Abstract
Post-transcriptional modifications in ribosomal RNA are believed to fine-tune the RNA functions. The present study describes the characterization of the post-transcriptional modifications in Clostridium acetobutylicum 16S rRNA, using high-pressure liquid chromatography (HPLC) coupled to electrospray ionization mass spectrometry and reverse transcriptase assays. The combination of these techniques allowed the identification of eleven modified nucleosides, which were mapped onto the rRNA sequence. The C. acetobutylicum modification map is similar to that of Escherichia coli, with the majority of the modifications near functionally important sites in the rRNA. Although, in general, the number of modifications in rRNA is smaller than in tRNA, the conservation of the modification sites seems to indicate that the post-transcriptional modifications in 16S rRNA provide a necessary prerequisite for the ribosomal function.
INTRODUCTION
One of the most intriguing aspects of non-coding RNAs (ncRNAs) is their post-transcriptional modification. Post-transcriptional modifications in RNAs are of three main types (1): (i) conversion of uridine to pseudouridine (5-ribosyluracil, Ψ); (ii) methylation of 2′ hydroxyls (Nm) and (iii) alterations to bases, generally methylations of different positions (mN). High-pressure liquid chromatography (HPLC) coupled to mass spectrometry (MS) is the first method of choice for the analysis of post-transcriptional modifications because it is both very sensitive and specific. Furthermore, nearly all modifications cause a change in mass, making detection with LC/MS fairly straightforward.
Up till now, post-transcriptional modifications have been studied mostly in tRNA and oligonucleotide models (2–4). On the other hand, 16S rRNA is much less investigated and consequently, the number of organisms for which the post-transcriptional modifications in the 16S rRNA are characterized is much smaller (5). Escherichia coli is the only mesophilic bacterium for which the rRNA (16S and 23S) appears to be fully characterized (6) and recently, the 16S rRNA modification map of Thermus thermophilus was published (7).
The collective importance of post-transcriptional modifications for efficient protein synthesis has been demonstrated by the superior performance of authentic rRNAs compared to unmodified 16S rRNA (8) and 23S rRNA (9) counterparts. Further analysis of the function of post-transcriptional modifications in 16S rRNA would most definitely benefit from a larger number of fully characterized organisms to allow comparison of the data (5). As a first step in the production of these data, we analyzed the C. acetobutylicum 16S rRNA. Clostridium acetobutylicum was selected in order to add a Gram-positive, anaerobic and mesophilic bacterium to the short list of bacteria for which the 16S rRNA modification map has been completed (5). Furthermore, the rather low-G content of the C. acetobutylicum 16S rRNA (30%) leads to larger oligonucleotides when the rRNA is hydrolyzed with the G-specific endonuclease T1 and this facilitates the localization of modifications.
At first, modified nucleosides were identified solely based on their chromatographic mobility (e.g. 32P and/or 14C-labeling and 2D electrophoresis combined with thin layer chromatography (TLC), anion exchange chromatography and HPLC). However, these methods suffer from poor specificity and reproducibility and identification becomes problematic as the number of modifications or RNA chain length increases. In contrast, MS is a better technique for the analysis of post-transcriptional modifications, because nearly all modifications produce a change in mass of the canonical nucleosides (10). The application of MS to nucleic acids has long been restricted due to the experimental difficulties associated with the ionization of polar compounds such as nucleosides, nucleotides and oligonucleotides. Since the development of electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI), these molecules can be ionized and analyzed by MS. ESI currently holds the advantage because of its greater accuracy and ease with which it can be coupled to chromatographic separation systems. Combining the high-resolving power of the HPLC system and the high specificity of the mass measurement offers a method which is superior to either method alone (10). More than a hundred different modifications are known (11) and the majority have been characterized by some form of MS.
The general setup for identification and sequence placement of post-transcriptional modifications in large RNAs is based on the LC/ESI-MS analysis of two enzymatic digests. First the rRNA strand is digested to the nucleoside-level and analyzed by LC/ESI-MS. The combination of chromatographic retention times and mass measurements allows identification of nearly all the modifications that are present in the original intact RNA strand. Distinction between isomers (e.g. m5C and m4C) is made based on comparison to tandem MS (pseudo MS3, see the Methods section) analysis of reference samples. In a second step, sequence-specific endonucleases are used to cleave the intact RNA at specific positions (e.g. RNase T1 cleaves the 3′-side of all unmodified guanosines and RNase A cleaves at the 3′ end of all pyrimidine nucleotides). This way, oligonucleotides restricted to one or two 3′-nucleotide types are produced, which are subsequently analyzed by LC/ESI-MS. Comparison of the observed mass data with data predicted from the gene sequence identifies oligonucleotides that contain modifications. These anomalous oligonucleotides are analyzed by MS/MS for exact sequence placement of the modification in the oligonucleotide and thus in the RNA sequence. Two types of MS/MS experiments were used: In a first condition without precursor selection in the quadrupole, a higher collision energy is used to release monomer ions from the oligonucleotide. These monomers are often base ions, but also nucleoside phosphates and cyclic phosphates can be used. The Time-of-Flight (TOF) analyzer screens both the low-mass region for the modified monomer and the high-mass region for the intact oligonucleotide. Corresponding signals in the reconstructed ion chromatograms (RICs) identify the nature of the modification in the oligonucleotide. In a second condition, MS/MS analysis, with precursor selection, is used for sequencing of the oligonucleotide and exact sequence placement of the modification that was identified in the base-release experiments.
Using LC/ESI-MS nearly all the post-transcriptional modifications can be mapped in the RNA sequence. Pseudouridine, however, does not allow straightforward analysis by MS because it is isobaric to the omnipresent uridine. Patteson et al. (12) and Mengel-Jörgenson et al. (13) reported addition of a mass tag to all Ψ by specific derivatization. We developed a method to increase Ψ detection sensitivity by derivatization with methyl vinyl sulfone (14). Pomerantz and McCloskey identified Ψ solely based on identification of signature masses for Ψ (15). Unfortunately, the presence of Ψ in the sequence GΨG (as in the case of C. acetobutylicum 16S rRNA) makes these methods not useful because they are all based on preliminary RNase T1 digestion, which will produce numerous UG isomers. Pomerantz and McCloskey propose in this case to replace RNase T1 by nuclease U2, but this enzyme is no longer commercially available. Therefore, a reverse transcriptase assay was used for placement of pseudouridines in the 16S rRNA sequence. Pseudouridine bases were selectively derivatized with N-cyclohexyl-N′-β-(4-methylmorpholinium) ethylcarbodiimide p-tosylate (CMCT). Upon subsequent reverse transcription, CMC derivatized Ψ blocks elongation of radioactively labeled primers, highlighting the position of the Ψ on a PAGE gel (16–18).
