Abstract
The mechanism of mitochondrial DNA replication is a subject of intense debate. One model proposes a strand-asynchronous replication in which both strands of the circular genome are replicated semi-independently while the other model proposes both a bidirectional coupled leading- and lagging-strand synthesis mode and a unidirectional mode in which the lagging-strand is initially laid-down as RNA by an unknown mechanism (RITOLS mode). Both the strand-asynchronous and RITOLS model have in common a delayed synthesis of the DNA-lagging strand. Mitochondrial DNA is replicated by a limited set of proteins including DNA polymerase gamma (POLG) and the helicase Twinkle. Here, we report the effects of expression of various catalytically deficient mutants of POLG1 and Twinkle in human cell culture. Both groups of mutants reduced mitochondrial DNA copy number by severe replication stalling. However, the analysis showed that while induction of POLG1 mutants still displayed delayed lagging-strand synthesis, Twinkle-induced stalling resulted in maturated, essentially fully double-stranded DNA intermediates. In the latter case, limited inhibition of POLG with dideoxycytidine restored the delay between leading- and lagging-strand synthesis. The observed cause-effect relationship suggests that Twinkle-induced stalling increases lagging-strand initiation events and/or maturation mimicking conventional strand-coupled replication.
INTRODUCTION
Human mitochondrial DNA (mtDNA) is a closed circular molecule of ∼16.5 kb and was sequenced 25 years ago (1,2). The two strands of mtDNA are denoted as the Heavy-(H)-strand and the Light-(L)-strand on the basis of their mobility in a denaturing caesium chloride gradient.
The strand-asynchronous or strand-displacement model for mammalian mitochondrial DNA replication was first proposed in the early 70s (3). In this model, synthesis of the nascent H-strand starts at a fixed point in the major non-coding region (NCR) of mtDNA denoted OH. OH was originally defined by mapping the 5′ ends of the so called D-loop and is located on the L-strand upstream of three conserved sequence blocks. Leading-strand (nascent H-strand) synthesis proceeds two-thirds of the way around the molecule, displacing the parental H-strand in the process with mitochondrial single-stranded DNA-binding protein (mtSSB) suggested to provide protection against the action of nucleases and other insults such as reactive oxygen species. Following exposure of the lagging-strand initiation site (OL) synthesis of the nascent L-strand begins (4,5).
More recently, Holt and co-workers proposed two models of mtDNA replication, one a more conventional strand-synchronous theta mode (6–9) where mtDNA replication initiates bidirectionally at various sites across an initiation zone (OriZ). In this case termination occurs at or near OH. The other mode of replication is similar to the strand-asynchronous mode of replication so that the nascent L-strand DNA was suggested also to be synthesized with a considerable delay. Initiation is essentially unidirectional and occurs in the NCR, importantly however RNA is deposited on the displaced H-strand rather than mtSSB, thus forming ribonucleotide incorporation throughout the lagging strand (RITOLS) intermediates, which is a crucial difference from the strand-asynchronous model (10). Although the high levels of mtSSB (11) could be seen as supporting the strand displacement model, also for example Escherichia coli is estimated to have several thousands of molecules of SSB (12) even though it contains a single copy genome and replicates via conventional theta replication. SSB is nevertheless essential, as it would be in mammalian mitochondria, not only at the replication fork but also in repair, recombination and other DNA maintenance processes. Given the various essential functions of SSB, the high levels might simply reflect a cell's precaution to ensure it is readily available.
The RITOLS model requires that the RNA is replaced by DNA to produce a dsDNA lagging-strand. It was shown that the RITOLS replication intermediates (RIs) are prone to RNaseH degradation during mtDNA purification leaving a single-stranded parental H-strand (7), thus producing RIs originally predicted by the strand-asynchronous model. Strand-asynchronous RIs are therefore considered purification/degradation artefacts. In rodent and chick liver and cultured human cells under normal culture conditions RITOLS intermediates are the predominant class (6,9,10). However, in cultured human cells recovering from mtDNA depletion, the majority of the replication intermediates are essentially double-stranded DNA suggesting a switch from the RITOLS replication mode to more conventional theta replication (9). Alternatively, initiation of lagging-strand DNA synthesis occurs more frequently resulting in an increased rate of conversion of RITOLS RIs to dsDNA RIs.
All proteins responsible for mammalian mtDNA maintenance are encoded in the nucleus, translated by cytosolic ribosomes and imported into the mitochondrial compartment. So far, a limited number of proteins has been identified. These include the mitochondrial DNA polymerase gamma (POLG1) and its accessory subunit (POLG2) [see, e.g. (13)], the mitochondrial DNA helicase Twinkle (14,15), mitochondrial single-stranded DNA-binding protein (mtSSB) (16) and various proteins with a more general role in mtDNA maintenance. The POLG holoenzyme, Twinkle and mtSSB can form a minimal mitochondrial replisome capable of genome length DNA synthesis on an artificial template (17). Some of the components of the mitochondrial replisome and transcription machinery show similarity to their counterparts in T-odd bacteriophages suggesting that a T-odd phage ancestor contributed to the early ‘mitochondrial’ endosymbiosis event (18). For example, Twinkle shows striking similarity to the T7 phage primase/helicase protein gp4 (T7 gp4) (14). The Metazoan primase domain of Twinkle has diverged from the ones of more primitive Eukaryotes and T-odd phages suggesting it has lost its primase function (19).
The genes for Twinkle, POLG1 and more recently POLG2 (20) have been implicated in human diseases. Autosomal dominant (ad) mutations in Twinkle are associated with Progressive External Ophthalmoplegia (adPEO)(14), while a single recessive mutation is associated with infantile onset spinocerebellar ataxia or IOSCA (21). Mutations in POLG1 are associated with a variety of disorders, including dominant and recessive PEO, various types of ataxia, Parkinsonism and the severe mtDNA depletion Alpers syndrome (see, e.g. (22) and references therein, and http://dir-apps.niehs.nih.gov/polg/).
The catalytic subunit of polymerase gamma, POLG1, is well-characterized biochemically (see (13), bearing similarity with prokaryotic A-type DNA polymerases such as E. coli DNA polymerase I and T7 DNA polymerase. Conserved regions include a C-terminal domain responsible for polymerase activity and an N-terminal 3′-5′ exonuclease domain involved in proofreading. Several disease associated POLG1 mutations have been studied using purified recombinant enzyme. These include the common autosomal dominant Y955C and other mutations, which result in a moderate to severe decrease in polymerase activity, reduced nucleotide selectivity or reduced processivity (23).
In vivo, the properties of POLG1 have also been partly characterized in yeast and in cultured human cells (24–27). In both cases, expression of a mutant form of the protein deficient in 3′-5′ exonuclease activity results in the accumulation of mtDNA mutations. An exonuclease deficient variant in mouse also results in a mutator phenotype and shows a whole-organism phenotype of reduced lifespan with a variety of tissue specific ageing associated defects (28,29). Expression of an adPEO associated Twinkle mutation in transgenic mice has shown a late onset phenotype with striking similarities to late onset PEO (30).
Although there is a need for further biochemical characterization of POLG1 and Twinkle mutants, and animal models can provide a wealth of information on disease aetiology and pathogenesis, both approaches have their limitations. We therefore chose an alternative approach of inducible expression of wild-type and mutant POLG1 and Twinkle in cultured human cells, allowing us to study protein function and mtDNA replication dynamics in vivo. Using this inducible system in combination with two-dimensional neutral/neutral agarose gel-electrophoresis (2DNAGE), we show here that the induced expression of either Twinkle or POLG1 mutants results in distinct replication stalling phenotypes suggesting defined roles for both proteins in mtDNA replication and in particular the frequency of initiation of lagging-strand maturation/synthesis.
