Abstract
A long RNA oligomer, a 110mer with the sequence of a precursor-microRNA candidate, has been chemically synthesized in a single synthesizer run by means of standard automated phosphoramidite chemistry. The synthetic method involved the use of 2-cyanoethoxymethyl (CEM), a 2′-hydroxyl protecting group recently developed in our laboratory. We improved the methodology, introducing better coupling and capping conditions. The overall isolated yield of highly pure 110mer was 5.5%. Such a yield on a 1-μmol scale corresponds to 1 mg of product and emphasizes the practicality of the CEM method for synthesizing oligomers of more than 100 nt in sufficient quantity for biological research. We confirmed the identity of the 110mer by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry, as well as HPLC, electrophoretic methods, and RNase-digestion experiments. The 110mer also showed sense-selective specific gene-silencing activity. As far as we know, this is the longest chemically synthesized RNA oligomer reported to date. Furthermore, the identity of the 110mer was confirmed by both physicochemical and biological methods.
INTRODUCTION
The discovery of RNA interference (RNAi) (1), a specific gene-silencing mechanism mediated by small RNA molecules such as small interfering RNA (siRNA) (2) and microRNA (miRNA) (3), has had a major impact on research in the life sciences, and, in recent years, great advances have been made in our understanding of these and other kinds of non-coding RNA (4,5). Application to basic research is exemplified by the work of Siolas et al. (6) on short hairpin RNA (shRNA) and Amarzguioui et al. (7) on siRNA. RNA is also expected to play an important role in drug discovery, and several candidate small RNA molecules have already entered clinical trials (8). As a result of the increased demand for RNA for use as research reagents and potential therapeutic agents, more importance than ever is being attached to practical new methods in the chemical synthesis of RNA.
From the earliest days of research on RNA synthesis, it has been appreciated that the single most demanding problem is the selection of a suitable protecting group for the 2′-hydroxyl function of the ribose of the RNA. This protecting group must be stable throughout the solid-phase synthetic cycle, yet it must be readily removable under mild conditions (9). t-Butyldimethylsilyl (TBDMS) (10) is a popular 2′-hydroxyl protecting group whose phosphoramidite is commercially available. While the TBDMS method gives RNA of reasonable purity in reasonable yield, both yield and purity are sensitive to small changes in the synthetic conditions. Furthermore, the TBDMS group is associated with relatively long coupling times (11) and insufficiently high coupling yields (12), though improvements based on the use of more powerful activators or less bulky 2′-hydroxyl protecting groups have been proposed (12). To resolve these pro-blems, three new protecting groups, bis(2-acetoxyethyl-oxy)methyl (ACE) (13), triisopropylsilyloxymethyl (TOM) (14), and more recently t-butyldithiomethyl (DTM) (15), have been developed. Though such protecting groups represent major improvements in the synthesis of RNA oligonucleotides (16), however, they still leave something to be desired in their practical application. Thus, the synthesis of ACE-amidites is relatively complex (9) (though they are now commercially available), and automated synthesizers require special modification for use with them because of the incompatibility of glass materials with triethylamine trihydrofluoride, the 5′-desilylation reagent. TOM-protected oligonucleotides, meanwhile, are not readily amenable to routine analysis and purification by HPLC because of the hydrophobic nature of the silyl group. Finally, DTM-amidites, particularly the G amidite, are somewhat unstable in acetonitrile solution (15).
Chemical rather than enzymatic synthesis of RNA is desirable because it avoids the various errors and inefficiencies associated with in vitro transcription by T7 RNA polymerase (17). Chemical synthesis is also desirable for the ease of incorporation of modified nucleosides, which is important for studies of RNA structure and function. For example, site-specific disulphide cross-links have been used as probes of RNA tertiary structure (18). For such purposes, it is useful if the synthetic method can be extended to the efficient production of very long RNA oligomers. However, despite the advances that have been made in automated solid-phase synthesis, the production of longer RNA oligomers is still difficult and time-consuming. Previously synthesized RNA oligonucleotides up to 77 nt long have been generally restricted to tRNA (19–23), although there is also an example of the synthesis of an 84mer RNA oligonucleotide by the TOM method (14). Sometimes it is necessary to assemble shorter oligoribonucleotides to the full-length RNA by enzymatic ligation, as in the synthesis of Ascaris suum tRNAMet by Ohtsuki et al. (24).
Therefore, many points remain to be resolved in the field of RNA synthesis, and because of the increased demand for RNA there is a need for a radically improved synthetic method that would allow RNA synthesis to be carried out as efficiently as DNA synthesis, scaled up to the levels now obtained in DNA synthesis, and applied to the production of very long RNA oligomers. To meet this need, we have developed an achiral 2′-hydroxyl protecting group, 2-cyanoethoxymethyl (CEM), that has low steric hindrance and that can be completely removed under mild conditions (25). CEM chemistry is also compatible with standard, unmodified DNA synthesizer equipment.
