Abstract
We have developed a novel DNA microarray-based approach for identification of the sequence-specificity of single-stranded nucleic-acid-binding proteins (SNABPs). For verification, we have shown that the major cold shock protein (CspB) from Bacillus subtilis binds with high affinity to pyrimidine-rich sequences, with a binding preference for the consensus sequence, 5′-GTCTTTG/T-3′. The sequence was modelled onto the known structure of CspB and a cytosine-binding pocket was identified, which explains the strong preference for a cytosine base at position 3. This microarray method offers a rapid high-throughput approach for determining the specificity and strength of ss DNA–protein interactions. Further screening of this newly emerging family of transcription factors will help provide an insight into their cellular function.
INTRODUCTION
We show that microarray technology can provide rapid high-throughput assays for the identification of sequence-specific ss DNA–protein interactions. The majority of transcription factors recognize target sequences in duplex form. However, single-stranded regions can be induced by torsional stress of double-stranded DNA, allowing single-stranded nucleic-acid-binding proteins (SNABPs) access to their binding sites (1). SNABPs have been shown to bind with high affinity, non-specifically (2) and specifically (3), to ss DNA, which has been shown to regulate gene expression both positively and negatively (1,4). Gene expression can also be regulated on a translational level, by SNABP binding to mRNA (5).
Genome sequencing has allowed SNABPs to be identified and characterized for a range of eukaryotic and prokaryotic organisms. To understand how binding of SNABPs to ss nucleic acids regulates transcription and translation, the regions of sequence specificity must be identified. Many techniques including electrophoretic mobility shift assay (EMSA) (6), nitrocellulose-binding assays (7), Southwestern blotting (8), phage display (9), UV cross-linking (10) and X-ray crystallography (11) were developed to study sequence-specific ss DNA–protein interactions. Available techniques including, fluorescence measurements (12), polymerase chain reaction (PCR), fluorescence resonance energy transfer (FRET) combined with a DNA foot-printing assay (13), surface plasmon resonance (SPR) and fluorescence polarization (14) have all been used to study effectively specific ss DNA–protein interactions. The most frequent approach used to study the sequence specificity of DNA-binding molecules is by systematic evolution of ligands by exponential enrichment (SELEX), this method allows for the identification of sequences which bind with high affinity to the molecule of interest (15). This method has been used mostly to select for double-stranded DNA molecules that bind to the target but it has been also used to screen ss DNA molecules (16,17). SELEX has advantages to the previous methods but still lacks in its ability for high-throughput analysis as numerous microarray experiments can be completed in a single day, thus providing a detailed analysis of binding-site recognition at an unparalleled rate.
These techniques made use of non-immobilized ss DNA in liquid phase to probe ss DNA interactions with other molecules such as proteins, drugs and ligands, all of which suffered from being time-consuming, laborious, expensive and incapable of high-throughput screening. Therefore, oligonucleotides immobilized to solid supports provide an important tool for the rapid high-throughput examination of sequence-specific DNA–protein interactions.
Two current studies (18,19) have used microarrays displaying all possible 8-mer and 10-mer DNA duplexes to study effectively the sequence-recognition of both transcription factors and small molecules. These methods illustrate the potential high-throughput use of k-mer arrays in examining the DNA–binding properties of duplex-binding molecules but leave the area of SNABP specificity unexplored.
The innovative high-throughput assay described here provides a parallel screening system for identifying the specificity of SNABP binding. The major cold shock protein from Bacillus subtilis (CspB) was used to develop this microarray-based assay. This protein influences transcription and translation in vitro (20) by binding to stretches of 6–7 nucleotides (21) of ss DNA with a high degree of specificity (21,22).
We have used an oligonucleotide chip, for the identification of high-affinity 6-mer binding motifs for CspB. The chip contains all possible 4096 ss hexadeoxynucleotides incorporated onto a standardized anchor. The oligonucleotides on the array were originally designed to hybridize to folded mRNA which requires a significant spacer between the array surface and the recognition hexamer (Figure D, supplementary).
The use of a competitor protein in this assay allowed the identification of high-affinity DNA-binding sites. The binding affinity of a competitor protein will limit the amount of ss DNA-binding sites available to the CspB. The competitor protein chosen was a single-stranded DNA-binding protein from the crenarchaeote Sulfolobus solfataricus (SsoSSB). SsoSSB has a molecular weight of 16 kDa and binds non-specifically to ss DNA with a binding density of 5 nt per monomer and an apparent dissociation constant (Kd) of ∼90 nM (2). Thus, the competitive binding of the SsoSSB protein provided a means of identifying high-affinity consensus binding motifs for CspB by reducing non-specific and weak CspB-ss DNA binding.