MATERIALS AND METHODS
Cloning of expression constructs
The full-length cDNA of POLG1 and Twinkle variants were originally cloned in the pcDNA3.1(−)/Myc-His A (Invitrogen, Carlsbad, CA, USA), as previously described (14,27). All constructs were re-cloned in the pcDNA5/FrT/TO vector (Invitrogen) taking advantage of two PmeI restriction sites flanking the multiple cloning sites of the original pcDNA3 vectors and target vector. The resulting fusion proteins contained the sequence of the respective proteins followed by the Myc-His. All resulting plasmid-constructs were confirmed by DNA sequencing.
Creation and maintenance of stable transfected inducible cell lines
The Flp-In™ T-REx™ 293 host cell-line (Invitrogen), a HEK293 variant containing a Flip recombination site at a transcriptionally active locus, was grown in DMEM medium (Cambrex Bioscience, Walkersville, MD, USA) with 2 mM L-glutamine (Cambrex Bioscience), 10% FCS (Euroclone, Milan, Italy) and 50 μg/ml uridine (Sigma, St. Louis, MO, USA) supplemented with 100 μg/ml Zeocin (Invivogen) and 15 μg/ml Blasticidin (Invivogen) in a 37°C incubator at 8.5% CO2. Two-day prior to transfection cells were split to 10 cm plates and grown to ∼80% confluence in medium lacking antibiotics. Cells were co-transfected with TransFectin (Bio-Rad, Hercules, CA, USA) according the manufacturer's protocol with the appropriate pcDNA5/FrT/TO construct (0.4 μg) and pOG44 (Invitrogen; 3.6 μg), a plasmid encoding the Flp-recombinase necessary for targeted stable integration. Six hour later, transfection medium was replaced with regular fresh medium lacking antibiotics. Twenty-four hour after transfection the selective antibiotics Hygromycin (150 μg/ml) (Invivogen) and Blasticidin (15 μg/ml) were added. Selective medium was replaced every 2 days for cell maintenance. All inducible cell lines were created according this method. To induce expression the indicated amount of doxycycline (Sigma) was added to the growth medium, and cells were processed for further analyses. With longer than 2 days induction medium was refreshed every 2 days.
Western blot analysis
Cell lysates were prepared and analyzed for protein expression by immunoblotting after SDS-PAGE (27). A primary monoclonal c-myc (Roche Molecular Biochemicals, Nutley, NJ, USA) antibody was used for detection of recombinant proteins. Peroxidase-coupled secondary antibody horse-anti-mouse was obtained from Vector Laboratories, Burlingame, KS, USA. Enhanced Chemiluminescence detection was done essentially as described (27).
Quantitative PCR
The copy number of mitochondrial DNA per cell was determined by real time PCR of cytochrome b using the gene for amyloid precursor protein APP as a nuclear standard as described (31). Briefly, crude nucleic acid extracts were obtained from cells by lysis, proteinase K digest and subsequent isopropanol precipitation, and copy numbers of cytochrome b and APP were determined in a duplex Taqman PCR on an Abiprism 7000 (Applied Biosciences, Foster City, CA, USA) using pCR 2.1-TOPO (Invitrogen) vectors containing the cytochrome b and APP amplicon as standards.
Immunocytochemistry
Immunofluorescent detection was done essentially as described previously (32). For the detection of mtDNA we used a monoclonal anti-DNA antibody AC-30-10 (PROGEN, Shingle Springs, CA, USA) as described previously (33). Secondary antibodies were anti-mouse IgG-Alexa Fluor®488 (Invitrogen; Myc) and anti-mouse IgM-Alexa Fluor®568 (DNA). Image acquisition using confocal microscopy was done as described (32).
Protein isolation and helicase assays
In vitro assays for determination of helicase activities were performed with highly enriched Twinkle preparations derived from 293 Flp-In™ T-REx™ cells. The cells were induced with 50 ng/ml doxycycline (Sigma) for 2 days, harvested and mitochondria isolated by hypotonic lysis and differential centrifugation (32). The mitochondrial pellet obtained was lysed in high salt buffer (50 mM KH2PO4 pH 7.0, 1 M NaCl, 1% Triton X-100, 1 × complete Protease inhibitors EDTA-free, Roche) and sonicated on ice (Sonics Vibra-cell, 1 min 40% amplitude, 1 s pulses with 2 s break). The insoluble DNA fraction was pelleted for 10 min at 12 000 g and 4°C. Supernatant was incubated with Talon metal-affinity resin (Clontech, Palo Alto, CA, USA) for 1–2 h at 4°C to allow binding of His-tagged proteins. Resin was washed twice with high salt buffer and twice with low salt buffer (25 mM Tris–HCl pH 7.6, 40 mM NaCl, 4.5 mM MgCl2, 10% glycerol, 100 mM L-Arginine) containing 20 mM Imidazole. Elution was carried out with low salt buffer containing 500 mM Imidazole. The supernatant of this step was shock-frozen in liquid nitrogen and stored at −80°C.
As standard substrate for helicase assays a radioactively end-labeled 60 nt oligonucleotide hybridized to M13 ssDNA was used (5′ACATGATAAGATACATGGATGAGTTTGGACAAACCACAACGTAAAACGACGGCCAGTGCC 3′), forming a 20 nt double-stranded stretch with a 40 nts 5′ overhang.
The assay was performed by incubating 1 ng Twinkle protein in 40 μl helicase buffer (25 mM Tris pH 7.6, 40 mM NaCl, 4.5 mM MgCl2, 100 mM L-Arginine-HCl pH 7.6, 10% glycerol, 3 mM UTP, 1 mM DTT, 5 μM unspecific oligonucleotide) with 2 amol substrate for 30 min at 37°C. The reaction was stopped by adding 10 μl loading buffer (90 mM EDTA, 6% SDS, 30% glycerol, 0.25% bromophenol blue). Twenty microliter reaction mixes were separated on a 15% acrylamide gel in 1 × TBE, dried on a vacuum gel drier and exposed to X-ray film or quantified by phosphoimager.
Mutation sequencing
Point mutation levels in the NCR and cytochrome b region of mtDNA of POLG1 cell lines were measured as previously described (34).
Brewer-Fangman 2D neutral/neutral Agarose electrophoresis
Mitochondrial nucleic acids were extracted using cytochalasine (Sigma-Aldrich) as described (9). Purified mtDNA was digested with HincII and where mentioned further treated with RNase H or S1 nuclease (Fermentas, Hanover, MD, USA) with the indicated amounts and time. The fragments were separated by 2DNAGE as described (35,36) and the gels were blotted and hybridized with a 32P-labeled DNA probe for human mtDNA nts 14 846–15 357 (9).
RESULTS
Establishing stable inducible cell lines expressing mtDNA replication factors
In order to study the mtDNA maintenance machinery in cultured human cells, inducible cell lines expressing wild-type and mutant variants of Twinkle, the Twinkle splice variant Twinky (14) or POLG1 were established using HEK293 Flp-In™ T-REx™ cells (see Table 1 for a list of all variants). Twinkle mutants included a lysine mutation (K421A) in the highly conserved Walker A motif implicated in ATP binding and hydrolysis; a mutation (G575D) in helicase motif H4 and implicated in DNA binding (37); a deletion of 31 amino acids (▵346–376) of the region that shows similarity with the T7 gp4 linker region that has been implicated in multimer formation [see e.g. (38)]; a large deletion (▵70–343) in the region of the protein that shows similarity with the T7 gp4 primase domain (19). POLG1 mutants included two polymerase deficient mutants (D890N and D1135A), one exonuclease deficient mutant D198A and a non-deleterious deletion mutant (ΔCAG) of 10 consecutive glutamines in the N-terminus, all as previously described (27). All cell lines and >99% of all cells expressed the recombinant proteins upon doxycycline (DC) induction and expressed proteins were all targeted to the mitochondrial compartment (Figure 3 and not shown).
Table 1.
Expressed proteins with their predicted sizes
| Variant | Size (kDa) |
|---|---|
| Twinklea wt | 74 |
| K421A, G575D | |
| Twinklea ▵AA346–376 (▵346–376) | 71 |
| Twinkya | 64 |
| Twinklea ▵AA70–343 (▵70–343) | 41 |
| POLG1 wt | 142 |
| D198A, D890N, D1135A | |
| POLG1 ▵CAG | 141 |
All expressed proteins had the MycHis epitope tag and this tag was included in the calculation of size in kiloDaltons (kDa).
aSize based on predicted processing at AA42.