In the present study, we have further developed the CEM method by significantly improving the capping and coupling conditions. To verify the potential of the improved method, we have undertaken the total chemical synthesis of a very long RNA oligomer. To the best of our knowledge, there has been no report of the chemical synthesis of an RNA oligomer longer than 100 nt. Here, we describe the synthesis of a 110mer precursor-miRNA (pre-miRNA) candidate by our improved new method. In contrast to previously synthesized long RNA oligonucleotides, which have generally been characterized by biochemical or biological means alone, we have confirmed the identity of the product by physicochemical methods as well as through assessing its biological activity by measuring its gene-silencing effect.
MATERIALS AND METHODS
General methods
Column chromatography was performed with Wako silica gel C-200, and TLC was carried out on Merck Silica gel 60 F254 TLC aluminium sheets. Analytical HPLC was performed on Shimadzu HPLC systems equipped with DNAPac PA100 (4 × 250 mm, Dionex) or PLRP-S 300 Å (4.6 × 150 mm; Polymer Laboratories, Church Stretton, UK) columns, preparative reverse-phase column chromatography (DMTr-on mode) was performed with PLRP-S 300 Å resin packed in 20 × 110 mm columns, and semipreparative anion-exchange HPLC (DMTr-off mode) was performed on a DNAPac PA100 (9 × 250 mm) column. Capillary gel electrophoresis was carried out on a P/ACE MDQ Molecular Characterization System with the ssDNA 100-R Kit (Beckman Coulter). UV spectra were recorded on a Hitachi U-3210 spectrometer, and 1H, 13C, and 31P NMR spectra on a Bruker DRX500 or DPX300 spectrometer. Chemical shifts are reported relative to tetramethylsilane and referenced to the residual proton signal of the following deuterated solvents: CDCl3 (7.25 p.p.m.) or DMSO-d6 (2.50 p.p.m.) for 1H NMR, CDCl3 (77.0 p.p.m.) or DMSO-d6 (39.5 p.p.m.) for 13C NMR, and external 85% phosphoric acid for 31P NMR. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectra were obtained with a Bruker Autoflex spectrometer (Bruker Daltonics, Billerica, MA). Reagents and solvents were purchased from commercial suppliers and used without further column chromatography. Anhydrous solvents were purchased from Kanto Kagaku (Tokyo, Japan), and tetrabutylammonium fluoride (TBAF) was purchased from Nacalai Tesque (Kyoto, Japan).
Synthesis of CEM-phosphoramidites
Full experimental procedures describing the synthesis of the CEM-phosphoramidites (henceforth termed ‘CEM-amidites’) are available in Supplementary Data.
Synthesis of 110mer RNA
The long oligomer selected for synthesis was the 110mer pre-miRNA hairpin candidate 5′-UGCUCGCUCA GCUGAUCUGU GGCUUAGGUA GUUUCAUGUU GUUGGGAUUG AGUUUUGAAC UCGGCAACAA GAAACUGCCU GAGUUACAUC AGUCGGUUUU CGUCGAGGGC-3′ (miR-196a-2 precursor; listed in miRBase) (26).
Using the CEM-amidites prepared as described in Supplementary Data, we synthesized the 110mer RNA on an Applied Biosystems Expedite Model 8909 nucleic acid synthesizer. In HPLC analyses and preparative purification, the following solvent systems were used: for reverse-phase HPLC, buffer A [5% acetonitrile in 50 mM triethylammonium acetate (TEAA), pH 7] and buffer B (90% acetonitrile in 50 mM TEAA, pH 7); and for anion-exchange HPLC, buffer C (10% acetonitrile in 25 mM Tris-HCl, pH 8.0) and buffer D (10% acetonitrile containing 700 mM NaClO4 in 25 mM Tris-HCl, pH 8.0). For preparative reverse-phase column chromatography (DMTr-on mode), the column was filled with PLRP-S 300 Å resin (50–70 μm), and elution was carried out with a gradient of buffer A and buffer B. For semipreparative anion-exchange HPLC (DMTr-off mode), elution was carried out with a gradient of buffer C and buffer D. Dialysis was carried out with a Spectra/Pore® membrane (molecular weight cut-off, 1000; Spectrum Laboratories, Laguna Hills, CA). Nuclease P1 was from Yamasa (Chiba, Japan), and alkaline phosphatase was from Promega (Madison, WI).
Analysis of 110mer RNA by MALDI-TOF mass spectrometry of RNase cleavage products
For RNase T1 digestion, RNA (60 pmol) was incubated with 0.3 U RNase T1 (Ambion, Austin, TX) in 50 mM Tris-HCl, pH 7.5, containing 1 mM EDTA and 25 μg/ml GpA in a total reaction volume of 30 μl at 37°C for 18 h. For MazF digestion, RNA (60 pmol) was incubated with 213 U MazF (Takara, Otsu, Japan) in 20 mM sodium phosphate, pH 6.0, containing 0.05% Tween-20 in a total reaction volume of 30 μl at 37°C for 1 h. The digestion products were purified by phenol/chloroform extraction and ZipTip C18 reverse-phase microcolumns (Millipore, Bedford, MA). MALDI-TOF mass spectra were acquired in positive-ion mode with a matrix solution consisting of 10 mg/ml 3-hyroxypicolinic acid and 1 mg/ml diammonium citrate. The potential for linear acceleration was 19 kV, and the potential for reflector acceleration was 20 kV. The IS/2 potential was set at 16.6 kV. Ion extraction was delayed by 130 ns, and 240 shots were accumulated for each sample.