MATERIALS AND METHODS
Microchip manufacture
Oligonucleotide chips were supplied by Nyrion Ltd and contained all possible 4096 ss hexadeoxynucleotides incorporated into a general structure, 5′-NH2-C12-Spacer-AAAAAAAAAA-NNNNNNNNN-XXXXXX-3′, where N was one of four bases and X was a specific hexadeoxynucleotide. Each chip is made up of a 4 × 4 meta-grid and each of these sub grids contains 18 columns × 15 rows of spots, which are 135 μm ± 15% in diameter. Oligonucleotides were immobilized to the chip surface using standard Exiqon amino-link chemistry. All arrays were manufactured by pin spotting, according to complete standard commercial practices. This was all done under contract by MWG Biotech custom arrays. For control purposes, arrays are batch tested using a standard mRNA template and a standard QC procedure expected to give a standard signal. This standard signal serves as a positive and negative control for all arrays. MWG Biotech spot biotin on the surface of the array, which also serves as a negative standard control (generates zero signal).
Expression and purification of recombinant SsoSSB
A mutant version of the SsoSSB protein from the crenarchaeote Sulfolobus solfataricus was constructed by changing the C-terminal glutamate residue to a cysteine (E145C mutant), allowing for the incorporation of spin labels and fluorescent probes on the C-terminal tail. This mutation minimizes the affect of labelling on DNA-binding activity as the C-terminal glutamate is not involved in ss DNA binding. The E145C mutant was constructed only as a precaution if the amine-reactive labelling methods were unsuccessful. Protein expression was induced by addition of 0.2 mM IPTG at 37°C for 3 h, after which cells were pelleted and frozen until required. Cell lysis, centrifugation and chromatography steps were carried out at 4°C. Cells (20 g) were thawed in 50 ml lysis buffer (50 mM Tris–HCl pH 7.5, 500 mM NaCl, 1 mM EDTA, 1 mM DTT) and immediately sonicated for 5 × 1 min with cooling. The lysate was centrifuged at 40 000 g for 45 min. DNase I [40 µg/ml] and RNase A [10 µg/ml]) were then added to the cell lysate and incubated at room temperature for 30 min with gentle agitation. The supernatant was heated to 70°C for 30 min in a water bath, and denatured proteins were precipitated by centrifugation at 40 000 g at 4°C. The supernatant was analysed by SDS–PAGE, and shown to contain recombinant SSB, which migrated as a band of ∼16 kDa as expected. The supernatant was diluted 5-fold with buffer A (50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 mM DTT) and applied to a Heparin-Sepharose (Amersham) column equilibrated with buffer A. SsoSSB was eluted over a linear gradient comprising 0–1 M NaCl. Fractions corresponding to a distinct absorbance peak were analysed by SDS–PAGE, pooled and concentrated. A subsequent gel filtration step (HR 10/30 Superdex-200) in a buffer containing, 10 mM Tris/HCl pH 7.5, 150 mM NaCl, 1 mM EDTA and 1 mM DTT, resulted in essentially homogeneous SsoSB, as determined by SDS-PAGE analysis. This method is an adaptation of the previously published method (23). SsoSSB was concentrated using a Viva Spin column (MWCO = 5 kDa) and quantified using both the Bradford method and the theoretical extinction coefficient, ε280 nm = 12660 M−1. cm−1.
Cloning, expression and purification of recombinant His6-CspB
Primers B.S_CSP fwd (5′-dAGCCATATG TTA GAA GGT AAA GTA AAA TAA -3′) and B.S_CSP rev (5′-dCGGATCC TAA CGC TTC TTT AGT AAC GTT AGC-3′) were used in a PCR with plasmid DNA (pET11-CspB vector provided by Michael Wunderlich (University of Bayreuth)) containing the CspB gene. Bases were added to the primers to introduce the NdeI and BamHI restriction sites (underlined). These sites were used to clone the PCR product in NdeI-BamHI-digested pET28a vector, resulting in the plasmid pET28a_B.S_CspB.
BL21 (DE3)pLysS Eschericia coli (E. coli) was transformed with pET28_B.S._CspB and transformants were grown in Luria-Bertani medium, containing 50 μg/ml kanamycin at 37°C with agitation. One-litre cultures were grown to an OD600 of 0.5–0.7 and IPTG (isopropyl-β-D-thiogalactosidase) was then added to a final concentration of 1 mM. Incubation was then continued for an additional 5–6 h and the cells were harvested at 8000 g in a JLA-9.1000 rotor for 12 min at 10°C. Pellets were then frozen in liquid nitrogen and stored at −80°C. Cell pellets were resuspended in lysis buffer (20 mM Tris HCl pH 8.0, 500 mM NaCl, 0.1% Triton X-100, 0.1 mM phenylmethylsulphonyl fluoride (PMSF), 1 mM EDTA and protease inhibitors) to a final volume of 30 ml/5 g of cells and then lysed using a French-press. DNase I [40 µg/ml] and RNase A [10 µg/ml]) were then added to the cell lysate and incubated at room temperature for 30 min with gentle agitation. As an initial step, the fusion protein was purified using a Ni-NTA resin affinity column, as per manufacturer's instructions and then purified to homogeneity as described previously (24). Briefly, to remove minor contaminants the fractions containing His6-CspB were pooled and dialysed overnight into a buffer containing 20 mM Tris/HCl pH 6.8, 1 mM DTT. The solution was applied to an HR 5/5 Mono-Q (1 ml) anion exchange column. Bound protein was eluted with a NaCl-gradient ranging from 0–1 M. CspB eluted at a concentration of 250 mM NaCl. A subsequent gel filtration step (HR 10/30 Superdex-75) in a buffer containing 10 mM Tris/HCl pH 7.5 and 100 mM NaCl) resulted in visually pure His6-CspB, as determined by SDS-PAGE analysis. His6-CspB was concentrated using a Viva Spin column (MWCO = 3.5 kDa) and quantified using both the Bradford method and the theoretical extinction coefficient, ε280 nm = 5690 M−1. cm−1.