Even without CMC derivatization, the reverse transcriptase approach can be used for sequence confirmation of modifications found by LC-MS, because most modifications block the reverse transcriptase elongation. Therefore, reverse transcriptase assays can be used to determine the sequence location of modifications when this is not possible by LC-MS analysis. Examples are the identification of a modified oligonucleotide in a group of isomers or when problems arise during MS/MS sequencing (for examples, see the result section below). In any case, the reverse transcriptase stops should be checked by simultaneous analysis of in vitro transcribed RNA, because stops can be caused by secondary structure of the RNA, even at elevated temperatures (18,19).
MATERIALS AND METHODS
Cells
Clostridium acetobutylicum (ATCC824) was grown in 2 × YT medium containing 16 g bacto-tryptone, 10 g yeast extract, 4 g NaCl and 10 g glucose per liter. Cultures were grown anaerobically in a modular atmosphere-controlled system (MACS, Don Whitley Scientific, Shipley, UK) at 37°C in 50 ml medium. Growth phase was monitored by turbidity measurement at 600 nm and cells were harvested at an OD of ±1.5 by centrifugation at 10 000 r.p.m. for 10 min. Multiple isolates of these 50 ml batches were combined and pellets were stored at −80°C.
Isolation of rRNA
16S rRNA was isolated in two steps. A first step consisted of the isolation of total RNA from the bacterial pellet using lysozyme digestion and phenol/chloroform extraction. In a second step, the 16S rRNA was purified from this total RNA mixture by rate zonal centrifugation through a 10–30% sucrose gradient. The samples were centrifuged at 94 000g on a Beckman L7-55 ultracentrifuge (SW 28 rotor) for 24 h, and 750 μl fractions were collected.
Purity of the isolated 16S rRNA was assessed by formaldehyde-agarose gel electrophoresis (1.85% formaldehyde, 1.3% agarose) using a Bio-Rad Mini Sub DNA Cell electrophoresis system.
Enzymatic digestion of rRNA
To produce the nucleoside mixture, 100 μg (3 μg/μl) 16S rRNA was hydrolyzed with 10 U nuclease P1 (Amersham, Chalfont, UK), 0.01 U snake venom phosphodiesterase I (E.C.3.1.15.1, Sigma, St. Louis, MO, USA), and 2.5 U shrimp alkaline phosphatase (E.C. 3.1.3.1., Amersham) as described by Crain (20). Digests were stored at −80°C until analysis by LC/ESI-MS.
To produce the oligonucleotide mixture, 100 μg 16S rRNA (10 μg/μl in 10 mM Tris–HCl, 1 mM EDTA acid, pH 7.4) was hydrolyzed with RNase T1 (E.C. 31.27.3, Amersham-GE Healthcare) at 37°C for 30 min. The amount of enzyme was 1 U RNase T1/3 μg rRNA and digests were also stored at –80°C until analysis by LC/ESI-MS. With these conditions, m22G and m7G modifications were resistant to RNase T1 hydrolysis.
LC/ESI-MS of the nucleosides
The instrument used for all studies was an orthogonal acceleration quadrupole time of flight, oa-qTOF (Q-Tof 2 Micromass, Manchester, UK), with an ESI source, interfaced to a capillary liquid chromatograph with a diode array detector (Waters, Milford, MA, USA). Both the mass spectrometer and the CapLC are controlled by the MassLynx 3.4 software (Micromass, Manchester, UK).
For the nucleosides mixture, 2–5 pmol of rRNA hydrolysate were injected directly onto an Atlantis dC18 column (0.32 × 150 mm, 3 μm diameter particles, Waters, Milford, USA). The column was eluted at a flow rate of 5 μl/min using an ammonium acetate (50 mM in Millipore Milli-Q water, pH 6.0)/acetonitrile gradient (21). Diode array UV absorbance data were acquired from 240–300 nm.
The chromatographic effluent was conducted without splitting into the mass spectrometer and two different scan functions were used. For the first scan function, the cone voltage and collision energy were set to 30 V and 10 eV, respectively, allowing mass measurement of the intact nucleoside. Qualitative interpretation of data was carried out as previously described (21). Data were acquired over a mass range of m/z 100–700. For the second scan function, pseudo MS3 was accomplished by fragmentation in the source, selection of the base anion in the quadrupole and fragmentation in the collision cell. The cone voltage used for MS3 was 30–40 V and the collision energy 30 eV. Data were acquired in positive mode over a mass range of m/z 50–250. With this second scan function, distinction between nucleoside isomers (e.g. m5C and m4C) is possible.
The capillary voltage for both scan functions was 3 kV, and data were acquired in continuum mode during 1.9 s (with 0.1 s interscan delay, giving a cycle time of 2 s).
LC/ESI-MS of an oligonucleotides mixture
The same instruments were used as for the analysis of the nucleoside mixture. 2–5 pmol of the oligonucleotide mixture were injected directly onto a PepMap C18 column (0.3 × 150 mm, 5 μm-diameter particles, LC-Packings, Amsterdam, The Netherlands). The column was eluted at a flow rate of 5 μl/min, using a 1,1,1,3,3,3-hexafluoro-2-propanol (HFiP, Acros, Geel, Belgium) based solvent system (60 mM HFiP in MQ water, adjusted to pH 7.5 with triethylamine) (22). The organic modifier was acetonitrile and a 2-step linear gradient (0.5%/min during 50 min and 5%/min during 10 min.) was used. Diode array UV absorbance data were acquired from 230 to 320 nm.