Compositional analysis of 110mer RNA by nuclease digestion
RNA oligomer (20 ODU at 260 nm) was incubated with nuclease P1 (0.3 units) at 37°C for 24 h. Then, alkaline phosphatase (3 units) and buffer (50 mM Tris-HCl, pH 9.3, containing 1 mM MgCl2, 0.1 mM ZnCl2, and 1 mM spermidine, all at final concentrations) were added to give a total volume of 115 μl, and the mixture was incubated at 37°C for 2 days. The reaction mixture was then analyzed by HPLC.
Plasmid construction
Reporter plasmids pHOXB-Luc and pHOXB-Luc-antisense were constructed by inserting DNA duplex into the Xba I site of the luciferase reporter vector pGL4.13[luc2/SV40] (Promega) according to the supplier's protocol. The sequences of the inserted fragments were as follows: pHOXB-Luc sense, 5′-CTAGATTGTG CTAAGTTCTC CCAACAACAT GAAACTGCCT ATTCACGCCG TAATTT-3′; pHOXB-Luc antisense, 5′-CTAGAAATTA CGGCGTGAAT AGGCAGTTTC ATGTTGTTGG GAGAACTTAG CAGAAT-3′; pHOXB-Luc-antisense sense, 5′-CTAGAAATTA CGGCGTGAAT AGGCAGTTTC ATGTTGTTGG GAGAACTTAG CAGAAT-3′; and pHOXB-Luc-antisense antisense, 5′-CTAGATTGTG CTAAGTTCTC CCAACAACAT GAAACTGCCT ATTCACGCCG TAATTT-3′.
Cell culture and transfection
Human embryonic kidney derived cell line G3T-hi was purchased from Takara and maintained in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% foetal bovine serum at 37°C in a humidified atmosphere. Transfections with plasmid DNA and/or RNA were performed with FuGENE 6 transfection reagent (Roche, Indianapolis, IN) according to the supplier's instructions.
Luciferase reporter assay
G3T-hi cells (1 × 104) were seeded on a 96-well plate and incubated for 20 h. Reporter plasmid DNA (10 ng) and effector RNA (30 or 100 nM each at final concentration) were transfected by means of FuGENE 6. Forty-eight hours after transfection, luciferase activity was measured with the Steady-Glo Luciferase Assay System (Promega) according to the supplier's protocol. Sense and antisense 22-nt single-stranded RNAs (Japan BioService, Saitama, Japan) were annealed in distilled water for use as mature miRNA duplex. Luminescence intensity was measured with a microplate scintillation and luminescence counter (Packard, Meriden, CT), and luciferase activity was calculated relative to the basal activity expressed by pHOXB-Luc in the absence of effector RNA.
RESULTS AND DISCUSSION
Synthesis of CEM-amidites
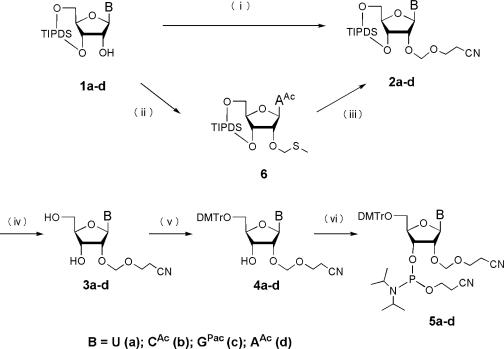
There was room for improvement in our previously reported method of synthesizing the monomer CEM-amidites (25). In that method, we started with the 5′-O-DMTr compound and alkylated the 2′-hydroxyl group to give intermediate 4. However, because we could not achieve selective alkylation of the 2′-hydroxyl group, the concurrently obtained 3′-O-CEM derivative had to be removed by column chromatography. Therefore, we sought an improved synthetic route for the CEM-amidites that could be carried out simply on a large scale, and we established the routes shown in Figure 1. In these routes, we introduce the CEM group into a 3′,5′-protected nucleoside (1a–d). With 2-cyanoethyl methylthiomethyl ether as the alkylating agent and N-iodosuccinimide (NIS) as the activator, the alkylation of the uridine, cytidine, and guanosine derivatives 1a–c proceeded efficiently at low temperature (−45°C). In the case of the adenosine derivative 1d only, the efficiency of alkylation was low. Therefore, compound 2d was produced by activating the nucleoside through methylthio derivative 6. Using these routes, we were able to synthesize all of the CEM-amidites with selective alkylation of the 2′-hydroxyl group with relative ease on a preparative scale. We are currently working on the further improvement of this method.
Figure 1.