Electrophoretic mobility shift assay (EMSA)
Three hundred picomoles of each ss DNA templates were 5′-end labelled by incubating templates with 0.03 mCi of [γ–32P]ATP, T4 polynucleotide kinase and T4 polynucleotide kinase buffer in a total volume of 60 μl at 37°C for 2.5 h. The reaction was stopped by heat inactivation (30 min at 65°C). Unincorporated [γ–32P]ATP was removed with QIAquick Nucleotide Removal Kit (Qiagen).
For a standard EMSA, 20 pmol of labelled ss DNA was mixed with increasing amounts of protein (total volume, 18 μl) at 4°C for 20 min in binding buffer (50 mM Tris, pH 8.0, 100 mM NaCl) unless stated otherwise. Two microlitres of dye solution (20% glycerol, 0.034% bromophenol blue) was added to the samples prior to gel electrophoresis.
Electrophoresis preformed in TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.0) buffer through a non-denaturing acrylamide gel (a 10% or 20% gel was used depending on the protein; e.g. for 50 ml of 20% gel; 25 ml of 40% acrylamide, 2.5 ml of 10 × TBE, 0.5 ml APS and 50 μl Temed) at 75 V until the samples had entered the gel and then at 100 V at 4°C (overnight for a 20% gel and 12 h for a 10% gel). Autoradiographs were obtained by exposing gels to Kodak BioMax MS film for 1–3 h at room temp.
Labelling of ss DNA-binding proteins with Cy5 dye
In the standard procedure, the contents of 1 vial (‘to label 1 mg of protein’) of cyanine 5 (Cy5) mono-functional dye the contents (Amersham) were dissolved in 50 μl of anhydrous DMSO. Proteins were dialysed into buffer B (150 mM, Na2CO3 (pH 9.3, pH was adjusted with H3PO4)) and concentrated using a Viva Spin column (MWCO = 5 kDa). Typical working concentrations of proteins were 1 mg/ml, unless stated otherwise. Ten microlitres of dye/DMSO was pipetted into 200 μl protein solution under slow vortexing. After a 30-min incubation at 25°C in the dark, the reaction was terminated by the addition of 300 μl of 100 mM NaH2PO4 (to suppress further labelling) to the sample. To separate the unbound dye, the sample was loaded onto a PD-10 column (10 ml bed of Sephadex G-25M), which had been pre-equilibrated in a buffer A (100 mM NaCl, 50 mM NaH2PO4 and 1 mM EDTA (pH 7.5, pH adjusted with NaOH). The column was then washed with buffer A (2 × 1 ml) and the labelled protein was then eluted by adding 2 ml of water to the column. The extent of the modification was assessed using MALDI-TOF mass-spectrometry. Protein concentration was determined before and after labelling, Cy5-protein concentration was calculated as per manufacturer's instructions.
Protein hybridization
Microarrays were pre-wet with phosphate-buffered saline (PBS) and 0.01% Triton X-100 and then blocked in 2% non-fat dried milk for 1 h. Blocked microarray slides were washed once with PBS and 0.1% Tween 20, and once with PBS and 0.01% Triton X-100. Protein (Cy5 labelled and unlabelled) binding to ss 25-mers (containing all possible [4096] 6-mer sequences) on a generic microchip was carried out in a hybridization chamber (Camlab, RTP/7870). Protein binding was performed in a humid chamber at 4°C with 80 μl of protein-binding reaction mix containing: 50 mM KCl, 20 mM Tris (pH 8.0), 2% (w/v) non-fat dried milk, 0.2 μg/μl bovine serum albumin (BSA) and 40 μM of test protein. Slides were covered with a siliconized cover-slip (BDH cover glass 22 × 50 mm, Cat. No. 406/0188/42, Borosilicate Glass) and incubated for 1 h at 4°C. The cover-slip was removed and the slide was washed (3x) in a slide chamber filled with PBS and 0.05% Tween-20, with PBS and 0.01% Triton X-100 (3×) and once with PBS for 3 min each. Excess water was removed from the slide surface (by flicking), which was allowed to dry before scanning. This method is an adaptation of the previously published method (25). Various methods (denaturing conditions; including high temperatures, various concentrations of detergents and pH range in combination with high NaCl concentration) were used to remove bound protein from the chip surface in order to reuse the array but were all unsuccessful as they either had detrimental affects on ONs bound to the surface of the array or were unable to remove bound protein.
Competitive assay
The method was essentially the same as above except that both His6-CspB and SsoSSB proteins were added to the binding reaction mix at the specified molar ratio. The binding reaction was carried out as before. The array was then incubated for 1 h in a humid chamber at 4°C with 100 μl of diluted (1:100 in blocking buffer) Alexa 532-conjugated polyclonal antibody to His5 (Molecular Probes). After incubation, the array was washed (3×) with PBS and 0.05% Tween-20 and once with PBS for 3 min each. Excess water was removed from the slide surface (by flicking), which was allowed to dry before scanning.