Different scan functions were used for data acquisition. During the first scan function, cone voltage and collision energy were kept at 30 and 10 eV, respectively. Thus, minimizing fragmentation, intact oligonucleotide molecular masses can be detected. Data were recorded in continuum mode over a m/z 500–1500 mass range. For the second scan function, the collision energy was increased to 40 eV to release monomers from oligonucleotides. Data were acquired over a m/z 100–1500 mass range and modified bases and intact precursor ions (the modification containing oligonucleotides) are recorded simultaneously. Oligonucleotide sequencing was performed in a third scan function at 30 V cone voltage and collision energies varying from 20 to 40 eV. During this scan function, precursor ions were selected in the quadrupole. Finally, to confirm the detection of sugar methylated nucleosides, a fourth scan function was used: The cone voltage was increased to 60 V in order to release modified nucleoside cyclic phosphate ions. After selection of these ions in the quadrupole, a collision energy of 20 eV was used to release the unmodified base and the methylated ribose ion (pseudo MS3).
During all scan functions, samples were recorded in negative mode and the capillary voltage was kept at −2850 V.
Software
Mongo Oligo Mass Calculator (http://medlib.med.utah.edu/massspec/mongo.htm) generated a mass-ordered list of all oligonucleotides produced by RNase T1 digestion of 16S rRNA of C. acetobutylicum. This list is calculated based on the gene sequence (GenBank accession number NC_003030) and the software allows calculation of the electrospray series and the selection of modified nucleosides.
In-house software was used to search the C. acetobutylicum 16S rRNA sequence for oligonucleotides corresponding to masses that were found in the mass data from the samples analyzed. It allows the user to restrict the search on the basis of the 5′ or 3′ end, number and residue masses of modifications, undercut residues and phosphorylation state.
Simple oligonucleotide sequencer (SOS) was used for the sequencing of modified oligonucleotides based on the fragmentation spectra produced by CID (23).
In vitro production of 16S rRNA
The gene sequence corresponding to C. acetobutylicum 16S rRNA was isolated from genomic DNA by a PCR with primer 27f: 5′-AGAGTTTGATCCTGGCTCAG-3′ and primer 1492r: 5′-GGCTACCTTGTTACGACT-3′ (Sigma Genosys). For this PCR, a mixture of 10:1 Taq: Pfu Turbo polymerases was used and primer concentrations were 0.5 μM. Buffer conditions were as provided by the TOPO cloning protocol. After 3 min at 94°C, 20 cycles of 1 min at 94°C, 1 min at 55°C and 3 min incubation at 72°C were used. Incubation of the samples for 7 min at 72°C guaranteed full-length PCR products with 3′-A overhangs, necessary for TOPO cloning. After purification with a PCR purification kit (Qiagen, Crawley, UK), the amplified 16S rRNA gene was cloned, downstream of the T7 promotor sequence, into a pCR4-TOPO vector (Invitrogen, Carlsbad, CA, USA) following the protocol provide by the supplier. After heat shock transformation of TOP10 cells, distinct colonies were incubated overnight in 0.01% ampicillin containing LB. For the selection of colonies with inserts in the correct direction, PCR was performed with two combinations of primers. For a first PCR (the positive control), these primers were 27f (sequence as above) and M13r: 5′-GGAAACAGCTATGACCATG-3′. If the insert is in the correct position, a band should be visible at 1.6 kB after agarose gel electrophoresis. A second PCR (negative control) with primers 27f and M13f: 5′-GTAAAACGACGGCCAGT-3′ was used for confirmation. If the insert is in the correct position, no band should be visible after agarose gel electrophoresis. Conditions were as above, except that primer concentrations were 0.25 μM and no final 7-min incubation step at 72°C was necessary. Correct clones were digested with PmeI, which produces blunt ends downstream of the rRNA sequence. After cleanup with a PCR purification kit (Qiagen), in vitro RNA was produced using the Ribomax T7 kit (Promega, Madison, WI, USA). After RNeasy Mini Kit (Qiagen) purification, 1–2 μg/μl 16S rRNA was obtained.
CMC derivatization and reverse transcription
Fifteen to twenty-five microgram of total RNA was derivatized as in ref. (17). To remove the CMC groups of U and G, the mixture was placed for 4.5 h at pH 10.4 on 37°C. After precipitation with sodium acetate and ethanol the RNA was reverse transcribed with Avian Myeloblastosis Virus (AMV) reverse transcriptase (GE Healthcare, Brick, NJ, USA) using 5′-33P-labeled primers. The primer complementary to residues 548–569 (E. coli numbering) was used for identification of Ψ-516. Other primers used for Ψ-screening were complementary to residues 1098–1219, 1221–1243, 1435–1475 and 1492–1510. For hybridization, the mixture containing 6 pmol RNA, 0.3 pmol primer and hybridization buffer (55.5 mM HEPES Potassium salt pH 7.0 and KCl 111 mM) in a total volume of 13.5 μl was heated at 80°C for 2 min and cooled gradually to 40°C in 30 min. For the extension reaction, 1 μl hybridization mixture and 1 μl 100 μM ddNTP were added to 3 μl extension mix containing: 0.166 mM Tris–HCl pH 8.4, 13.3 mM MgCl2, 13.3 mM DTT, 0.066 mM each dGTP, dATP, dCTP and dTTP (GE Healthcare) and 0.33 U AMV reverse transcriptase. Samples were incubated 5 min at room temperature and 35 min at 42°C. After reaction and precipitation with sodium acetate and ethanol, the samples were dissolved in 6 μl urea loading buffer containing 8 M urea, 20 mM Tris–HCl pH 7.8, 1 mM EDTA and 0.02% xylene cyanol and bromophenol blue dyes. The samples were loaded on 7 M urea − 8% polyacrylamide gels and run in 1× TBE buffer at 45 W until the bromophenol blue dye reached the bottom of the gel. Radioactive products were visualized using a PhosphorImager.
Except for the CMC derivatization, sequence location of m5C at position 1409 (E. coli numbering) was performed by reverse transcription of radiolabeled primers as described above. The radiolabeled primer was: 5′-CCAAAAGGTTACCTCACGGG-3′ complementary to nucleotides 1435–1475 (E. coli numbering).