Synthesis of CEM-admities. (i) 2-cyanoethyl methylthiomethyl ether, molecular sieves 4A (MS 4A), NIS, CF3SO3H, THF, −45°C; (ii) DMSO, acetic anhydride, acetic acid; (iii) 3-hydroxypropionitrile, MS 4A, NIS, CF3SO3H, THF, −45°C; (iv) NH4F, MeOH, 50°C or TEA·3HF, THF, 45°C; (v) 4,4′-dimethoxytrityl chloride, THF, pyridine, MS 4A, room temperature; (vi) diisopropylammonium tetrazolide, bis(N,N-diisopropylamino) cyanoethylphosphite, CH3CN, 40°C.
Synthesis, deprotection and purification of 110mer RNA
The RNA was synthesized on a 0.8-μmol scale by the phosphoramidite method modified for CEM chemistry. The conditions of the synthetic cycle are shown in Table 1.
Table 1.
Synthetic conditions for 110mer RNA
| Step | Operation | Reagent | Time (s) |
|---|---|---|---|
| 1 | Deblocking | 4% CCl3COOH in CH2Cl2 | 60 |
| 2 | Coupling | 0.075 M amidite in CH3CN + 0.25 M BMT in CH3CN | 150 |
| 3 | Capping | 0.1M Pac2O in THF + 6.5% 2-DMAP, 2% NMI and 10% 2,6-lutidine in THF | 170 |
| 4 | Oxidation | 0.1 M I2 in THF/pyridine/water (7/1/2) | 5 |
| 5 | Capping | 0.1 M Pac2O in THF + 6.5% 2-DMAP, 2% NMI and 10% 2,6-lutidine in THF | 30 |
Trichloroacetic acid in dichloromethane was used as the detritylation reagent. For the coupling reaction, amidites were used as 0.075 M solutions in acetonitrile, and 5-benzylmercaptotetrazole (BMT) was used as the activator instead of the 5-ethylthio-1H-tetrazole (ETT) used in the original method (25). BMT was adopted as the activator after a trial of about a dozen activators in the synthesis of short oligomers. The alternative activators tried included 4,5-(dicyano)imidazole, benzimidazolium triflate, 1H-tetrazole, and pyridinium trifluoroacetate. In addition to being a very powerful activator, BMT was found to give the best coupling yields and the cleanest crude product. A mixture of phenoxyacetic anhydride (Pac2O) in THF and 2-dimethylaminopyridine (2-DMAP), N-methylimidazole (NMI), and 2,6-lutidine in THF was used as the capping reagent to suppress the formation of 2,6-diaminopurine (2,6-DAP). [In the original method (25), NMI and pyridine were used as the base catalysts.] The oxidizing agent was 0.1 M iodine in THF/pyridine/water. Commercially available controlled-pore glass (CPG) with a pore size of 2000 Å derivatized with N4-benzoyl-2′-O-TBDMS-ribocytidine as the leader nucleoside was used as the solid support instead of 500-Å or 1000-Å CPG. Although the leader nucleoside bears a different base-protecting group and 2′-hydroxyl protecting group, their removal from a single residue at the 3′ end of the oligomer presented no problem under the conditions of the CEM method. A 2000-Å resin was used because, in our experience, a large-pore resin gives better yields in the synthesis of very long oligomers. Thus, in several respects, as described above, the synthetic conditions have been improved over those used in the original method (25).
After synthesis, the RNA was purified on a 0.7-μmol scale by two rounds of HPLC (DMTr-on and DMTr-off modes). First, the DMTr-on CPG solid support was treated with 50% triethylamine in acetonitrile to remove the cyanoethyl protecting group from the phosphate groups. Cleavage from the CPG solid support and removal of the base-protecting group was then carried out by treatment with a mixture of 28% ammonia solution and EtOH [3:1 (v/v)] for 24 h at 35°C. Removal of the phosphate-protecting and base-protecting groups can in principle be carried out in a single step. However, a minor side-reaction, adduct formation by the cyanoethyl group, may become significant in the synthesis of very long oligomers. To avoid this possibility, the two-step protocol was used. In the next step, the CEM protecting group was removed by treatment with 0.5 M TBAF in DMSO, with 0.5% nitromethane as an acrylonitrile scavenger, for 5 h at room temperature to remove the CEM protecting group. To achieve smooth and complete deprotection, the TBAF solution needs to be dried with molecular sieves 4A before use. After quenching the deprotection reaction mixture with Tris-HCl buffer, the crude RNA was obtained by EtOH precipitation.
Capillary gel electrophoresis and HPLC of the crude product after the removal of all protecting groups showed a single main peak with only tiny amounts of shorter products, though HPLC did resolve a couple of impurity peaks corresponding to long products (Supplementary Figures 1 and 2). The RNA was purified by reverse-phase column chromatography (PLRP-S 300 Å) in the DMTr-on mode, and the DMTr-on product was then treated with 80% AcOH solution to remove the DMTr group. DMTr-off purification was carried out by semipreparative HPLC (DNAPac PA100), and pure fractions were desalted by dialysis. The desired RNA product was obtained in good overall yield (5.5%; 41.5 ODU, 38.2 nmol).