Microarray analysis: data collection
All microarray slides were scanned using an ArrayWorx microarray scanner at a range of laser settings, the highest of which produced a saturated signal for the majority of spots. The Alexa-532 (Cy3 equivalent) fluorophore was excited at 532 nm and the emission was recorded at 570 nm. The Cy5 fluorophore was excited at 633 nm and the emission was recorded at 675 nm. The data were filtered initially using a series of quality-control criteria so that only high-quality spots were used in our analysis. For each array we removed any flagged spots, these were spots that had dust flakes, scratches and irregular spots (spots that outmatched the average size). The average size of a spot is 135 μm ± 15% in size, any spot that did not correspond to this size constraint was excluded from the data. This size constraint also provided a crude method of approximating the DNA concentration of each spot, which allowed only spots with an optimum DNA concentration into the data collected. All microarray TIF images were quantified using Imagene Version 5.0 software.
Microarray analysis: data processing
The extent of background fluorescence was initially determined from an array experiment using BSA. The level of background fluorescence from the spots and array surface was found to be similar. Therefore, the average fluorescence between spots on the array surface was used as the background value throughout the experiments, which was minimal in comparison to the average signal intensity. Background subtracted median intensities were calculated for each spot on the microarray and the data was normalized according to the total signal intensity, so that the average spot intensity was the same for each replicate slide (×3). The normalized data of each competitive array experiment was used to generate a list of the high-intensity sequences/spots (the highest to lowest intensity), i.e. spots which were above a threshold level of intensity (Supplementary Data, Figure Aa). High-intensity spots/sequences which occurred in all three replicates were carried forward for further data analysis, this procedure minimized the occurrence of any false positives* or negatives* (*False positives = spots which fluoresce highly on one array but not on all three arrays. Total = 6.5%; False negatives = spots which did not fluoresce on one array but fluoresced highly on two arrays. Total = 2%) in the overall data collection. The average intensity was calculated and the sequences were ranked accordingly. The list of sequences generated were condensed to include only the best binding sequences, these were spots that had intensity above 55% normalized fluorescence and at least 6 standard deviations away from the global mean intensity (Figure Ab, Supplementary Data). The final list contained a total of 50 high affinity-binding sequences for His6-CspB.
Isothermal titration calorimetry (ITC)
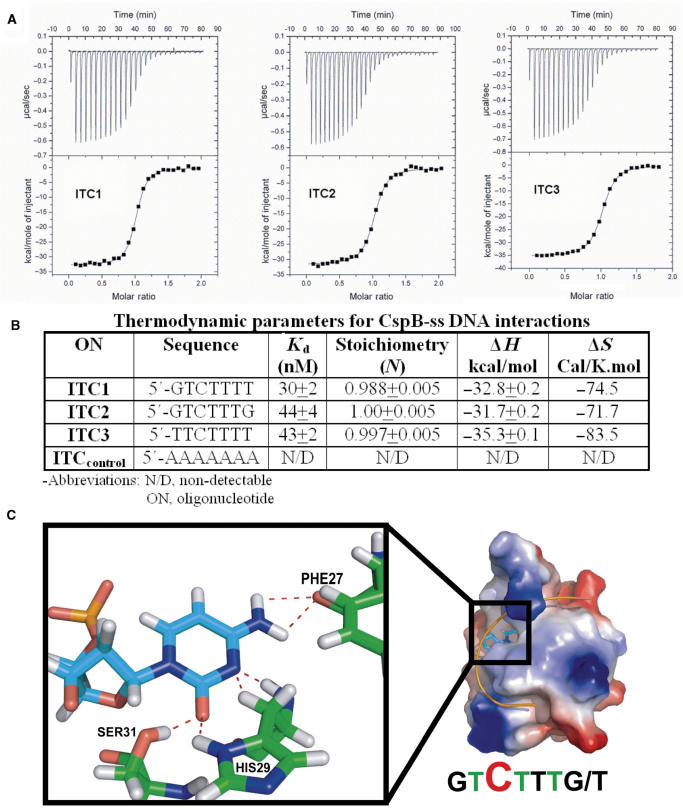
ITC experiments were carried out as described previously (21) with minor modifications. Four oligonucleotides were used for the experiments (Figure 2B), both possibilities of the consensus-binding sequence (ITC1 and ITC2), a positive (ITC3(26)) and negative control (ITCcontrol). Each exothermic heat pulse (Figure 2A, upper panels) corresponds to an injection of 5 μl of each oligonucleotide (100 μM) into the cell containing 5 μM CspB at 28°C. Integrated heat data (Figure 2A, lower panels) constitutes a differential binding curve, which was fitted to a single-site binding model to give, the stoichiometry of binding (N), binding affinity (Kd) and enthalpy of binding (ΔH) for each heptanucleotide.
Figure 2.