RESULTS
Mapping of modified nucleosides by LC/MS
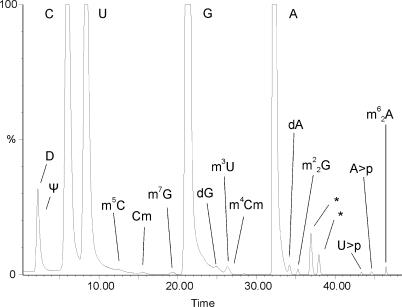
Modified nucleosides were identified by LC/ESI-MS analysis of the C. acetobutylicum 16S rRNA. The chromatographic separation of a total nucleoside digest is shown in Figure 1. The presence of nine different modified nucleosides in the 16S rRNA of C. acetobutylicum is indicated, Ψ, D, m5C, Cm, m7G, m3U, m4Cm, m22G and m62A. Most of the assignments could be derived from relative retention times and correspondence of RICs for the protonated base and nucleoside masses. For m5C, m4Cm and m3U, specific placement of the modifications on the base or sugar residue was done based on LC/ESI-pseudo MS3 comparison to standard samples (data not shown). The absence of specific tRNA nucleosides (in particular N6-threonylcarbamoyl-adenosine) indicates that no tRNA contamination is present (24).
Figure 1.
HPLC chromatogram after total nucleoside digestion of purified C. acetobutylicum 16S rRNA. UV trace at 260 nm with annotation of modified nucleosides and other components. (*) Are non-nucleoside artefacts.
The mass silent modification pseudouridine (Ψ) was not detected by LC/ESI-MS of the nucleosides digest and it was added in Figure 1 based on literature data (21). Even with the method described in (14), no reliable identification was possible, probably because of the very low concentration, short retention time on the reversed phase column and higher number of fragment ions (25). The structures of the modified nucleosides that were characterized in this study can be viewed at http://medlib.med.utah.edu/RNAmods (2).
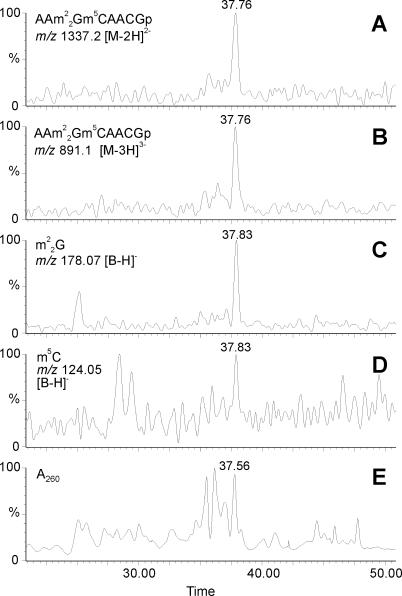
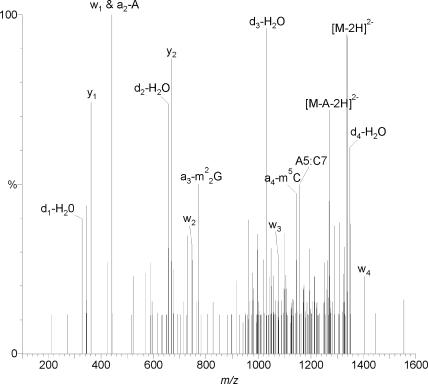
The modified nucleosides depicted in Figure 1 were mapped onto the 16S rRNA sequence by LC/ESI-MS analysis of an RNase T1 digest of the intact purified 16S rRNA. Comparison of the measured data to data predicted from the gene sequence identified eight modified oligonucleotides. These oligonucleotides and the m/z values for modified monomers inside these oligonucleotides are shown in Table 1. The modified monomers are released during LC/ESI-MS/MS analysis with fragmentation in the collision cell, without precursor selection in the quadrupole. This experimental setup allows detection of the released modified bases and nucleotide monomers at the same time as the molecular ion signals of the oligonucleotides from which they originate. Figure 2 illustrates these experiments for the oligonucleotide AAm22Gm5CAACGp. Exact sequence placement of these modifications and of the sugar modifications was done in a third step where fragmentation with precursor selection was used to sequence the modified oligonucleotides. Using sequencing software (23) unambiguous mapping of the modification in the modification containing oligonucleotides and thus in the rRNA could be established (Figure 3).
Table 1.
Assignments of oligonucleotides from RNase T1 digestion of C. acetobutylicum 16S rRNA
| Oligonucleotide | Sequence locationb | Mass | ||
|---|---|---|---|---|
| Calculatedc | Measured | Monomer ions (m/z)d | ||
| CCm7GCGa | 525–529 | 1623.2 | 1637.1 | 164 [B-H]− |
| AAm22Gm5CAACGa | 964–971 | 2634.4 | 2676.2 | 178, 124 [B-H]− |
| UCmAAAUCAUCAUGe | 1194–1206 | 4147.5 | 4161.1 | 336 [Np-H]− |
| CCCCUUAUGf | 1207–1215 | 2830.4 | 2832.4 | 307 [N > p-H]− |
| m4CmCCG | 1402–1405 | 1278.2 | 1306.1 | 124 [B-H]− |
| UCAm5CACCAUG | 1406–1415 | 3182.4 | 3196.1 | 318 [N > p-H]− |
| m5UAACAAG | 1498–1504 | 2290.3 | 2304.2 | 125 [B-H]− |
| m62Am62ACCUG | 1518–1523 | 1937.3 | 1993.2 | 164 [B-H]− |
aUnmodified oligonucleotides contain guanosines and should have been hydrolyzed by RNase T1.
bSequence location utilizes E. coli numbering.
cPredicted, monoisotopic masses, calculated from the 16S rRNA gene sequence.
dm/z Values for monomers released in the collision cell at higher collision energy, [B-H]−: base, [Np-H]−: nucleoside phosphate, [N > p-H]−: nucleoside cyclic phosphate.
eCm1195 is identified by Pseudo MS3 with fragmentation of N > p (m/z) 318.
fThe exact location of the dihydrouridine could not be determined, it is position 1211 or 1212.
Figure 2.
HPLC-MS analysis of an RNase T1 hydrolysis mixture of C. acetobutylicum 16S rRNA. Analysis was performed at 40 eV collision energy without precursor selection. (A, B, C and D) Reconstructed ion chromatograms for the oligonucleotide AAGCAACGp + 3 methyl groups, showing co-elution of signature ions for the intact oligonucleotide and base ions for m22G and m5C. (E) UV trace at 260 nm.
Figure 3.