In a comparison of the synthesis of a mixed-base 55mer by the CEM and the TBDMS methods, both with BMT as the activator, the HPLC profiles of the crude product were quite different. The CEM profile shows a single major peak with only tiny amounts of shorter impurities (25), while the TBDMS profile showed substantial amounts of shorter impurities (data not shown). Furthermore, though the CEM and TOM methods were not compared under the same conditions in the synthesis of the same sequence, published data on the TOM method shows substantial amounts of shorter impurities on capillary gel electrophoresis of the crude product [84mer; Ref. (14)], whereas the CEM method showed only tiny amounts (82mer; unpublished data).
While this study was in progress, another 2′-hydroxyl protecting group, 2-(4-tolylsulphonyl)ethoxymethyl (TEM), was reported by Zhou et al. (27). So far, however, only the synthesis of a 38mer uridine homo-oligomer and mixed-base oligomers of up to 21 nucleotides has been described. In the CEM method, meanwhile, despite the loss of <5% of CEM during ammonia treatment, no chain cleavage is observed (25), and no limitation in the use of the CEM method for the synthesis of RNA oligomers of up to 110 nt was noted in the present study.
Physicochemical identification of 110mer RNA
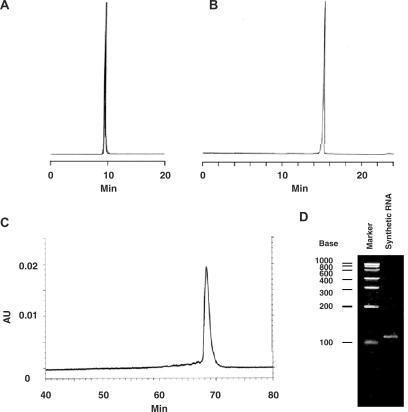
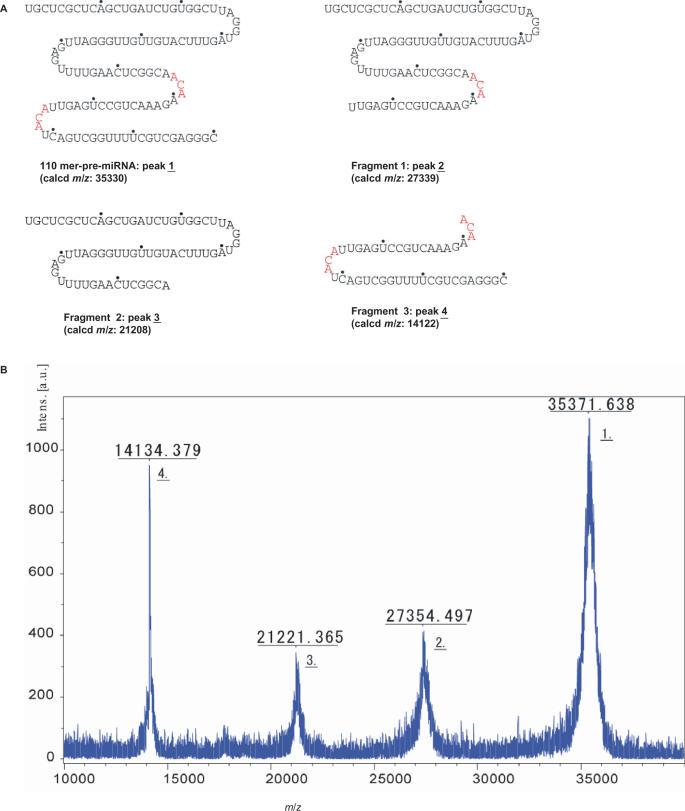
Reverse-phase and anion-exchange HPLC and capillary gel electrophoresis analysis of the final product showed only a single peak (Figure 2A–C), confirming the purity of the RNA obtained. Polyacrylamide gel electrophoresis analysis (Figure 2D) is consistent with the synthesis of a 110mer. We also confirmed the structure of the product by mass spectrometry. A MALDI-TOF mass-spectrometric analytical method is now available to confirm the molecular weight of oligonucleotides, though the analysis of oligomers longer than 50 nt is reported to be difficult (28). Therefore, we carried out MALDI-TOF mass-spectrometric analysis of the partial-digestion products of MazF, an endonuclease that cleaves specifically at ACA sequences (29). The highest m/z peak corresponds to the undigested RNA oligomer, and the small difference between the calculated and observed m/z values is consistent with a length of precisely 110 nt (Figure 3). We also observed peaks corresponding to the expected 44mer, 66mer and 85mer fragments. The smallest expected fragment, a 25mer, was observed in other digests (data not shown).
Figure 2.