Figure 3.
Subcellular localization of Twinkle variants. (A) Localization of all Twinkle variants was determined by immunocytochemistry. Two examples compare the obtained signals without (−DC) induction with induction (+DC) at 3 ng/ml DC. For this comparison images were taken with exactly the same settings for exposure, laser intensity, etc. Images were in addition processed for brightness and contrast in the same manner. The nucleus in most images in Figure A is also faintly visible due to the inclusion of DAPI in the mounting medium. In the first panel (n) indicates the nuclei. (B) Two examples of co-localization of mutant Twinkle variants visualized using anti-Myc tag antibody (−αMyc) with mtDNA using anti-DNA antibody (−αDNA) are shown. Arrows indicate a selection of foci that co-localize but by now means show all co-localizing foci as most protein foci did co-localize with more or less clear foci stained for mtDNA.
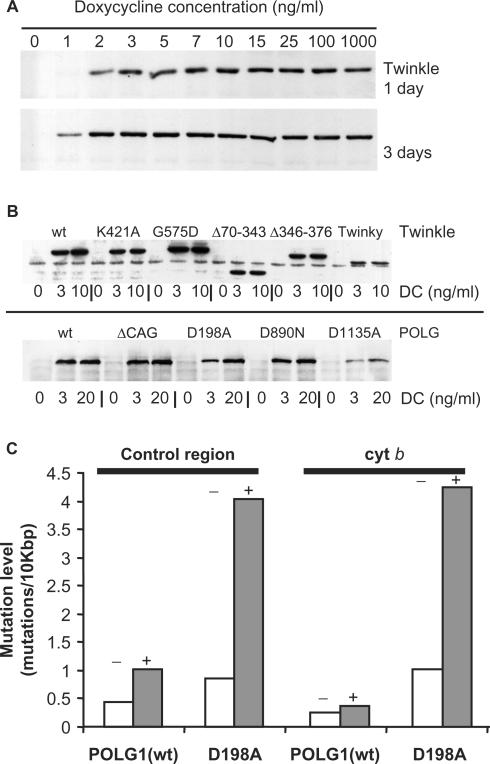
Since DC is a mitochondrial protein synthesis inhibitor at μg/ml concentrations, we first determined the lowest possible levels of DC to achieve full induction. Figure 1A shows an increase of protein expression of wild-type (wt) Twinkle in cells with increasing DC concentrations (0–1000 ng/ml.) Both after one or three days, maximum induction levels were reached at low ng/ml concentrations, but at slightly lower concentrations after three days induction.
Figure 1.
Expression of mtDNA maintenance proteins in HEK293 Flp-In™ T-REx™ cells. (A) Twinkle wild-type (wt) expressing cells were induced with various concentrations of doxycycline (DC) and analyzed by western blot analysis after 1 or 3 days of induction. Maximum expression levels were reached within the low ng/ml range. Similar results were obtained for all other cell lines. (B) Expression of all proteins used in this study was confirmed by western blot analysis comparing no induction with 3 ng/ml DC and full (10 or 20 ng/ml DC) induction for 3 days. Note that full induction with the POLG1 cell lines was chosen as 20 ng/ml, only in order to make sure maximum expression was reached as expression of POLG1 was generally much weaker when compared with the expression of Twinkle variants. (C) POLG1 wild type (wt) and POLG1 D198A expressing cells were grown for 2 months either without (−) or with (+) 3 ng/ml DC. MtDNA mutation levels were determined for part of the cytochrome b gene (cyt b) and part of the non-coding control region using a PCR-based approach (see Materials and Methods section). Results show that the D198A accumulated ∼5–10-fold increased level of mutations compared to non-induced D198A and induced POLG1 wt. The mutation pattern was similar as reported earlier for this POLG1 variant (27).
Figure 1B shows the expression of all proteins used in this study confirmed by immunoblotting, following 72 h of treatment with 0, 3 and 10 or 20 ng/ml DC. All proteins were detected using their respective epitope tag and gave bands of the expected size (Table 1) upon induction. In the absence of DC we could detect leaky expression of most Twinkle variants, but only when films were overexposed considerably (not shown). However, immunofluorescence in the absence of DC induction did not result in any mitochondrial signal above background fluorescence suggesting the expression levels were very low (see also below and Figure 3A). Some of the analyzed Twinkle variants, such as Twinky, showed reproducibly lower protein levels with full induction of expression, indicating differences in protein (or mRNA) stability of these variants. Similarly, the POLG1 D1135A mutant showed lower expression levels than the other POLG1 variants, suggesting that the mutant protein is less stable.
As a final test for the inducible expression system, we created a cell-line expressing a POLG1 variant (D198A) in which exonuclease activity is abolished. We have shown previously that constitutive expression of D198A in cultured human cells results in the accumulation of point mutations in mtDNA (27). To validate the obtained inducible cell-line we determined the point mutation levels in two regions of mtDNA (Figure 1C). After 60 days of induction, both the cytochrome b and control region showed elevated mutation levels in the D198A cell-line while non-induced D198A cells showed low mutation levels similar to cells expressing POLG1 wt.
Expression of several Twinkle and POLG1 mutants results in mtDNA copy-number depletion
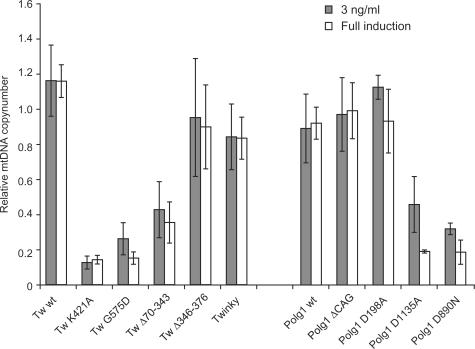
The relative mtDNA copy number in the various inducible cell lines was compared by quantitative real-time PCR (QPCR) using the nuclear amyloid precursor protein (APP) gene as a standard (31) (Figure 2). The absolute copy number determined by us for the HEK293 Flp-In™ T-REx™ and the majority of non-induced transgenic cells was ∼3000 copies/cell (2798 ± 450 (n = 4) for the non-induced parental cell line). Induced overexpression of POLG1 wt or Twinkle wt did not significantly change mtDNA copy number per cell, indicating that abundance of these proteins is not rate-determining for mtDNA replication at least in cell culture. More importantly it also showed that overexpression per se did not otherwise interfere with mtDNA replication. Similarly, overexpression of Twinky, Twinkle Δ346–376, POLG1-D198A and POLG1-ΔCAG did not influence steady-state mtDNA levels. In contrast even low-level expression of Twinkle mutants K421A or G575D and POLG1 mutants D890N or D1135A lead to a dramatic decrease of mtDNA levels within a few days. The Twinkle K421A and G575D cell lines showed a significant steady-state reduction in mtDNA copy number of ∼60% even prior to induction, presumably caused by the slightly leaky expression of the Twinkle variants. This suggests these mutants are strongly dominant in nature. In contrast, the POLG1 D890N and D1135A mutants did not show a significant copy-number reduction without induction (not shown). Notwithstanding the minor leakiness, depletion upon induction was dose-dependent, as higher expression levels lead to a faster depletion than low-level expression (data not shown). For the Twinkle K421A and G575D as well as the POLG1 D890N and D1135A mutants, the mtDNA levels after three days of full DC induction were ∼20–30% compared to non-induced cells, indicating a complete abolishment of successful replication and dilution of mtDNA by cell division.
Figure 2.