(A) Interaction of CspB with ss DNA templates as monitored by ITC. Titration data for 27 injections of 5 μl of each oligonucleotide (100 μM) into a cell containing 5 μM CspB. Oligonucleotides used for ITC are listed below. All data have been corrected for heats of dilution. (B) Thermodynamic parameters for CspB-ss DNA interactions. Non-detectable = Kd ≤ 5 μM. (C) A molecular model of a potential cytosine-binding pocket on CspB. The electrostatic surface potential of CspB and the backbone trace of the bound consensus heptanucleotide and the hydrogen bonds (depicted as dotted lines) between CspB and a cytosine nucleobase in the proposed binding pocket.
RESULTS AND DISCUSSION
Cy5 labelled SsoSSB binds ss DNA
SsoSSB was covalently labelled with the mono-functional dye Cyanine 5 (Cy5; Amersham). Electrophoretic mobility shift assays (EMSA) were conducted to verify that SsoSSB retained its DNA-binding activity subsequent to labelling with Cy5. A single-stranded 25-mer (ONc:5′–dATCCTACTGATTGGCCAAGGTGCTG-3′), labelled at the 5′-end with γ-32P, was used to compare the binding affinity of unlabelled and Cy5-labelled SsoSSB. Figure Cc (Supplementary Data) shows a gel-shift experiment performed with ONc in the presence of increasing amounts of unlabelled (lanes 1–4) or labelled (lanes 5–8) protein. The similarity in the EMSA for both unlabelled and Cy5-labelled protein suggests that labelling did not significantly affect the binding affinity of the protein. The double banding seen (Figure Cc, Supplementary Data) is a result of the gradual dissociation of labelled DNA from SsoSSB. The fact that previous SsoSSB-ss DNA-binding studies using ITC have shown that SsoSSB bound to ss DNA with a ratio of 5 nt/monmer (2) suggests that the double banding seen in the gel is probably a result of the EMSA technique used and the on/off-rates of SsoSSB-ss DNA complex formation.
SsoSSB-Cy5 binds non-specifically to all ss DNA sequences
A generic chip (Figure D, Supplementary Data) was constructed, which contained all possible 4096 ss hexadeoxynucleotide sequences found in DNA and incorporated into a general construct, 5′-NH-A10-N9-X6-3′ (X = G, C, A, or T and the stretch of 9 Ns is composed of random bases). The binding of SsoSSB-Cy5 to the generic chip was analysed and all the spots on the array fluoresced with similar intensities, consistent with non-specific binding of SsoSSB-Cy5 (2).
CspB and His6-CspB have similar affinity for ss DNA
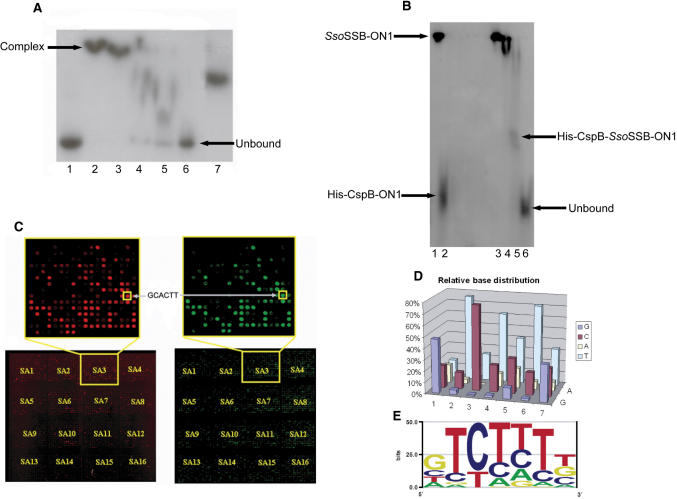
A ss 25-mer (3) (ONc: 5′–dATCCTACTGATTGGCCAAGGTGCTG-3′), labelled at the 5′-end with γ–32P was used to compare the binding affinities and specificities of His6-CspB and CspB. Figure 1A shows a gel-shift experiment performed for ONc in the presence of decreasing amounts of His6-CspB (lanes 2–6) and non-His6-tagged CspB (lane 7). Lanes 3, 4 and 5 show decreasing migration patterns for the His6-CspB ss DNA complex as less protein molecules bind. This is most likely a result of deviation in the affinity of CspB for specific binding sites within ONc. The data show that the addition of the His-tag does not significantly affect the binding affinity or specificity of the protein. The effect of flanking DNA at the 3′ end of the oligonucleotides, on CspB binding, was also examined by EMSA using a series of oligonucleotides that were structurally consistent with the oligonucleotides found on the array; the only difference was that a varying number of bases were added to the 3′ end. The results from these experiments show that flanking bases at the 3′ end or a lack of them did not seem to affect the binding of CspB (T1 and Figure B, Supplementary Data).
Figure 1.