HPLC and electrophoretic analysis of purified 110mer RNA. (A) A PLRP-S 300 Å HPLC reverse-phase column was operated at a flow rate of 1 ml/min and maintained at a temperature of 80°C. The solvent system was buffers A and B, and the RNA was eluted with a linear gradient from 0 to 50% buffer B in 20 min. (B) A DNAPac PA100 HPLC anion-exchange column was operated at a flow rate of 1.5 ml/min and maintained at a temperature of 70°C. The solvent system was buffers C and D, and the RNA was eluted with a linear gradient from 5 to 50% buffer D in 20 min. (C) Capillary gel electrophoresis. (D) Polyacrylamide gel electrophoresis. The synthetic RNA was analyzed on a 5% polyacrylamide gel and stained with the cyanine dye SYBR Green II.
Figure 3.
Mass-spectrometric analysis of MazF cleavage products of the 110mer RNA. (A) Sequence of 110mer and the predicted cleavage products. (B) MALDI-TOF mass spectrum of MazF cleavage products.
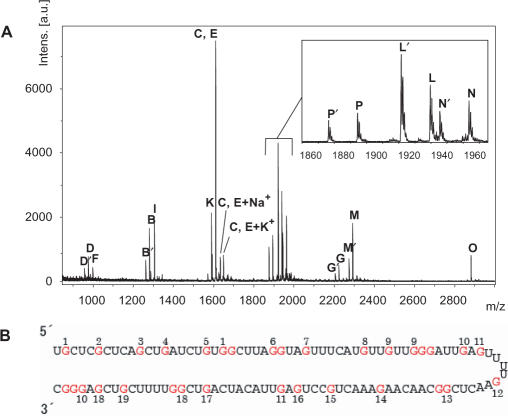
To check the identity of the 110mer by an independent method, we digested it with RNase T1, an endonuclease that cleaves specifically on the 3′ side of G, leaving a 3′-phosphate or 2′,3′-cyclic phosphate, and analyzed the fragments by mass spectrometry. Except for monomers and dimers, which were hard to distinguish among the noise of the spectrum, all of the expected fragments were identified (Figure 4A and B and Table 2), consistent with the synthesis of the target 110mer. RNase T1 digestion yielded pairs of fragments with 3′-phosphate or 2′,3′-cyclic phosphate, differing in molecular weight by 18. For example, fragments L and L′ (AACUCG; sequence 13, Table 2) yielded observed molecular weights of 1939.5 (fragment L; calcd 1939.2) and 1921.5 (fragment L′; calcd 1921.2). All other fragments were similarly identified.
Figure 4.
Mass-spectrometric analysis of RNase T1 cleavage products of the 110mer RNA. (A) MALDI-TOF mass spectrum of RNase T1 cleavage products. The inset shows an expanded view of the mass range 1860–1980. (B) Sequence of fragments produced by RNAse T1 digestion. The numbers shown represent the entry numbers of the fragments given in Table 2. Cleavage occurs on the 3′ side of G, and each number is placed at the G residue where cleavage occurs.
Table 2.
Sequences and masses of fragments produced by RNase T1 digestion
| 3′-Phosphate | 2′,3′-Cyclic phosphate | ||||||
|---|---|---|---|---|---|---|---|
| Entry numbera | Sequence (5′→3′) | Peak | Calculated mass | Observed mass | Peak | Calculated mass | Observed mass |
| 1 | UG | A | 670.4 | N.I.b | A′ | 652.4 | N.I. |
| 2 | CUCG | B | 1280.8 | 1280.9 | B′ | 1262.8 | 1262.9 |
| 3 | CUCAG | C | 1610.0 | 1610.2 | C′ | 1592.0 | N.D.c |
| 4 | CUG | D | 975.6 | 975.6 | D′ | 957.6 | 957.6 |
| 5 | AUCUG | E | 1611.0 | 1611.2 | E′ | 1593.0 | 1593.2 |
| 6 | CUUAG | E | 1611.0 | 1611.2 | E′ | 1593.0 | 1593.2 |
| 7 | UAG | F | 999.6 | 999.7 | F′ | 981.6 | N.D. |
| 8 | UUUCAUG | G | 2223.3 | 2223.6 | G′ | 2205.3 | 2205.6 |
| 9 | UUG | H | 976.6 | N.I. | H′ | 958.6 | N.I. |
| 10 | AUUG | I | 1305.8 | 1306.0 | I′ | 1287.8 | 1287.9 |
| 11 | AG | J | 693.5 | N.I. | J′ | 675.5 | N.I. |
| 12 | UUUUG | K | 1588.9 | 1589.1 | K′ | 1570.9 | N.D. |
| 13 | AACUCG | L | 1939.2 | 1939.5 | L′ | 1921.2 | 1921.5 |
| 14 | CAACAAG | M | 2291.4 | 2291.8 | M′ | 2273.4 | 2273.7 |
| 15 | AAACUG | N | 1963.2 | 1963.5 | N′ | 1945.2 | 1945.5 |
| 16 | CCUG | B | 1280.8 | 1280.9 | B′ | 1262.8 | 1262.9 |
| 17 | UUACAUCAG | O | 2880.7 | 2881.1 | O′ | 2862.7 | N.D. |
| 18 | UCG | D | 975.6 | 975.6 | D′ | 957.6 | 957.6 |
| 19 | UUUUCG | P | 1894.1 | 1894.4 | P′ | 1876.1 | 1876.4 |
aThe entry numbers correspond to the numbered fragments shown in Figure 4B.
bN.I., not identified.
cN.D., not detected.