POLG1 and Twinkle mutants can cause mitochondrial DNA copy-number depletion. MtDNA copy number was determined using duplex Taqman PCR. Shown are the copy numbers for each cell line relative to their copy number in non-induced cells following either 3 ng/ml or full induction (10 ng/ml for Twinkle variants; 20 ng/ml for POLG1 variants). Note that although Twinkle K421A and G575D cell lines had a reduced steady-state copy number in non-induced cells, induction resulted in a further strong drop in copy number (see also main text). The same general trends as observed here were obtained by Southern analysis (not shown). Copy number decline was highly significant in Twinkle K421A, G575D and ▵70–343 lines as well as in the POLG1 D890N and D1135A mutant lines.
Several Twinkle mutants show altered nucleoid localization
The localization of Twinkle variants was analyzed by immunofluorescence using the myc-tag of the recombinant proteins (Figure 3A). Overexpressed Twinkle wt showed the typical punctuated pattern within mitochondria, indicating the normal localization in mtDNA nucleoids. Twinky and ▵346–376 both showed diffuse mitochondrial staining. The N-terminal deletion variant ▵70–343 showed almost normal punctuate nucleoid-like localization. The variant G575D could be detected in punctate foci, but in addition showed enhanced diffuse staining in mitochondria. The K421A variant having a mutation in the WalkerA motif was detected in few enlarged spots, indicating either abnormal nucleoid segregation or protein aggregation. To differentiate between these possibilities we used an anti-DNA antibody to see to what extent this and other Twinkle mutants co-localized with mtDNA. As previously shown (32), Twinkle wt showed excellent co-localization with DNA as did ▵70–343 (not shown). The K421A variant, despite its abnormal appearance of enlarged foci, did co-localize with mtDNA (Figure 3B). The number of DNA foci, however, was severely reduced compared to non-expressing or Twinkle wt expressing cells. For Twinkle G575D, the number of nucleoids was again severely reduced (Figure 3B) but most of the protein-foci co-localized with mtDNA. Both Twinky and the ▵346–376 variant showed normal nucleoid numbers judging from detection with the anti-DNA antibody.
Helicase activity of Twinkle mutants is reduced or absent
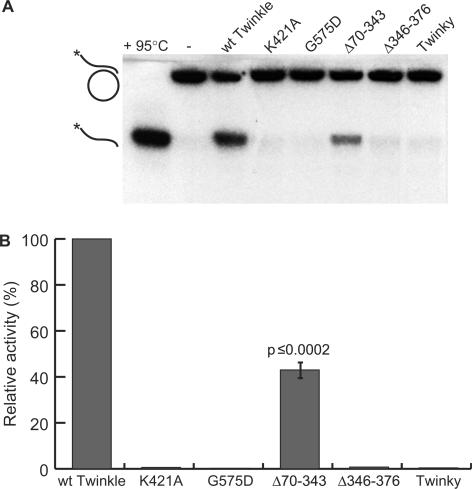
The helicase activity of Twinkle variants was compared using His-affinity purified protein in an in vitro helicase assay. Twinkle wt showed the expected helicase activity and was able to unwind a DNA substrate with a 5′ overhang of >20 bases, as long as the double-stranded part was less than 25 base pairs (Figure 4 and data not shown). No such helicase activity could be detected with ▵346–376 or Twinky. Similarly, proteins bearing the mutation K421A in the Walker A motif or the G575D mutation in the helicase motif H4 also had less than 5% residual unwinding activity. ▵70–343 had ∼40% helicase activity of the wild-type protein.
Figure 4.
Helicase activities of purified Twinkle protein fractions. (A) An example of a helicase activity assay using the various Twinkle variants purified to near homogeneity. As indicated on the left-hand site, the substrate consisted of M13 plasmid with an annealed 60-nt 5′end-labeled oligo with a long (40 nt) single-stranded 5′ overhang. (−) indicates the purified substrate whereas the denatured substrate (95°C) indicates the released labeled product, which is also released when a purified protein contains helicase activity. (B) This panel shows the averaged results of six independent measurements, using several independent protein preparations. Except for the ▵70–343 mutant, which still had activity though significantly (Student's t-test) reduced, all other mutants had essentially lost all helicase activity.
Twinkle and POLG1 mutants show different replication stalling phenotypes
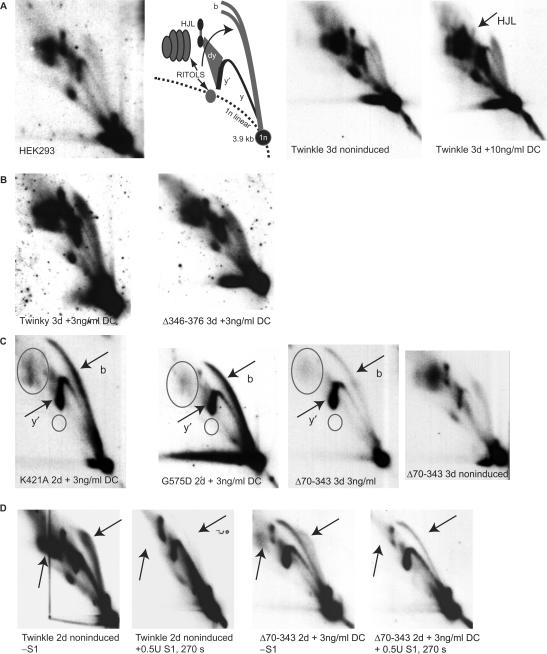
The effect of overexpression of the various mutant proteins on replication was studied using two-dimensional neutral/neutral agarose gel electrophoresis (2DNAGE) and Southern blotting, as first established by Brewer and Fangman (36,39). This method allows visualization of RIs, as DNA fragments are separated both by size and shape.
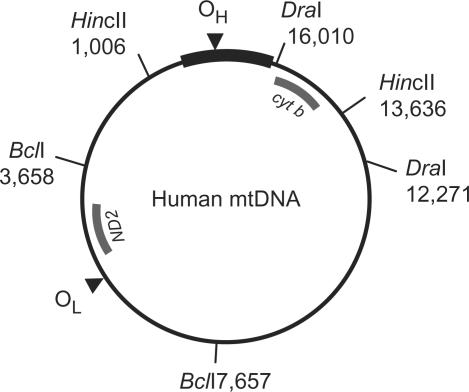
When applied to the analysis of mtDNA isolated from cultured cells, Holt and co-workers showed that various types of RIs can be detected [see e.g. (9)]. Figure 5 shows a schematic figure of human mtDNA, indicating the appropriate restriction fragments and probes used for detection of these fragments on 2DNAGE gel blots. The first 2 panels of Figure 6A show the example of a HincII digest of purified HEK293 mtDNA run on a 2DNAGE gel and probed for the 3.9 kb mtDNA fragment (nucleotide number [nt] 13 636–1006) that includes the whole NCR (for a detailed explanation of the various RIs see Supplementary Data). All the analyses shown in the subsequent figures consider this HincII fragment. Analysis of a second region of the genome was also performed (Supplementary Figure 1).
Figure 5.
Digests and probes for human mtDNA. A schematic map of human mtDNA is shown with the positions of the fragments detected by 2DNAGE as in Figures 6–8 and Supplementary Figures 2 and 3 (HincII digests-fragment nt 13 636–1006 detected with a cytochrome b probe (cyt b, nt 14 846–15 357)), Supplementary Figure 1 (BclI digest- fragment nt 3658–7657 detected with an ND2 probe, nt 4480–4984) and Supplementary Figure 5 (DraI digest-fragment nt 12 271–16 010 also detected with the cyt b probe). Probes are indicated with names and a thick gray line. The NCR is indicated with a thick black line. Please note that for clarity only the restriction sites of both ends of the probed fragments are shown here, even though all three enzymes cut at multiple sites on human mtDNA.
Figure 6.