Product ion mass spectrum from CID of AAGCAACGp + 3 methyl groups (m/z 1337.2) at 45 eV. Signature ion series are indicated and place the modifications at AAm22Gm5CAACGp (for ion series nomenclature see (26).
Sequencing analysis by MS/MS of the oligonucleotide UCACACCAUGp (mass: 3196.4), for sequence placement of the extra methyl group, was problematic. Both precursor ions with sufficient intensity for MS/MS fragmentation (m/z values 1064.5 and 798.1) co-eluted with other oligonucleotides with nearby m/z values (e.g. the 3′-terminal AUCACCUCCUUUCU, without 3′ phosphate, with mass 4262.5 and m/z 1064.6 and the oligonucleotide AUUAAUACCGp at pos. 155 with mass 3204.4 and m/z 800.9). The masses of the precursor ions could not be selected separately without sacrificing sensitivity. Fortunately, m5C blocks reverse transcription and sequence location of this modification could be confirmed by a reverse transcriptase assay (result not shown).
Mapping of pseudouridine in the 16S rRNA sequence by CMC derivatization and reverse transcription
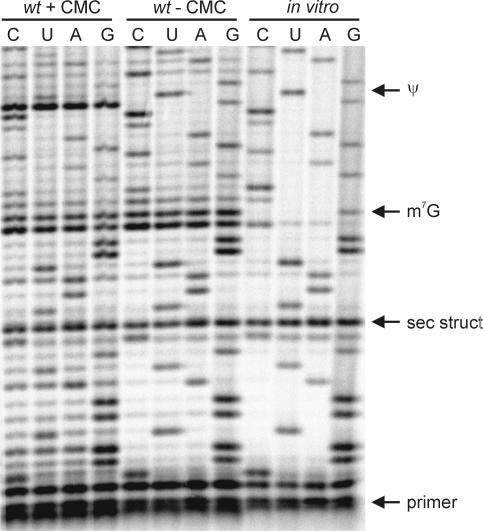
The phosphorimage of reversed transcriptase products identifying Ψ516 in C. acetobutylicum 16S rRNA is depicted in Figure 4. This figure also shows a band caused by m7G, which is also important because of the redundancy of the CCGCGp oligonucleotide produced by RNase T1 (three occurrences). With the reversed transcriptase technique, ∼30% of the 16S rRNA, containing the Ψ modification sites in other organisms (27), were scanned. No other Ψs were detected in C. acetobutylicum.
Figure 4.
Phosphorimage of reverse transcriptase products of the radiolabeled primer complementary to residues 548–569 (E. coli numbering). C, U, A and G indicate (dideoxy-)sequencing lanes, wt. + CMC: CMC derivatized and alkali treated 16S rRNA purified from C. acetobutylicum bacteria. wt. − CMC: alkali treated 16S rRNA purified from C. acetobutylicum bacteria. In vitro: T7 RNA polymerase transcribed 16S rRNA from the cloned 16S rRNA gene. Arrows indicate bands corresponding to modified nucleosides or secondary structure.
DISCUSSION
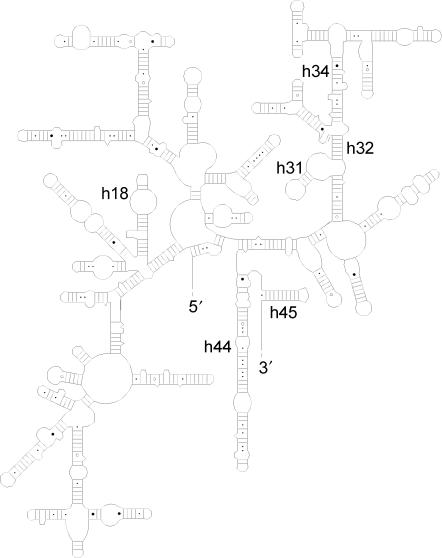
Of the 11 post-transcriptionally modified nucleosides that were reported in E. coli 16S rRNA, seven were also detected in C. acetobutylicum 16S rRNA, while position 966 contains a m2G in E. coli and a m22G in C. acetobutylicum. Interestingly, the same seven modifications, and the m22G at position 966, were recently reported in the 16S rRNA of T. thermophilus (7), indicating that these modifications are perhaps indispensable for the ribosomal biosynthesis or maturation. Indeed, they are situated in or near helices 18, 31, 34, 44, 45 (Figure 5), which are functionally important sites in the small subunit (SSU), where contacts to tRNA and the large subunit (LSU) are made (1,29). This location would allow them to directly influence these interactions or play a role in the 3D folding of these important domains. As shown in Table 2, three modifications in E. coli are not present in C. acetobutylicum, while three modifications in C. acetobutylicum are absent in E. coli. Some of these differences are situated in helix 34 in the head domain of the SSU. The Cm at position 1195 (E. coli numbering) in C. acetobutylicum is not present in E. coli, although the base sequence is nearly the same: … AGACGUCAAGUCA … in E. coli and … UGACGUCmAAAUCA … in C. acetobutylicum. The m2G modification at position 1207 in E. coli is not present in C. acetobutylicum, but this is not a surprise as the primary sequence of the 16S rRNA is different at this position (C in C. acetobutylicum, G in E. coli). Helix 34 interacts with A site tRNA, but the influence of these differences in modification maps on ribosomal function is unclear.
Figure 5.
Secondary structure from bacterial 16S rRNA, important helices are numbered (28).
Table 2.