(A) Electrophoretic mobility shift assay of His6-CspB and its complex with ONc. 15–20 pmol of 5′-end γ–32P-labelled ONc was mixed with 0, 500, 300, 100, 50 and 25 pmol (lanes 1–6) of His6-Csp. Lane 7 contains 500 pmol of CspB (untagged) as a positive control. Protein-DNA complexes were separated on a 20% native polyacrylamide gel. (B) Electrophoretic mobility shift assay of SsoSSB, CspB and the complexes formed with ON1. 15–20 pmol of 5′-end γ-32P-labelled ON1 was used at all times. Lanes; (1) 300 pmol SsoSSB; (2) 500 pmol CspB; (3) 300 pmol SsoSSB & 100 pmol CspB; (4) 300 pmol SsoSSB and 200 pmol CspB; (5) 300 pmol SsoSSB and 300 pmol CspB; (6) Unbound ON1 probe. Protein-DNA complexes were separated on a 10% native polyacrylamide gel. (C) Competitive microarray assay. Two individual microarray results for the competitive protein binding assay between His6-CspB (specific binding protein) and SsoSSB (non-specific binding protein). The fluorescent signal is a result of His6-CspB being detected on the surface using alexa 532-conjugated polyclonal antibody to His5. Microarray images shown here were results from two different batches of arrays and proteins. Sub-array three from each competitive protein-binding experiments has been enlarged to highlight the repeated pattern seen on all arrays. The oligonucleotide, GCACTT, appears as a bright spot on both arrays. The microarrays have been artificially coloured as a visual aid only. Abbreviations, SA = Sub-array (D) CspB consensus binding site. A 3-D bar chart showing the relative base distribution of each base within the proposed heptanucleotide-binding site. (E) Sequence logo (generated using the EnoLOGOS program), corresponding to the frequency of each base within the heptanucleotide-binding site. Both graphic visualizations show the resulting consensus sequence, 5′-GTCTTTG/T-3′.
CspB is competitive with SsoSSB
A competitive EMSA was used to show that His6-CspB binds more strongly than SsoSSB to the previously reported high affinity Y-box-binding motif (27) (ATTGG). A 25-mer, ON1 (5′–dA19-GATTGG-3′), which contains the Y-box-binding motif, was labelled at the 5′-end with γ–32P. ON1 is similar in composition to the oligonucleotides found on the generic chip. Figure 1B shows the result of a gel-shift experiment performed with ON1 in the presence of varying amounts of His6-CspB and SsoSSB proteins. An intermediate band can be seen in lane 5, corresponding to the formation of the His6-CspB-SsoSSB-ON1 complex. This band occurs only when the SsoSSB/CspB proteins are in a molar ratio of approximately 1:1.
The competitive binding assay can be transferred to microarray format
The fluorescent signal from His6-CspB bound to the chip was analysed (Figure E, Supplementary Data). About 20% of the spots had signals greater than threshold level (which was set at 40% of the maximum fluorescent signal). To eliminate weakly bound CspB, an equimolar mixture (as determined by gel-shift, Figure 1B) of a competitor SsoSSB, and His6-CspB was incubated with an oligonucleotide chip and bound His6-CspB was detected (Figure 1C). High-intensity fluorescence spots were observed in repeated patterns on the arrays, which is indicative of selective binding affinity (25).
High-affinity motifs identified by microarray analysis, are validated by electrophoretic mobility shift assay and isothermal calorimetry
An EMSA was carried out to confirm the binding-site data generated from the oligonucleotide chip analysis. The high affinity-binding motif, GCACTT, was chosen from the data (Figure 1C) to examine if the His6-CspB could compete successfully with the SsoSSB protein for this binding site. A 25-mer, ON-Microarray test (ON-Mt:5′–dAAAAAAAAAA-GCACTT-AAAAAAAAA-3′), containing the high affinity-binding motif was labelled at the 5′-end with γ–32P.
Figure F (Supplementary Data) shows a gel-shift experiment performed with ON-Mt in the presence of varying amounts of His6-CspB and SsoSSB proteins. An intermediate band (His-Csp-SsoSSB-ONc complex) can be seen in lanes 3–11 for ON-MTest, indicating that the CspB protein competed with SsoSSB for binding to the (microarray determined) high-affinity-binding site, GCACTT.
The highest intensity spots identified from the microarray analysis indicate that the strongest CspB-binding sites are pyrimidine rich (Figure Ga, Supplementary Data). The high incidence of thymine bases within the high-affinity 6-mer sequences agrees with previous reports that CspB has a preference for T-rich stretches of ss DNA (28). The presence of a stretch of 10 adenines in the linker region of each oligo (Figure Dd, Supplementary Data) is likely to lead to hairpin formation for T-rich sequences and may be expected to down weight the occurrence of poly T sequences. There is indeed a low intensity for TTTTTN sequences, which may in part be caused by the formation of such hairpins. Despite this effect, the averaging procedure still generates a T-rich consensus sequence. The standard motif alignment method (Genedoc) was used to align the resulting top fifty (as described in Materials and Methods) high-affinity CspB binding sequences (Figure Gb, Supplementary Data). A sequence alignment window 10 bases in length was used; only seven (coloured columns) out of those 10 positions are significantly (>40%) populated. Analysis of the relative distribution of each base within this proposed heptanucleotide-binding site gives a CspB consensus-binding sequence of 5′-GTCTTTG/T-3′ (Figure 1D and E).