Some Twinkle mutants cause replication stalling. 2DNAGE samples for all panels consisted of purified mtDNA digested with HincII and probed with a radiolabeled cytochrome b gene fragment (nt 14 846–15 357). The detected fragment includes the non-coding region of mtDNA also including the cytochrome b, ND6, part of the ND5 gene and intervening tRNA genes (nt 13 636–1006). (A) The first two panels on the left show the RIs of HEK293 cells and the interpretation based primarily on earlier 2DNAGE analysis of mtDNA RIs (7,9). Abbreviations: 1n, 3.9 kb non-replicating HincII fragment. (b) bubble arcs. MtDNA bubble arcs are usually very sensitive to RNase H due to the presence of patches of RNA–DNA especially on the lagging strand; these therefore also fall in the category of RITOLS as do various other RIs. Here, y and y′ indicate ascending and descending parts of the y arc and (dy) indicates double-Y structures. These will eventually form resolution intermediates resembling Holliday junctions (HJL-Holliday junction like molecules). The two panels on the right show a comparison of RIs of non-induced and fully induced cells expressing Twinkle wt. The only notable difference in this case is a reproducible reduction in one of the HJL RIs as indicated. (B) A normal pattern of RIs similar to non-expressing cells was observed in cells expressing Twinky or the ▵Linker variant. (C) K421A, G575D and ▵70–343 show similar patterns of replication stalling, with increased bubble (b) and descending Y-arc (y′) intensities, a sharpening and lengthening of the bubble arc and loss of RITOLS (ovals). The right-most panel shows the same exposure 2D gel pattern of the non-induced ▵70–343 line showing the typical HincII fragment pattern, including abundant RITOLS. (D) A limited S1 digestion illustrates that stalled RIs observed in panel C are S1 insensitive (right two panels). The effectiveness of the S1 treatment is illustrated by the left two panels, showing the effect on Twinkle non-induced cells. Similar to the S1 treatment RNase H treatment shows that the stalling RIs observed with Twinkle mutants are largely insensitive to this enzyme (Supplementary Figure 2), showing that the observed stalled RIs in panel C are essentially dsDNA. Although the intensities in subfigures A–D cannot be directly compared due to differences in exposure time, each panel contains appropriate controls of similar exposure. For example, the exposures of the left two panels in C have been chosen to be similar in comparison to the right-most panels to properly illustrate the severity of the stalling phenotype.
We applied the 2DNAGE methodology to analyze the effects of overexpression of POLG1 and Twinkle variants on mtDNA RIs and in particular to test the hypothesis that the variants depleting mtDNA do so by causing non-site specific, general replication stalling or pausing. First we examined the effect of overexpression of wild-type Twinkle by comparing mtDNA RIs with and without induction (Figure 6A, right two panels). Very little change in quality or quantity of RIs was observed, with the exception of a presumed resolution intermediate which markedly decreased after induction with >3 ng/ml DC. Similar results were observed with induced expression of untagged wild-type Twinkle (not shown). Increased expression of ▵346–376 or Twinky showed no effect on overall replication (Figure 6B) or RIs.
When overexpressing the Twinkle mutant K421A or G575D, a considerable increase in y- and bubble RIs was observed (Figure 6C). Since at the same time the total amount of mtDNA decreased strongly, these results indicate a severe, non-site specific, slowing down of replication fork movement. In parallel there was a concomitant decrease in RNA containing RIs, RITOLS or partially single-stranded RIs. Thus, the bubble arc was not only sharper and longer than normal (Figure 6C) but was substantially resistant to RNase H treatment (Supplementary Figure 2). In K421A and especially G575D expressing cells the majority of mtDNA molecules were found on the bubble arc, indicating stalling occurred in the early phase of replication as replicating molecules forming bubble arcs on 2DNAGE harbor initiation site(s) in the fragment. The fraction of molecules in a replicative state was ∼30% in the 3.9 kb HincII fragment, compared to <5% in control cells. In contrast, the ▵70–343 mutant showed a somewhat milder stalling phenotype (Figure 6C), with the majority of RIs in the upper part of the bubble arc and on the descending area of the y-arc suggesting that replication is not aborted so frequently in the very early stages as was the case for the K421A and G575D mutants. Nevertheless, in this case also RITOLS and single-stranded RIs were considerably reduced. Finally, S1 nuclease treatment for various lengths of time with a fixed amount of enzyme resulted only in very minor changes in the abundance of the stalled RIs in Twinkle stalling mutants (Figure 6D shows the treatment of ▵70–343 isolated mtDNA). Only with the longest treatment was there some reduction in the bubble arc intensity with a slight concomitant increase in the ascending part of the y-arc, suggesting some single-strandedness in the bubbles resulting in broken bubbles upon S1 nuclease treatment. This is not unexpected as some single-strandedness is always expected close to the junction point of the bubble structure of replication intermediates. The ▵70–343 stalling in this case still showed some RITOLS intermediates and these were efficiently removed by the S1 treatment (compare regions marked by arrows in the right two panels of Figure 6D). Nevertheless, most RIs were insensitive to the S1 nuclease showing that they are essentially dsDNA (this was further supported by the analyses shown in Supplementary Figures 5 and 6). In contrast, simultaneous S1 treatment in samples with abundant RITOLS showed a strong reduction in RITOLS RIs, illustrating the effectiveness of the S1 treatment (compare regions marked by arrows in the left two panels in Figure 6D).
Overexpression of POLG1 wt (Figure 7A) and POLG1 ▵CAG (not shown) did not result in any obvious effect on the RIs whereas even modest expression of the mutant variants D890N and D1135A lead to a clear increase in y- and bubble arcs, suggesting these POLG1 mutants also caused replication stalling (Figure 7B). However, unlike the stalling Twinkle variants, RITOLS and single-stranded RIs persisted in the case of D890N and D1135A POLG1 mutants (Figure 7B), even after full induction of the mutant proteins for three days (not shown).
Figure 7.
Replication stalling by POLG1 mutants. (A) Induction of wt POLG1 does not modify RIs compared to non-induced cells. (B) Induction of POLG1 mutants D890N and D1135A results in a similar pattern of replication stalling with an increased intensity both of bubble (b) and descending y (y′) RIs. RITOLS are not affected in sharp contrast with Twinkle-mutant induced stalling (Figure 6). All panels in A and B are cropped images from a single autoradiograph. (C) Fully induced expression of POLG1 D198A results in a modest change in RIs similar to but less severe than low-level induction of D890N and D1135A. This did, however, not result in copy number depletion (Figure 2). All panels in C are cropped images from a single autoradiograph.
The proofreading deficient D198A POLG1 showed little change in the appearance of RIs. Three ng/ml DC slightly enhanced both the Y- and bubble arc (Figure 7C). Only after full induction did we observe a phenotype suggestive of stalling, similar to but clearly less severe than that observed with D890N and D1135A overexpression at low induction levels.
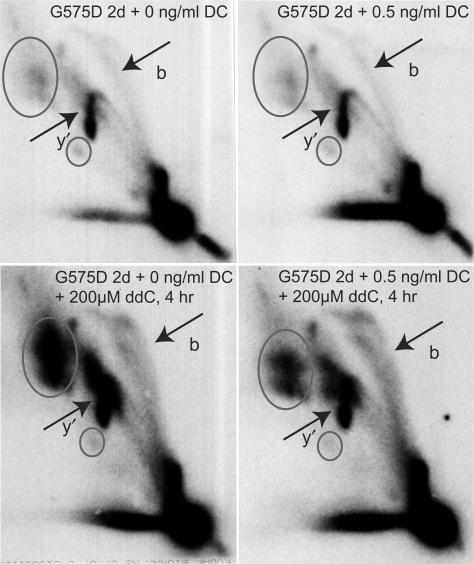
Comparison of Twinkle induced replication stalling with POLG induced replication stalling showed a considerable quantitative difference in RITOLS and single-stranded RIs (see also Supplementary Figures 3 and 5). We hypothesized (see also Discussion) that this difference could be explained by involvement of POLG1 in initiation of lagging-strand DNA synthesis or maturation. In this scenario, expression of POLG1 mutants would not only result in stalling of the leading-strand but also in delayed lagging-strand DNA synthesis. To test whether inhibition of POLG1 function could delay lagging-strand DNA synthesis in cells overexpressing either Twinkle wt or the stalling mutations ▵70–343, G575D and K421A, cells were treated with the cytidine analogue dideoxycytidine (ddC), a competitive inhibitor of POLG. Incorporation instead of deoxycytidine can result in chain termination. Surprisingly, already a short 3–4 h treatment with a high dose of ddC (200 μM) in cells showing modest stalling due to Twinkle mutants like G575D, resulted in the reappearance of considerable levels of RITOLS, while accumulated y- and bubble-arc RIs were as prominent as in untreated cells (Figure 8). The appearance of RIs under the applied conditions was highly similar to the appearance observed with POLG1 stalling (compare lower panels of Figure 8 with those in Figure 7B). Due to the slightly leaky nature of the Flp-In™ T-REx™ system, the strongest Twinkle stalling mutants G575D and K421A already showed signs of stalling without induction. In particular, the G575D mutant had already lost most RITOLS in the absence of DC. Treatment of these cells with ddC showed a similar reappearance of RITOLS (Figure 8). The same regime of ddC treatment did not result in obvious changes in RIs in cells expressing Twinkle wt or in non-induced Twinkle wt cells (not shown). Finally, ddC treatment of cells showing a strong stalling phenotype caused by higher level expression of Twinkle mutants did not result in a clear increase in RITOLS (not shown), suggesting that replication was completely abolished under these conditions (see Discussion section).