Comparison of modifications in 16S rRNA of C. acetobutylicum with E. coli and T. thermophilus
| Pos.a | E. coli | C. acetobutylicum | T. Thermophilus |
|---|---|---|---|
| 516 | Pseudouridine (ψ) | Pseudouridine (ψ) | Pseudouridine (ψ) |
| 527 | 7-methylguanosine (m7G) | 7-methylguanosine (m7G) | 7-methylguanosine (m7G) |
| 966 | N2-methylguanosine (m2G) | N2,N2-dimethylguanosine (m22G) | N2,N2-dimethylguanosine (m22G) |
| 967 | 5-methylcytidine (m5C) | 5-methylcytidine (m5C) | 5-methylcytidine (m5C) |
| 1195 | no modification present | 2′-O-dimethylcytidine (Cm) | No modification present |
| 1207 | N2-methylguanosine (m2G) | No modification present | N2-methylguanosine (m2G) |
| Xb | no modification present | Dihydrouridine | No modification present |
| 1400 | no modification present | No modification present | 5-methylcytidine (m5C) |
| 1402 | N4,2′-O-dimethylcytidine (m4Cm) | N4,2′-O-dimethylcytidine (m4Cm) | N4,2′-O-dimethylcytidine (m4Cm) |
| 1404 | no modification present | No modification present | 5-methylcytidine (m5C) |
| 1407 | 5-methylcytidine (m5C) | No modification present | 5-methylcytidine (m5C) |
| 1409 | no modification present | 5-methylcytidine (m5C) | No modification present |
| 1498 | 3-methyluridine (m3U) | 3-methyluridine (m3U) | 3-methyluridine (m3U) |
| 1516 | N2-methylguanosine (m2G) | No modification present | No modification present |
| 1518 | N6,N6-dimethyladenosine (m62A) | N6,N6-dimethyladenosine (m62A) | N6,N6-dimethyladenosine (m62A) |
| 1519 | N6,N6-dimethyladenosine (m62A) | N6,N6-dimethyladenosine (m62A) | N6,N6-dimethyladenosine (m62A) |
| 1540 | no modification present | No modification present | Pseudouridine |
| 1541 | no modification present | No modification present | Pseudouridine |
aSequence position utilizes E. coli numbering.
bX: 1211 or 1212.
Our data also indicate the presence of a dihydrouridine nucleoside at position 1211 or 1212, in between helices 32 and 34. However, both the LC/MS/MS data and RT assays do not allow identification of the exact location site. This is the first report on the presence of this modification in 16S rRNA, but further research is necessary to confirm the exact location of this modification and to determine its function.
Other differences are located in the 3′ minor domain. In helix 44 of E. coli 16S rRNA, a m5C is present at position 1407, while in C. acetobutylicum this modification is present at position 1409 (E. coli numbering). Alignment of incomplete modification maps of all organisms in the SSU database indicates that both are rather conserved modification sites (5). Why these two adjacent cytosines are so often modified is not known, both are situated in the functionally important helix 44, at either site of the bulged out A1406. G1516 in helix 45 of E. coli 16S rRNA, is methylated at position 2 of the base, while C. acetobutylicum contains an unmodified A at this position. Probably the purine at this position requires a rather hydrophobic site at position 2. Again, no further insight in the role of this modification is known.
In general, the level of modification appears to be equal in E. coli and C. acetobutylicum 16S rRNA (each 11 modification sites), while T. thermophilus 16S rRNA is more extensively modified (14 modification sites). However, it appears that E. coli can only represent mesophilic bacteria to a certain extent as some C. acetobutylicum modifications are absent from E. coli 16S rRNA, while some E. coli modifications are absent from C. acetobutylicum 16S rRNA.
The bacterial ribosomal SSU is a target for many antibiotics. Both aminoglycoside (30) and cyclic peptide antibiotics (31) target the top of helix 44 and tetracyclines bind multiple sites in the SSU, e.g. helix 34. It has been reported that some post-transcriptional modifications alter the resistance to these ribosome-targeting antibiotics. Therefore, knowledge of the natural modification maps might be interesting for the analysis of this type of resistance mechanism. As C. acetobutylicum is an important industrial bacterium, novel antibiotic resistance genes might be used for plasmid maintenance during genetic engineering. Furthermore, more information on this resistance mechanism is also interesting from a therapeutic point of view, because the Clostridium genus contains several pathogenic species. The influence of post-transcriptional modifications on the sensitivity to aminoglycosides has been the best studied, however, anaerobic Clostridium species are resistant to these antibiotics because they lack active membrane transport. Furthermore, resistance to the cyclic peptide antibiotics (capreomycin and viomycin) has been reported to be caused by the loss of a 2′-O-methyl group from C1409 (E. coli numbering) in Mycobacteria (31), other species without this modification (e.g. E. coli) are less sensitive. The influence of the 5-methylation of C1409 in C. acetobutylicum or the loss of this methylation on the sensitivity to capreomycin or viomycin remains to be studied. Clostridium acetobutylicum is sensitive to tetracyclines, but no links between resistance to these antibiotics and rRNA methylation has been reported yet (30).
The results for C. acetobutylicum were compared to the T1 catalog data, detected by Tanner et al. (32) and compiled in the SSU database (5) These short sequences were used in early studies to establish patterns of phylogenetic relatedness. Although C. acetobutylicum itself was not determined, good correspondence was found with the other gram-positive anaerobic species (Table 3). Not surprisingly, the best correspondence was found with C. butyricum, phylogenetically closest to C. acetobutylicum.
Table 3.
SSU database modifications list for the gram-positive anaerobes listed
| Gram-positive anaerobes | |
|---|---|
| 1: Clostridium barkeri, 2: Eubacterium limosum, 3: Acetobacterium woodii, 4: Clostridium lituseburense, 5: Eubacterium tenue, 6: Clostridium butyricum | |
| Oligonucleotide | Species |
| aaGa | 1–5 |
| AAgb | 1, 3–5 |
| gCCGa | 1–6 |
| CCgCGc | 1–6 |
| cAACGb | 1–6 |
| AuUAGa | 1–6 |
| aaCCUGc | 6 |
| uAACAAGc | 1–6 |
| UACACA(c,C)Ga | 1–3 |
| CCCC(u,U)AUGa | 6 |
| UCAcACCACGa | 1–4 |
| UCAcACCAUGc | 5, 6 |
| UcAAAUCAUCAUGc | 3–6 |
Predicted modified nucleosides are in bold and small caps.
aWere not detected in the C. acetobutylicum samples.
bWere detected as the oligonucleotide AAGCAACG (see Table 1).
cWere detected in the C. acetobutylicum samples.