Nucleoside-composition analysis of 110mer RNA by nuclease digestion
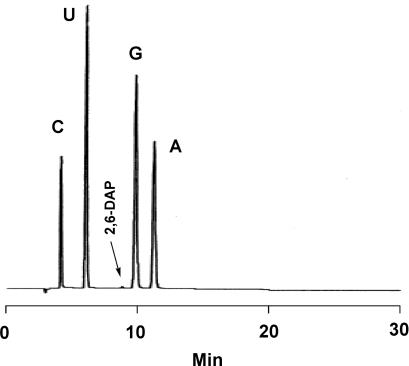
The RNA was treated with nuclease P1 and then alkaline phosphatase, and the digestion products were analyzed by HPLC (Figure 5). With the exception of 2,6-DAP noted below, no modified ribonucleosides were observed, showing that complete removal of the CEM protecting groups and base-protecting groups had been achieved. Furthermore, no dimers (2′,5′-phosphodiester-linked dinucleotides resulting from internucleotide migration or branched structures resulting from side reactions occurring during the condensation cycle) were observed. In our previously reported CEM synthetic method (25), the formation of 2,6-DAP was not observed in the synthesis of oligomers up to 55 nt in length. However, with continued repetition of the capping step during the synthesis of very long oligonucleotides, the formation of 2,6-DAP due to modification of G became detectable. (When pyridine or 4-dimethylaminopyridine (4-DMAP) is used in the capping reagent, displacement of the O6-phosphate triester adduct on G leads to the formation of 2,6-DAP after ammonia treatment during deprotection. When 2,6-lutidine or 2-DMAP is used instead, this displacement reaction is hindered by the presence of two methyl groups or a dimethylamino group ortho to the pyridine-ring nitrogen.) Though it was possible to completely suppress the formation of 2,6-DAP by using Pac2O/2-DMAP/2,6-lutidine as the capping reagent, under these conditions the efficiency of the capping reaction was slightly reduced, which is a significant disadvantage in the synthesis of very long oligomers. It was therefore decided to find compromise conditions in which very low levels of 2,6-DAP formation could be tolerated for the sake of a high capping efficiency. By adjusting the ratio of NMI to 2-DMAP in the capping reagent, we could achieve a very high capping efficiency, though not quite perfect suppression of 2,6-DAP formation.
Figure 5.
HPLC analysis of enzymatically digested 110mer RNA. A Develosil ODS-UG-5 reverse-phase column (4.6 × 250 mm) was operated at a flow rate of 1 ml/min and maintained at a temperature of 40°C. The buffer was 50 mM KH2PO4, pH 3.0, with H3PO4/MeOH, 20:1 (v/v).
Biological activity of 110mer RNA
To assess the biological activity of the chemically synthesized 110mer pre-miRNA candidate, we measured its specific gene-silencing effect. Generally, small dsRNAs suppress the expression of their target genes, whose mRNA possess sequences complementary to those of the small RNAs, by causing cleavage of the mRNA or inhibiting its translation (30). To determine the silencing effect of miRNAs, we constructed a reporter assay system. The sequence of the RNA synthesized was that of pre-miR-196a-2, so we used a reporter gene whose expression can be regulated by miR-196a. There is predicted to be a miR-196a target sequence in the 3′ untranslated region of human homeobox gene b-8 (HOXB8), and HOXB8 expression has been suggested to be regulated by miR-196a (31). We inserted DNA duplex corresponding to this target sequence and its flanking region into a luciferase-expressing reporter plasmid. In this system, a silencing effect by miR-196a results in a reduction in luciferase expression.
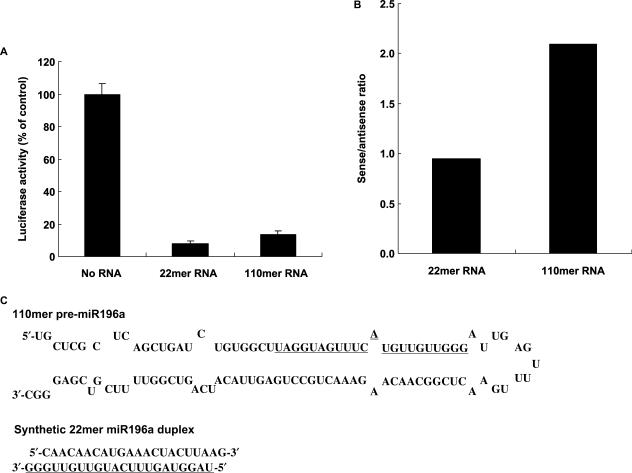
Transfection with chemically synthesized 110mer RNA decreased the expression of the target gene about as effectively as 22mer mature miR-196a RNA duplex (Figure 6A). The suppression of luciferase activity by 110mer RNA is considered to have resulted from the inhibition of luciferase protein expression, possibly at the transcriptional or translational level. Because pre-miRNA has to be processed to a smaller size to exert its gene-silencing effect, we conclude that our synthetic 110mer RNA was successfully processed and that the mature miRNA derived from the 110mer functioned as a silencing miRNA.