Figure 8.
Inhibition of POLG1 by ddC results in the reappearance of RITOLS RIs following stalling induced by Twinkle G575D. Non-induced and induced (0.5 ng/ml DC) Twinkle G575D cells were treated for 4 h prior to mtDNA isolation with 200 μM ddC. Note that exposures of these four samples are identical as all four samples were run on a single gel and blot. Markings are similar as in Figure 6.
DISCUSSION
Because mtDNA encodes some of the central components required for cellular energy metabolism, its maintenance is essential for development and overall cell function. While human and mouse mtDNA were sequenced 25 years ago, for a long time the knowledge of the replication and repair machinery was lacking. This made it very difficult to test predictions of replication models for example by reconstituting the various components needed for replication or by manipulating the individual enzymes.
Using inducible overexpression of wild type or mutant variants of Twinkle and POLG1 in cell culture, we show here that replication stalling results in changed patterns of RIs that can best be understood by considering the proposed mechanisms of lagging-strand synthesis. Most notably our results show that stalling induced by deficient Twinkle results in RIs that mimic conventional strand-coupled RIs and suggest that initiation of lagging-strand DNA synthesis or maturation occurs at multiple sites across the genome. We furthermore propose that this maturation involves POLG1.
Catalytically deficient POLG1 and Twinkle mutants
In this study we have used the HEK293 Flp-In™ T-Rex™ inducible expression system to study mutants of POLG1 and Twinkle expected to result in severe catalytic defects of these enzymes.
As the Flp-In™ T-Rex™ system is a transgenic system, endogenous copies of the genes under study are still being expressed. Thus, in order to discuss here the severity of the mutations studied and to better value the use of the inducible Flp-In™ T-Rex™ system we tried to estimate relative levels of endogenous and recombinant proteins. The results (presented in Supplementary Figures 7 and 8) showed that at 3 ng/ml DC following 2 days induction, POLG1 recombinant and endogenous protein levels were comparable, suggesting this regime recreates the effects of expression of a ‘heterozygous’ mutant. The situation was different for Twinkle. Both northern blot analysis and western blot analysis suggested that at 1 ng/ml DC for 2 days, the wt recombinant Twinkle protein level is 4–5-fold the level of endogenous Twinkle, whereas at 3 ng/ml for 2 days recombinant Twinkle is ∼8–10-fold the endogenous level.
We have previously shown that the polymerase mutants D890N and D1135A of POLG1 are deficient in a commonly used reverse transcriptase assay. Transient expression in cell culture showed modest mtDNA depletion (27). The corresponding aspartate 890 and 1135 residues in E. coli polymerase I (Asp705 and Asp882) have both been shown to be essential for catalysis (40,41). A model of POLG1 based on the family A polymerase structures including E. coli polymerase I places both these residues at critical sites for catalysis (23), which is corroborated by the fact that cell lines for stable constitutive expression of these variants could not be established. More recently, HEK293 T-Rex cells were established expressing the D1135A mutation (42). The authors showed, as we repeated here, that induced expression of this variant resulted in fast mtDNA copy-number depletion. Similarly we now show that the D890N mutation resulted in similar rapid mtDNA depletion upon induction. For both variants we observed a reduction in copy number that suggests that upon full induction mtDNA is diluted by half with each cell division and thus can no longer be replicated. The generally low expression levels of recombinant POLG1 and the 1:1 ratio to endogenous PolG1 at 3 ng/ml DC imply that both the D890N and D1135A mutation behave as dominant and furthermore suggest that the results are unlikely due to the capture of the entire pool of the POLG1 accessory subunit POLG2 by excessive expression of catalytically inactive mutants.
Like the POLG1 polymerase domain mutations, the Twinkle K421A mutation is predicted to abolish catalytic activity. This mutation is an invariable residue of the Walker A motif that based on the T7 helicase crystal structures is essential for nucleotide binding and hydrolysis (43,44). Indeed, using partially purified Twinkle K421A, we showed that this mutation abolished helicase activity of Twinkle whereas wild-type purified protein showed robust helicase activity and similar specificity (not shown) as published (15). The splice variant Twinky showed no evidence of being able to unwind the helicase substrate, even though most of the core helicase motifs except for the last few amino acids of motif H4 are left intact. The absence of helicase activity is nevertheless not unexpected as we showed previously that this variant is mostly monomeric (14), whereas the helicase activity is expected to require hexamer formation. In contrast to full-length Twinkle, overexpressed Twinky does not co-localize with mtDNA in nucleoids within the mitochondrial network. Likewise, the 31 amino acid deletion of the region, that by analogy with the region in the T7 primase/helicase we tentatively call the linker region, did not show nucleoid localization and was deficient in the unwinding assay. However, using glutaraldehyde cross-linking (Supplementary Figure 4) this mutant was still able to form multimers, unlike Twinky. The analogous mutation to G575D has previously been studied in the T7 primase/helicase (G488D) and was shown to be severely defective in its unwinding as well as its primase activity (37), although the remaining primase activity was still sufficient to allow for phage growth. It was furthermore shown to have weak DNA-binding capacity, in part explaining its weak enzymatic activities. In our hands, the Twinkle G575D mutant did not show helicase activity and its nucleoid localization was partially compromised consistent with weak DNA binding. Of the variants tested here, G575D and K421A behaved as dominant mutations since they reduced mtDNA copy number even at very low levels of expression, which occurred without induction by DC caused by leakiness of protein expression. Based on the comparison above and the difficulties to detect the protein without induction we are confident that under these conditions all recombinant Twinkle mutants are present at levels well below that of the endogenous protein. Twinky and ▵346–376 did not reduce copy number and are thus suggested not to compromise the activity of the endogenous Twinkle still expressed in these cell lines. Based on recent mechanistic insight in the T7 primase/helicase (45) and the fact that the purified ▵346–376 enzymes is inactive, this mutant is unlikely to form hetero-multimers with the endogenous enzyme. Finally, the ▵70–343 mutant reduced mtDNA copy number and helicase activity. Although this showed the importance of this region for Twinkle function, these results and the analysis by 2DNAGE have not yet established a specific function for this part of the protein such as a primase activity.
Replication stalling phenotypes are compatible with delayed lagging-strand synthesis
One of the powers of 2DNAGE is that it allows accurate predictions of results on the basis of a given replication model. The method is therefore very useful to generate and test hypotheses as demonstrated by the adaptation of the method in a computer model (46). One of the unusual features of the strand-displacement model (4) is the prediction of extensive single-stranded RIs. Although these RIs have been observed by electron microscopy and more recently by AFM after mtDNA coating by SSB (47), the RITOLS replication model considers them artefacts of degradation of the RNA patches of the initial lagging-strand. Indeed, single-stranded RIs predicted depending on the origin of replication and the restriction enzymes used (7,48) are not readily observed by 2DNAGE. It has been suggested however that the failure to observe these RIs is due to extensive branch-migration (47), thus arguing that the 2DNAGE technique is flawed and that the majority of RIs predicted by the strand-displacement model are not detected as such by this technique. This however does not provide an explanation for the observation of abundant RITOLS RIs.
