Google Scholar]
- 5.Myllykallio H, Lopez P, Lopez-Garcia P, Heilig R, Saurin W, Zivanovic Y, Philippe H, Forterre P. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000;288:2212–2215. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- 6.Matsunaga F, Forterre P, Ishino Y, Myllykallio H. In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc. Natl Acad. Sci. USA. 2001;98:11152–11157. doi: 10.1073/pnas.191387498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD. Identification of two origins of replication in the single chromosome of the archaeon. Sulfolobus solfataricus Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 8.Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl Acad. Sci. USA. 2004;101:7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson NP, Blood KA, McCallum SA, Edwards PA, Bell SD. Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. Embo. J. 2007;26:816–824. doi: 10.1038/sj.emboj.7601529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contursi P, Pisani FM, Grigoriev A, Cannio R, Bartolucci S, Rossi M. Identification and autonomous replication capability of a chromosomal replication origin from the archaeon Sulfolobus solfataricus. Extremophiles. 2004;8:385–391. doi: 10.1007/s00792-004-0399-y. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy SP, Ng WV, Salzberg SL, Hood L, DasSarma S. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome. Res. 2001;11:1641–1650. doi: 10.1101/gr.190201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang R, Zhang CT. Multiple replication origins of the archaeon Halobacterium species NRC-1. Biochem. Biophys. Res. Commun. 2003;302:728–734. doi: 10.1016/s0006-291x(03)00252-3. [DOI] [PubMed] [Google Scholar]
- 13.Berquist BR, DasSarma S. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bacteriol. 2003;185:5959–5966. doi: 10.1128/JB.185.20.5959-5966.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maisnier-Patin S, Malandrin L, Birkeland NK, Bernander R. Chromosome replication patterns in the hyperthermophilic euryarchaea Archaeoglobus fulgidus and Methanocaldococcus (Methanococcus) jannaschii. Mol. Microbiol. 2002;45:1443–1450. doi: 10.1046/j.1365-2958.2002.03111.x. [DOI] [PubMed] [Google Scholar]
- 15.Grainge I, Gaudier M, Schuwirth BS, Westcott SL, Sandall J, Atanassova N, Wigley DB. Biochemical analysis of a DNA replication origin in the archaeon Aeropyrum pernix. J. Mol. Biol. 2006;363:355–369. doi: 10.1016/j.jmb.2006.07.076. [DOI] [PubMed] [Google Scholar]
- 16.Lopez P, Philippe H, Myllykallio H, Forterre P. Identification of putative chromosomal origins of replication in Archaea. Mol. Microbiol. 1999;32:883–886. doi: 10.1046/j.1365-2958.1999.01370.x. [DOI] [PubMed] [Google Scholar]
- 17.Capaldi SA, Berger JM. Biochemical characterization of Cdc6/Orc1 binding to the replication origin of the euryarchaeon Methanothermobacter thermoautotrophicus. Nucleic Acids Res. 2004;32:4821–4832. doi: 10.1093/nar/gkh819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsunaga F, Norais C, Forterre P, Myllykallio H. Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 2003;4:154–158. doi: 10.1038/sj.embor.embor732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- 20.MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes. Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu W, Aparicio JG, Aparicio OM, Tavare S. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics. 2006;7:276. doi: 10.1186/1471-2164-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rickman DS, Herbert CJ, Aggerbeck LP. Optimizing spotting solutions for increased reproducibility of cDNA microarrays. Nucleic Acids Res. 2003;31:e109. doi: 10.1093/nar/gng109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome. Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee DG, Makhov AM, Klemm RD, Griffith JD, Bell SP. Regulation of origin recognition complex conformation and ATPase activity: differential effects of single-stranded and double-stranded DNA binding. Embo. J. 2000;19:4774–4782. doi: 10.1093/emboj/19.17.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grainge I, Scaife S, Wigley DB. Biochemical analysis of components of the pre-replication complex of Archaeoglobus fulgidus. Nucleic Acids Res. 2003;31:4888–4898. doi: 10.1093/nar/gkg662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Felice M, Esposito L, Pucci B, De Falco M, Rossi M, Pisani FM. A CDC6-like factor from the archaea Sulfolobus solfataricus promotes binding of the mini-chromosome maintenance complex to DNA. J. Biol. Chem. 2004;279:43008–43012. doi: 10.1074/jbc.M406693200. [DOI] [PubMed] [Google Scholar]
- 28.Singleton MR, Morales R, Grainge I, Cook N, Isupov MN, Wigley DB. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J. Mol. Biol. 2004;343:547–557. doi: 10.1016/j.jmb.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 29.Messer W, Weigel C. DnaA initiator—also a transcription factor. Mol. Microbiol. 1997;24:1–6. doi: 10.1046/j.1365-2958.1997.3171678.x. [DOI] [PubMed] [Google Scholar]
- 30.Moriya S, Imai Y, Hassan AK, Ogasawara N. Regulation of initiation of Bacillus subtilis chromosome replication. Plasmid. 1999;41:17–29. doi: 10.1006/plas.1998.1381. [DOI] [PubMed] [Google Scholar]
- 31.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du YC, Stillman B. Yph1p,an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–848. doi: 10.1016/s0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- 33.Snyder M, He W, Zhang JJ. The DNA replication factor MCM5 is essential for Stat1-mediated transcriptional activation. Proc. Natl Acad. Sci. USA. 2005;102:14539–14544. doi: 10.1073/pnas.0507479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.