During the last decade, several methods have been published that use MS for the analysis of post-transcriptional modifications (33–36). Quadrupole instruments are very popular because of the flexibility of their scan functions. Unfortunately, some of these scan functions are not available on a oa-qTof instrument. On the other hand, to determine the nature of modified bases in an oligonucleotide, the oa-qTof allows the use of a different scan function from the quadrupole scan functions. In quadrupole instruments, an increased cone voltage is applied to release modified bases or nucleotides from the oligonucleotides. Careful time alignment of chromatic profiles of runs with and without increased cone voltages is needed to identify modified oligonucleotides from which the modified bases or fragments originated. In contrast, in oa-qTof instruments, the release of these fragments can be initiated by increasing the collision voltage in the collision cell, thus producing monomer ions, which are measured in the TOF analyzer. The broad mass range of the TOF analyzer allows the simultaneous detection of the small base ions (m/z 100–150) and the large oligonucleotide molecular ions (m/z 500–1500) from which the base ions originate. With this different method of collision cell fragmentation without precursor selection, time alignment of chromatographic profiles is no longer necessary and nearly unambiguous connection of modified base ions and modification containing oligonucleotides is possible. It is another extension of the already wide variety of MS based methods for the analysis of post-transcriptional modifications in large RNAs. In source fragmentation instead of collision cell fragmentation is not an option on the oa-qTof, because the quadrupole filter, even when used in rf-only mode, restricts the transmission of a wide m/z range. This makes it impossible to simultaneously detect base and oligonucleotide anions.
Although LC/ESI-MS is extremely powerful in the analysis of post-transcriptional modifications, it seems that in some cases the combination with reverse transcriptase assays is necessary. For example, if both RNase T1 and RNase A digests produce redundant oligonucleotides (e.g. CCGCGp) or when MS/MS sequencing is problematic because of co-eluting oligonucleotides with the same m/z values (e.g. m/z 1064.5 for UCACACCAUGp, z = −3, and the 3′ terminal oligonucleotide AUCACCUCCUUUCU, without 3′ phosphate with m/z 1064.6 and z = −4). As was illustrated in this study for the oligonucleotide CCm7GCGp, reverse transcriptase is stopped at base modified nucleosides and, although exact identification of the modification is not possible, it can be used to discriminate redundant modified oligonucleotides, when comparison to data predicted from the gene sequence can not reveal the exact location of the modification.
Furthermore, analysis of pseudouridine by LC/MS is not straightforward because it is isobaric to uridine. Given the fact that the only known pseudouridylation site in Bacillus subtilis and E. coli 16S rRNA is present at position 516 (E. coli numbering) (37) and given the great correspondence between modifications in E. coli and modifications found in C. acetobutylicum, Ψ was expected to be present at position 516 in C. acetobutylicum 16S rRNA as well. Therefore, all methods based on preliminary RNase T1 digestion can not be used as they will produce 35 UG isomers from the C. acetobutylicum 16S rRNA. Pomerantz and McCloskey propose in this case to replace RNase T1 by nuclease U2, but this enzyme is no longer commercially available. We have chosen the approach described by Bakin and Ofengand (18): All U-like and G-like residues were derivatized with CMC followed by an alkaline removal of all CMC groups except those linked to the N3 of Ψ. CMC derivatized Ψ blocks the reverse transcription reaction of radiolabeled primers, highlighting the position of the Ψ on a PAGE gel.
For both the analysis of pseudouridine and other modifications by reverse transcription assays, comparison of wild-type 16S rRNA to in vitro produced 16S rRNA is necessary to differentiate between bands on the gel that are caused by secondary structure of the 16S rRNA and bands that are caused by post-transcriptional modifications. Thus unambiguous identification of modification sites is possible.
ACKNOWLEDGEMENT
The authors thank the Fonds voor Wetenschappelijk Onderzoek—Vlaanderen for financial support through grant 1.5.014.02 and Prof. B. Vester for advice with the reverse transcription reaction. Funding to pay the Open Access publication charges for this article was provided by FWO-Vlaanderen.
Conflict of interest statement. None declared.
REFERENCES
- 1.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 2.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis DR. Biophysical and conformational properties of modified nucleosides in RNA. In: Nuclear Magnetic Resonance Studies). (Grosjean H, Benne R, editors. Modification and Editing of RNA. Washington: ASM Press; 1998. pp. 85–102. [Google Scholar]
- 4.Björk GR, Ericson JU, Gustafsson CED, Hagervall TG, Jönsson YH, Wikström PM. Transfer RNA modification. Annu. Rev. Biochem. 1987;56:263–287. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- 5.McCloskey JA, Rozenski J. The small subunit rRNA modification database. Nucleic Acids Res. 2005;33:D135–D138. doi: 10.1093/nar/gki015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakin A, Kowalak JA, Ofengand J. The single pseudouridine residue in Escherichia coli 16S RNA is located at position 516. Nucleic Acids Res. 1994;22:3681–3684. doi: 10.1093/nar/22.18.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guymon R, Pomerantz SC, Crain PF, McCloskey JA. Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: the complete modification map of 16S rRNA of Thermus thermophilus. Biochemistry. 2006;45:4888–4899. doi: 10.1021/bi052579p. [DOI] [PubMed] [Google Scholar]
- 8.Krzyzosiak W, Denman R, Nurse K, Hellmann W, Boublik M, Gehrke CW, Agris PF, Ofengand J. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into a functional 30S ribosome. Biochemistry. 1987;26:2353–2364. doi: 10.1021/bi00382a042. [DOI] [PubMed] [Google Scholar]
- 9.Green R, Noller HF. Reconstitution of functional 50S ribosomes from in vitro transcripts of Bacillus stearothermophilus 23S rRNA. Biochemistry. 1999;38:1772–1779. doi: 10.1021/bi982246a. [DOI] [PubMed] [Google Scholar]