Our observations of mtDNA copy-number depletion and a concomitant overall increase in RIs observed by 2DNAGE are evidence of extensive replication stalling or slowing down of replication fork movement in a non-site-specific manner. Replication stalling induced by the Twinkle K421A and G575D mutations has direct bearing on the question of delayed lagging-strand DNA synthesis as proposed by both the strand-displacement and RITOLS models. Using 2DNAGE we have shown here that accumulated RIs were visible using 3 different restriction enzymes (HincII in Figures 6–8, BclI in Supplementary Figure 1 and DraI in Supplementary Figure 5A) and probes for fragments that cover half of the genome. The results show that reduced fork progression occurs throughout mtDNA and not at a few specific sites since we did not observe appearance of discrete spots on replication arcs but a general increase of the intensities of arcs of RIs. Probing for the 3.9 kb HincII fragment from mtDNA nt 13 636–1006, which includes OH as well as most of the initiation zone (OriZ) covering the cytochrome b, ND6, part of the ND5 gene region (8), showed both an intense bubble and descending y-arc typical of a fragment containing multiple initiation sites within that fragment. However, since the ascending y-arc signal in this fragment was generally weak even when the bubble arc signal was very strong, the results nevertheless suggest that most initiation occurred in the NCR region, possibly at or near OH and not in OriZ, which would result in a more uniform y-arc (8). 2DNAGE analysis of a DraI fragment spanning most or all of OriZ but not including OH further supports this hypothesis as it showed a strong y-arc signal in Twinkle stalling mutants but only a very weak bubble arc (Supplementary Figure 5A). These results thus suggest that despite the apparent lack of RITOLS on 2DNAGE gels in the Twinkle stalling mutants, the predominant initiation occurs at OH as was proposed for both the RITOLS (10) and the strand-displacement model (4). Since the RIs observed with the Twinkle stalling mutants were RNaseH and S1 nuclease insensitive they were most likely double-stranded DNA and thus resemble strand-coupled RIs. The double-stranded nature of the RIs in question was further confirmed by comparing RIs obtained by digestion with DraI and analysis of a fragment close to but not including the NCR origin (Supplementary Figure 5A and B). As explained in detail in Supplementary Figure 5C the prediction is that double-stranded RIs would yield a conventional Y-arc, whereas RITOLS and single-stranded intermediates being non-digestible on the nascent lagging-strand would yield a retarded arc resembling a Y-arc (SMY or slow-moving Y). Also by comparison with Supplementary Figure 5A, the results show that the vast majority of RIs resulting from Twinkle stalling mutants are on a conventional Y-arc whereas by comparison RIs from cells expressing Twinkle wt or expressing the POLG1 D1135A mutant are on the predicted SMY (7,10). As an alternative we purified replication intermediates from preparative Twinkle G575D 2DNAGE gels and analyzed the purified RIs by restriction digestion using two dsDNA-specific enzymes (XhoI and DraI) and AccI that is capable of digesting ds and ssDNA. As demonstrated and illustrated in Supplementary Figure 6, these digests also fully support the conclusion that RIs observed with the Twinkle stalling mutants are essentially double-stranded DNA. As it is highly unlikely that leading- and lagging- strand synthesis have become coupled due to fork stalling, the observed double-stranded RIs can only be reasonably explained by an increased rate of lagging-strand initiation and/or maturation relative to the rate of fork movement. The results furthermore imply this to occur at sites other than the proposed lagging-strand initiation site(s) because the 3.9 kb HincII is unlikely to contain any, based on the mapping of the mouse second preferred maturation start-site proposed to be between nt 12 966 and 12 671 (10). Although not excluded at the moment, in the strand displacement model this would require novel priming events by unknown proteins and mechanisms. In the case of RITOLS intermediates, processing of incorporated RNA can be more easily envisaged to yield fully dsDNA intermediates.
The conclusions above are further strengthened by the ddC experiments. In our study overexpression of POLG1 mutants induced replication stalling but maintained RITOLS RIs. In contrast, Twinkle stalling mutants invariably resulted in an extensive loss of RITOLS RIs on 2DNAGE and in an associated increase in dsDNA RIs. The suggested cause-effect that dsDNA RIs increase as RITOLS decrease is further reinforced by the demonstration that a short inhibition of POLG by ddC led to a re-emergence of the RITOLS RIs under mild stalling conditions induced by Twinkle mutants and demonstrates that RITOLS intermediates precede dsDNA replication intermediates. Importantly, higher expression levels of, e.g. G575D for two or three days followed by ddC treatment did not result in the reappearance of RITOLS. We interpret this as a sign of a complete halt in ongoing replication and initiation whilst visible stalled RIs have already been maturated. Because under these conditions no replication (re)initiation occurs, also no new RITOLS intermediates are observed upon inhibition of POLG1 by ddC. In contrast, low G575D expression levels allow for a continued fork progression albeit at a reduced rate. Accordingly, very low (leaky) G575D expression resulted in a reduced mtDNA copy number, but a steady-state level was maintained over many cell generations (not shown). As the only plausible target for ddC treatment is POLG, the results seem to imply that the lagging-strand POLG is more sensitive to ddC treatment than the leading-strand polymerase, suggesting a different composition of leading and lagging-strand polymerase holoenzymes. Although the proposal of RITOLS replication (10) leaves open many questions regarding the synthesis of the initial RNA-rich lagging-strand, our data suggest that POLG is involved in lagging-strand maturation. Furthermore, as proposed previously (7) our data confirm that this is one of the slower steps in mitochondrial replication. Although various RNA-associated activities of POLG1 have recently been characterized (49), to our knowledge a displacement synthesis where POLG1 would displace annealed RNA from a template strand has not yet been demonstrated. Alternatively, POLG1 exonuclease activity might be involved in removing nascent RNA before initiating DNA synthesis. Our data however seem to exclude this idea, as overexpression of the exonuclease deficient POLG1 D198A did not seriously affect mtDNA copy number or the abundance of RNA-rich versus RNA-poor RIs. Nevertheless, moderate changes in replication intermediates at high D198A expression levels were observed by 2DNAGE and warrant further investigation.