Analysis of the microarray binding results indicates that CspB can accommodate the binding of a heptanucleotide with a strong binding preference for cytosine at position 3 and thymine at positions 2, 4 and 6. This is in agreement with another recent study (26), where the sequence-specific binding of heptapyrimidines to CspB was analysed by tryptophan fluorescence quenching experiments. Interestingly, the microarray results show that neither the Y-Box recognition motif (22), 5′-ATTGG-3′, nor its reverse complement 5′-CCAAT-3′, bind strongly to CspB. The ATTGG sequence has recently been shown to bind with a low affinity for CspB at 15°C (Kd = 5.3 μM (29)), which is similar to the results described here for CspB binding to sequences containing ATTGG at 4°C.
Both variants of the preferential binding sequences (ITC-1 and ITC-2) were analysed by ITC and gave binding constants in the low nanomolar range (Figure 2B). There is an order of magnitude difference in the Kds for the oligonucleotides described here and the Kds described previously (26), for similar/identical oligonucleotides. This is likely due to a difference in temperature, buffer conditions and the method used (tryptophan fluorescence quenching). The control sequence (TTCTTTT-ITC3), used here and in the previous study allows us to compare and scale the results from the two methods.
Thus, both the ITC and EMSA results confirm that the microarray assay does indeed select and identify tight binding sequences. This screening procedure could, therefore, be used as a general method for the rapid identification of high-affinity binding sites for SNABPs.
CspB-cytosine model explains the preference for a cytosine at position 3
The X-ray structure for CspB has been reported previously (24). Figure 2C shows the positively charged face of CspB, highlighting amino acid residues known to be involved in DNA binding (3). Molecular modelling of CspB with the consensus oligonucleotide ITC1, using the programme WITNOTP, identified a pocket in the centre of the DNA-binding face, which provides an ideal shape and hydrogen-bonding complementarity for binding cytosine. Three hydrogen bonds are formed between the docked cytosine base and amino acid side chains Ser31, His29 and the backbone of Phe27 (Figure 2C), providing specificity for cytosine over the other bases.
In the recently published structure of CspB in complex with hexathymidine (26), the ligand binds to two protein molecules. The nucleobase of T2, T3 and T4 make contact with one protein molecule, T5 bridges between protein molecules and T6 binds to the next protein molecule. The contacts made by hexathymidine provide the necessary scaffold for the complex to crystallize but the sequence fails to bind across the face of the CspB protein, as this sequence lacks the key cytosine nucleobase at position 3 (26), which, as we have shown here, is required for optimum ss DNA docking (Figure 2).
SNABPs have been reported to activate transcription by binding to a specific recognition sequence upstream (30) or within (31) a promoter, resulting in activation or repression of transcription. In the present study, CspB of B. subtilis which is capable of binding single-stranded nucleic acids (21) and affects expression of over 100 genes under cold shock conditions (32), was used as the test protein. Database analysis of the B. subtilis genome reveals that 89 copies of the consensus-binding sequence (5′-GTCTTTT-3′) exist within potential SNABP promoter regions (100 bases upstream of the ATG start for all genes), of which only 24 have an assigned function. The use of an unbiased genomic assay to identify optimal-binding sequences for SNABPs in vitro, may provide insight into their role in regulating cellular functions. The full implications of these sequences on gene expression and binding of CspB remain to be determined.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank Prof. M White (University of St. Andrews) for providing purified SsoSSB protein and recombinant plasmid DNA. This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC). The open access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rothman-Denes LB, Dai X, Davydova E, Carter R, Kazmierczak K. Transcriptional regulation by DNA structural transitions and single-stranded DNA-binding proteins. Cold Spring Harb. Symp. Quant. Biol. 1998;63:63–73. doi: 10.1101/sqb.1998.63.63. [DOI] [PubMed] [Google Scholar]
- 2.Kerr ID, Wadsworth RI, Cubeddu L, Blankenfeldt W, Naismith JH, White MF. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. EMBO J. 2003;22:2561–2570. doi: 10.1093/emboj/cdg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeeb M, Balbach J. Single-stranded DNA binding of the cold-shock protein CspB from Bacillus subtilis: NMR mapping and mutational characterization. Protein Sci. 2003;12:112–123. doi: 10.1110/ps.0219703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swamynathan SK, Nambiar A, Guntaka RV. Role of single-stranded DNA regions and Y-box proteins in transcriptional regulation of viral and cellular genes. FASEB J. 1998;12:515–522. doi: 10.1096/fasebj.12.7.515. [DOI] [PubMed] [Google Scholar]
- 5.Minich WB, Ovchinnikov LP. Role of cytoplasmic mRNP proteins in translation. Biochimie. 1992;74:477–483. doi: 10.1016/0300-9084(92)90088-v. [DOI] [PubMed] [Google Scholar]
- 6.Karpel RL, Henderson LE, Oroszlan S. Interactions of retroviral structural proteins with single-stranded nucleic acids. J. Biol. Chem. 1987;262:4961–4967. [PubMed] [Google Scholar]