- 10.Crain PF. Detection and structure analysis of modified nucleosides in RNA by mass spectrometry. In: Grosjean H, Benne R, editors. Modification and Editing of RNA. Washington, DC: ASM Press; 1998. pp. 47–57. [Google Scholar]
- 11.Rozenski J, Crain PF, McCloskey JA. The RNA modification database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patteson KG, Rodicio LP, Limbach PA. Identification of the mass-silent post-transcriptionally modified nucleoside pseudouridine in RNA by matrix-assisted laser desorption/ionization mass spectrometry. Nucleic Acids Res. 2001;29:E49–E49. doi: 10.1093/nar/29.10.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mengel-Jorgensen J, Kirpekar F. Detection of pseudouridine and other modifications in tRNA by cyanoethylation and MALDI mass spectrometry. Nucleic Acids Res. 200;30:e135. doi: 10.1093/nar/gnf135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emmerechts G, Herdewijn P, Rozenski J. Pseudouridine detection improvement by derivatization with methyl vinyl sulfone and capillary HPLC-mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;825:233–238. doi: 10.1016/j.jchromb.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 15.Pomerantz SC, McCloskey JA. Detection of the common RNA nucleoside pseudouridine in mixtures of oligonucleotides by mass spectrometry. Anal. Chem. 2005;77:4687–4697. doi: 10.1021/ac058023p. [DOI] [PubMed] [Google Scholar]
- 16.Ho NWY, Gilham PT. Reaction of pseudouridine and inosine with N-Cyclohexyl-N’-b-(4-methylmorpholinium)ethylcarbodiimide. Biochemistry. 1971;10:3651–3657. [PubMed] [Google Scholar]
- 17.Ofengand J, Del Campo M, Kaya Y. Mapping pseudouridines in RNA molecules. Methods. 2001;25:373. doi: 10.1006/meth.2001.1249. [DOI] [PubMed] [Google Scholar]
- 18.Bakin A, Ofengand J. Four newly located pseudouridylate residues in E. coli 23S Ribosomal RNA are alle at the peptidyltransferase center: analysis by application of new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 19.Hansen MA, Kirpekar F, Ritterbusch W, Vester B. Posttranscriptional modifications in the A-loop of 23S rRNAs from selected archaea and eubacteria. RNA. 2002;8:202–213. doi: 10.1017/s1355838202013365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crain PF. Preparation and enzymatic hydrolisis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 21.Pomerantz SC, McCloskey JA. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol. 1990;193:796–824. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- 22.Apffel A, Chakel JA, Fischer S, Lichtenwalter K, Hancock WS. Analysis of Oligonucleotides by HPLC-electrospray ionization mass spectrometry. Anal. Chem. 1997;69:1320–1325. doi: 10.1021/ac960916h. [DOI] [PubMed] [Google Scholar]
- 23.Rozenski J, McCloskey JA. SOS: a simple interactive program for ab initio oligonucleotide sequencing by mass spectrometry. J. Am. Soc. Mass Spectrom. 2002;13:200–203. doi: 10.1016/S1044-0305(01)00354-3. [DOI] [PubMed] [Google Scholar]
- 24.Noon KR, Bruenger E, Mccloskey Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J. Bacteriol. 1998;180:2883–2888. doi: 10.1128/jb.180.11.2883-2888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudley E, Tuytten R, Bond A, Lemière F, Gareth Brenton A, Esmans E, Newton RP. Study of the mass spectrometric fragmentation of pseudouridine: comparison of fragmentation data obtained by matrix-assisted laser desorption/ionisation post-source decay, electrospray ion trap multistage mass spectrometry, and by a method utilising electrospray quadrupole time-of-flight tandem mass spectrometry and in-source fragmentation. Rapid Commun. Mass Spectrom. 2005;19:3075–3085. doi: 10.1002/rcm.2151. [DOI] [PubMed] [Google Scholar]
- 26.McLuckey SA, Habibi-Goudarzi S. Decompositions of multiple charged oligonucleotide anions. J. Am. Chem. Soc. 1993;115:12085–12095. [Google Scholar]
- 27.Ofengand J, Fournier MJ. The pseudouridine residues of rRNA: number, location, biosynthesis, and function. In: Grosjean H, Benne R, editors. Modification and Editing of RNA. Washington, DC: ASM Press; 1998. pp. 229–253. [Google Scholar]
- 28.Gutell RR, Lee JC, Cannone JJ. The accuracy of ribosomal RNA comparative structure models. Curr. Opin. Struct. Biol. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 29.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal structure of the Ribosome at 5.5 Å Resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 30.Douthwaite S, Fourmy D, Yoshizawa S. Nucleotide methylations in rRNA that confer resistance to ribosome-targeting antibiotics. In: Grosjean H, editor. Fine-Tuning of RNA Functions by Modification and Editing. Berlin, Heidelberg: Springer; 2005. pp. 285–307. [Google Scholar]
- 31.Johansen SK, Maus CE, Plikaytis BB, Douthwaite S. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNA. Mol. Cell. 2006;23:173–182. doi: 10.1016/j.molcel.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 32.Tanner RS, Stackebrandt E, Fox GE, Woese CR. A phylogenetic analysis of acetobacterium woodii, Clostridium barkeri, Clostridium butyricum, Clostridium lituseburense, Eubacterium limosum, and Eubacterium tenue. Curr. Microbiol. 1981;5:35–38. [Google Scholar]
- 33.McCloskey JA, Whitehill AB, Rozenski J, Crain PF. New techniques for the rapid characterization of oligonucleotides by mass spectrometry. Nucleosides Nucleotides. 1999;18:1549–1553. doi: 10.1080/07328319908044782. [DOI] [PubMed] [Google Scholar]
- 34.Crain PF, Gregson JM, McCloskey JA, Nelson CC, Peltier JMPDR, Pomerantz SC, Reddy DM. Characterization of posttranscriptional modification in nucleic acid by tandem mass spectrometry. In: Burlingame AL, Carr SA, editors. Mass Spectrometry in the Biological Sciences. Clifton, UK: Humana Press; 1996. pp. 497–517. [Google Scholar]
- 35.Crain PF, McCloskey JA. Applications of mass spectrometry to the characterization of oligonucleotides and nucleic acids. Curr. Opin. Biotechnol. 1998;9:25–34. doi: 10.1016/s0958-1669(98)80080-3. [DOI] [PubMed] [Google Scholar]
- 36.Crain PF, Ruffner DE, Ho Y, Qiu F, Rozenski J, McCloskey JA. Problems and prospects in the characterization of posttranscriptional modifications in large RNAs. In: Burlingame AL, Carr SA, Michael A, editors. Mass Spectrometry in Biology and Medicine. Clifton, UK: Humana Press; 2000. pp. 531–551. [Google Scholar]
- 37.Wrzesinski J, Bakin A, Nurse K, Lane BG, Ofengand J. Purification, cloning, and properties of the 16S RNA Pseudouridine 516 Synthase from Escherichia coli. Biochemistry. 1995;34:8904–8913. doi: 10.1021/bi00027a043. [DOI] [PubMed] [Google Scholar]