The system we have established here provides an excellent tool to study the function of other proteins in mtDNA maintenance, to establish their enzymatic or structural roles in, e.g. replication or repair and to address questions concerned with the mechanisms of DNA replication. It will also be invaluable to determine the in vivo effects of mutations in Twinkle and POLG1 associated with human disorders of mtDNA maintenance. Low-level expression of the severe Twinkle mutations K421A and G575D due to the leakiness of the expression system also in the absence of DC caused reduced mtDNA steady state levels of ∼60%. Medium to high induction of the same mutations ultimately lead to the loss of protein expression, probably due to growth arrest or death of cells that lost their mtDNA completely and outgrowth of a few remaining cells that either had lost protein expression or did not have expression to start with, very similar to our earlier observations with constitutive expression of the POLG1 D198A mutant in 293T cells (27). In contrast expression of the null mutations did not have any effect on mtDNA levels also after 7 days and no negative selection occurred. These considerations also illustrate that the system is best used for transient expression of mutant proteins, that care must be taken when long-term effects are studied and that long-term maintenance of master plates without DC is not without hazard.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We like to acknowledge Outi Kurronen and Merja Jokela for their skilful technical assistance, and Ian Holt and Howy Jacobs for many fruitful discussions. This research was financially supported by the Academy of Finland (Grants 110689, 103213 and CoE funding), the Juselius Foundation, Tampere University Hospital Medical Research Fund, the University of Tampere and the EU sixth Framework Programme for Research, Priority 1 ‘Life sciences, genomics and biotechnology for health, contract number LSHM-CT-2004-503116’ supporting EUMITOCOMBAT. T.Y. was supported by the UK Medical Research Council and was the recipient of a Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science (April 2003–April 2005) and is currently funded by the UK Muscular Dystrophy Campaign. We gratefully acknowledge Drs Massimo Zeviani and Valeria Tiranti for providing us with a polyclonal antibody against Twinkle. Funding to pay the Open Access publication charges for this article was provided by EUMITOCOMBAT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Anderson S, Bankier AT, De Bruijn M.HL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 3.Robberson DL, Kasamatsu H, Vinograd J. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc. Natl Acad. Sci. USA. 1972;69:737–741. doi: 10.1073/pnas.69.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 5.Spelbrink JN. Replication, repair and recombination of mitochondrial DNA. In: Holt IJ, editor. Genetics of Mitochondrial Diseases. Oxford: Oxford University Press; 2003. [Google Scholar]
- 6.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 7.Yang MY, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 8.Bowmaker M, Yang MY, Yasukawa T, Reyes A, Jacobs HT, Huberman JA, Holt IJ. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem. 2003;278:50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- 9.Yasukawa T, Yang MY, Jacobs HT, Holt IJ. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell. 2005;18:651–662. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;18:18. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu. Rev. Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 14.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 15.Korhonen JA, Gaspari M, Falkenberg M. Twinkle has 5′ to 3′ DNA helicase activity and is specifically stimulated by mtSSB. J. Biol. Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 16.Tiranti V, Rocchi M, DiDonato S, Zeviani M. Cloning of human and rat cDNAs encoding the mitochondrial single- stranded DNA-binding protein (SSB) Gene. 1993;126:219–225. doi: 10.1016/0378-1119(93)90370-i. [DOI] [PubMed] [Google Scholar]
- 17.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. Embo J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shutt TE, Gray MW. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006;22:90–95. doi: 10.1016/j.tig.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Shutt TE, Gray MW. Twinkle, the Mitochondrial Replicative DNA Helicase, Is Widespread in the Eukaryotic Radiation and May Also Be the Mitochondrial DNA Primase in Most Eukaryotes. J. Mol. Evol. 2006;62:588–599. doi: 10.1007/s00239-005-0162-8. [DOI] [PubMed] [Google Scholar]
- 20.Longley MJ, Clark S, Yu Wai Man C, Hudson G, Durham SE, Taylor RW, Nightingale S, Turnbull DM, Copeland WC, et al. Mutant POLG2 disrupts DNA polymerase gamma subunits and causes progressive external ophthalmoplegia. Am. J. Hum. Genet. 2006;78:1026–1034. doi: 10.1086/504303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikali K, Suomalainen A, Saharinen J, Kuokkanen M, Spelbrink JN, Lonnqvist T, Peltonen L. Infantile onset spinocerebellar ataxia is caused by recessive mutations in mitochondrial proteins Twinkle and Twinky. Hum. Mol. Genet. 2005;14:2981–2990. doi: 10.1093/hmg/ddi328. [DOI] [PubMed] [Google Scholar]
- 22.Longley MJ, Graziewicz MA, Bienstock RJ, Copeland WC. Consequences of mutations in human DNA polymerase gamma. Gene. 2005;354:125–131. doi: 10.1016/j.gene.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 23.Graziewicz MA, Longley MJ, Bienstock RJ, Zeviani M, Copeland WC. Structure-function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nat. Struct. Mol. Biol. 2004;11:770–776. doi: 10.1038/nsmb805. [DOI] [PubMed] [Google Scholar]
- 24.Foury F, Vanderstraeten S. Yeast mitochondrial DNA mutators with deficient proofreading exonucleolytic activity. EMBO J. 1992;11:2717–2726. doi: 10.1002/j.1460-2075.1992.tb05337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, Vanderstraeten S, Foury F. Isolation and characterization of ten mutator alleles of the mitochondrial DNA polymerase- encoding MIP1 gene from Saccharomyces cerevisiae. Gene. 1995;160:105–110. doi: 10.1016/0378-1119(95)00215-r. [DOI] [PubMed] [Google Scholar]
- 26.Vanderstraeten S, Van den Brule S, Hu J, Foury F. The role of 3′-5′ exonucleolytic proofreading and mismatch repair in yeast mitochondrial DNA error avoidance. J. Biol. Chem. 1998;273:23690–23697. doi: 10.1074/jbc.273.37.23690. [DOI] [PubMed] [Google Scholar]
- 27.Spelbrink JN, Toivonen JM, Hakkaart GA, Kurkela JM, Cooper HM, Lehtinen SK, Lecrenier N, Back JW, Speijer D, et al. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J. Biol. Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- 28.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlof S, Oldfors A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 29.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 30.Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, Jalanko A, Spelbrink JN, Paetau A, Suomalainen A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl Acad. Sci. USA. 2005;102:17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, et al. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 32.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, Van Der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Legros F, Malka F, Frachon P, Lombes A, Rojo M. Organization and dynamics of human mitochondrial DNA. J. Cell Sci. 2004;117:2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- 34.Wanrooij S, Luoma P, van Goethem G, van Broeckhoven C, Suomalainen A, Spelbrink JN. Twinkle and POLG defects enhance age-dependent accumulation of mutations in the control region of mtDNA. Nucleic Acids Res. 2004;32:3053–3064. doi: 10.1093/nar/gkh634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 36.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 37.Washington MT, Rosenberg AH, Griffin K, Studier FW, Patel SS. Biochemical analysis of mutant T7 primase/helicase proteins defective in DNA binding, nucleotide hydrolysis, and the coupling of hydrolysis with DNA unwinding. J. Biol. Chem. 1996;271:26825–26834. doi: 10.1074/jbc.271.43.26825. [DOI] [PubMed] [Google Scholar]
- 38.Guo S, Tabor S, Richardson CC. The linker region between the helicase and primase domains of the bacteriophage T7 gene 4 protein is critical for hexamer formation. J. Biol. Chem. 1999;274:30303–30309. doi: 10.1074/jbc.274.42.30303. [DOI] [PubMed] [Google Scholar]
- 39.Brewer BJ, Fangman WL. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 40.Polesky AH, Steitz TA, Grindley N.DF, Joyce CM. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J. Biol. Chem. 1990;265:14579–14591. [PubMed] [Google Scholar]
- 41.Polesky AH, Dahlberg ME, Benkovic SJ, Grindley N.DF, Joyce CM. Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J. Biol. Chem. 1992;267:8417–8428. [PubMed] [Google Scholar]
- 42.Jazayeri M, Andreyev A, Will Y, Ward M, Anderson CM, Clevenger W. Inducible Expression of a Dominant Negative DNA Polymerase-gamma Depletes Mitochondrial DNA and Produces a rho 0 Phenotype. J. Biol. Chem. 2003;278:9823–9830. doi: 10.1074/jbc.m211730200. [DOI] [PubMed] [Google Scholar]
- 43.Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. Crystal structure of the helicase domain from the replicative helicase- primase of bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 44.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 45.Crampton DJ, Mukherjee S, Richardson CC. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol. Cell. 2006;21:165–174. doi: 10.1016/j.molcel.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 46.Viguera E, Rodriguez A, Hernandez P, Krimer DB, Trellez O, Schvartzman JB. A computer model for the analysis of DNA replication intermediates by two-dimensional agarose gel electrophoresis. Gene. 1998;217:41–49. doi: 10.1016/s0378-1119(98)00375-8. [DOI] [PubMed] [Google Scholar]
- 47.Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19:2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belanger KG, Mirzayan C, Kreuzer HE, Alberts BM, Kreuzer KN. Two-dimensional gel analysis of rolling circle replication in the presence and absence of bacteriophage T4 primase. Nucleic Acids Res. 1996;24:2166–2175. doi: 10.1093/nar/24.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murakami E, Feng JY, Lee H, Hanes J, Johnson KA, Anderson KS. Characterization of novel reverse transcriptase and other RNA-associated catalytic activities by human DNA polymerase gamma: importance in mitochondrial DNA replication. J. Biol. Chem. 2003;278:36403–36409. doi: 10.1074/jbc.M306236200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.