- 7.Namsaraev EA, Berg P. Branch migration during Rad51-promoted strand exchange proceeds in either direction. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10477–10481. doi: 10.1073/pnas.95.18.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelm RJ, Jr, Wang SX, Polikandriotis JA, Strauch AR. Structure/function analysis of mouse Purbeta, a single-stranded DNA-binding repressor of vascular smooth muscle alpha-actin gene transcription. J. Biol. Chem. 2003;278:38749–38757. doi: 10.1074/jbc.M306163200. [DOI] [PubMed] [Google Scholar]
- 9.Simon MD, Sato K, Weiss GA, Shokat KM. A phage display selection of engrailed homeodomain mutants and the importance of residue Q50. Nucleic Acids Res. 2004;32:3623–3631. doi: 10.1093/nar/gkh690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papatsenko DA, Priporova IV, Belikov SV, Karpov VL. Mapping of DNA-binding proteins along the yeast genome by UV-induced DNA-protein crosslinking. FEBS Lett. 1996;381:103–105. doi: 10.1016/0014-5793(96)00091-9. [DOI] [PubMed] [Google Scholar]
- 11.Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez MM, Makhatadze GI. Major cold shock proteins, CspA from Escherichia coli and CspB from Bacillus subtilis, interact differently with single-stranded DNA templates. Biochim. Biophys. Acta. 2000;1479:196–202. doi: 10.1016/s0167-4838(00)00048-0. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Li T, Guo X, Lu Z. Exonuclease III protection assay with FRET probe for detecting DNA-binding proteins. Nucleic Acids Res. 2005;33:e23. doi: 10.1093/nar/gni021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker GT, Linn CP, Nadeau JG. DNA detection by strand displacement amplification and fluorescence polarization with signal enhancement using a DNA binding protein. Nucleic Acids Res. 1996;24:348–353. doi: 10.1093/nar/24.2.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klug SJ, Famulok M. All you wanted to know about SELEX. Mol. Biol. Rep. 1994;20:97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 16.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Xu F, Jin YX, Wang DB. SELEX Screening and Characterization of Small RNA Molecules That specifically bind the reactive blue dye. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 1999;31:504–508. [PubMed] [Google Scholar]
- 18.Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Jr, Ansari AZ. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl. Acad. Sci. U.S.A. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofweber R, Horn G, Langmann T, Balbach J, Kremer W, Schmitz G, Kalbitzer HR. The influence of cold shock proteins on transcription and translation studied in cell-free model systems. FEBS J. 2005;272:4691–4702. doi: 10.1111/j.1742-4658.2005.04885.x. [DOI] [PubMed] [Google Scholar]
- 21.Lopez MM, Yutani K, Makhatadze GI. Interactions of the major cold shock protein of Bacillus subtilis CspB with single-stranded DNA templates of different base composition. J. Biol. Chem. 1999;274:33601–33608. doi: 10.1074/jbc.274.47.33601. [DOI] [PubMed] [Google Scholar]
- 22.Graumann P, Marahiel MA. The major cold shock protein of Bacillus subtilis CspB binds with high affinity to the ATTGG- and CCAAT sequences in single stranded oligonucleotides. FEBS Lett. 1994;338:157–160. doi: 10.1016/0014-5793(94)80355-2. [DOI] [PubMed] [Google Scholar]
- 23.Wadsworth RI, White MF. Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res. 2001;29:914–920. doi: 10.1093/nar/29.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schindelin H, Herrler M, Willimsky G, Marahiel MA, Heinemann U. Overproduction, crystallization, and preliminary X-ray diffraction studies of the major cold shock protein from Bacillus subtilis, CspB. Proteins. 1992;14:120–124. doi: 10.1002/prot.340140113. [DOI] [PubMed] [Google Scholar]
- 25.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Max KE, Zeeb M, Bienert R, Balbach J, Heinemann U. T-rich DNA single strands bind to a preformed site on the bacterial cold shock protein Bs-CspB. J. Mol. Biol. 2006;360:702–714. doi: 10.1016/j.jmb.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 27.Schindelin H, Marahiel MA, Heinemann U. Universal nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold-shock protein. Nature. 1993;364:164–168. doi: 10.1038/364164a0. [DOI] [PubMed] [Google Scholar]
- 28.Lopez MM, Yutani K, Makhatadze GI. Interactions of the cold shock protein CspB from Bacillus subtilis with single-stranded DNA. Importance of the T base content and position within the template. J. Biol. Chem. 2001;276:15511–15518. doi: 10.1074/jbc.M010474200. [DOI] [PubMed] [Google Scholar]
- 29.Zeeb M, Max KE, Weininger U, Low C, Sticht H, Balbach J. Recognition of T-rich single-stranded DNA by the cold shock protein Bs-CspB in solution. Nucleic Acids Res. 2006;34:4561–4571. doi: 10.1093/nar/gkl376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santoro IM, Yi TM, Walsh K. Identification of single-stranded-DNA-binding proteins that interact with muscle gene elements. Mol. Cell. Biol. 1991;11:1944–1953. doi: 10.1128/mcb.11.4.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerrero MR, McEvilly RJ, Turner E, Lin CR, O’Connell S, Jenne KJ, Hobbs MV, Rosenfeld MG. Brn-3.0: a POU-domain protein expressed in the sensory, immune, and endocrine systems that functions on elements distinct from known octamer motifs. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10841–10845. doi: 10.1073/pnas.90.22.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaan T, Homuth G, Mader U, Bandow J, Schweder T. Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiology. 2002;148:3441–3455. doi: 10.1099/00221287-148-11-3441. [DOI] [PubMed] [Google Scholar]