Figure 6.
Gene-silencing effect of chemically synthesized 110mer RNA. (A) Effect of 110mer RNA on expression of luciferase target gene. G3T-hi cells were transfected with pHOXB-Luc reporter plasmid and effector RNA (100 nM) and reporter assays performed 48 h after transfection. Each value shown is the average of three independent experiments, and standard deviations are indicated as error bars. (B) Comparison of sense/antisense suppression activity ratio for 110mer RNA. G3T-hi cells were transfected with pHOXB-Luc or pHOXB-Luc-antisense reporter plasmid and effector RNA (30 nM) and reporter assays performed 48 h after transfection. The results are presented as the ratio of the average of percent suppression of luciferase-containing sense and antisense target by 110mer RNA (pre-miRNA) or 22mer mature miRNA. (C) Sequences of the effector RNAs used. The underlined part of the 110mer pre-miR-196a-2 and the synthetic duplex 22mer miR-196a represent the sequence of the mature miRNA-196a.
Although the guide (antisense) strand of an siRNA molecule is normally specifically incorporated into the RNA-induced silencing complex (RISC), it sometimes happens that the passenger (sense) strand is incorporated, giving rise to off-target effects (32,33). A pre-miRNA molecule, in contrast, is expected to interact more specifically with RISC, allowing more specific incorporation of the antisense strand and exclusion of the sense strand during processing of the pre-miRNA, with resulting preferential cleavage of the target mRNA (32–34). In the case of pre-miR-196a, we found that the silencing effect against the sense target was about twice as strong as that against the antisense target, whereas 22mer mature miR-196a showed about the same silencing effect against both targets (Figure 6B). The observation of an equal silencing effect against the sense and antisense target mRNA by the 22mer is probably due to the sequences at the ends, which might prevent preferential incorporation of the guide strand. The 110mer pre-miRNA, therefore, showed a strand selectivity in the gene-silencing effect that was not shown by the mature miRNA. This observation supports the idea that the use of pre-miRNA instead of mature miRNA can be useful in gene silencing to confer selectivity of the antisense strand and to reduce the likelihood of off-target effects.
CONCLUSIONS
We have devised an improved method for the synthesis of CEM-amidites that is suitable for large-scale synthesis. We have also improved the conditions for the solid-phase synthesis of oligomers. With our improved CEM method, it is possible to synthesize very long oligomers that have until now been extremely difficult to synthesize. We have demonstrated the potential of the method by synthesizing, for the first time, an RNA oligomer longer than 100 nt. The 110mer we synthesized was produced by total chemical synthesis in a single synthesizer run, without the need for enzymatic ligation of separately synthesized fragments. Its structure was confirmed by physicochemical methods, including mass spectrometry, and it was also shown to have biological activity. To our knowledge, it is by far the longest chemically synthesized RNA whose structure has been determined by physicochemical methods. We believe that our new improved CEM method is sufficiently practical to become the standard RNA synthetic method, with special application to the synthesis of very long RNA oligomers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This research was supported in part by grants from the New Energy and Industrial Technology Development Organization (NEDO) of Japan for its Functional RNA Project. We thank Dr G.E. Smyth, Discovery Research Laboratories, Nippon Shinyaku Co. Ltd, for helpful discussions and suggestions during the preparation of the manuscript. Funding to pay the Open Access publication charges for this article was provided by Nippon Shinyaku Co. Ltd.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA Interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Hammond SM. MicroRNA therapeutics: a new niche for antisense nucleic acids. Trends Mol. Med. 2006;12:99–101. doi: 10.1016/j.molmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 4.The FANTOM Consortium and RIKEN Genome Exploration Research Group and Genome Science Group. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 5.RIKEN Genome Exploration Research Group and Genome Science Group and the FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 6.Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 7.Amarzguioui M, Lundberg P, Cantin E, Hagstrom J, Behlke MA, Rossi JJ. Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat. Protoc. 2006;1:508–517. doi: 10.1038/nprot.2006.72. [DOI] [PubMed] [Google Scholar]
- 8.Uprichard SL. The therapeutic potential of RNA interference. FEBS Lett. 2005;579:5996–6007. doi: 10.1016/j.febslet.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reese CB. The chemical synthesis of oligo- and poly-nucleotides: a personal commentary. Tetrahedron. 2002;58:8893–8920. [Google Scholar]
- 10.Usman N, Ogilvie KK, Jiang M.-Y, Cedergren RJ. Automated chemical synthesis of long oligoribonucleotides using 2′-O-silylated ribonucleotides 3′-O-phosphoramidites on a controlled-pore glass support: synthesis of a 43-nucleotide sequence similar to the 3′-half molecule of an Escherichia coli formylmethionine tRNA. J. Am. Chem. Soc. 1987;109:7845–7854. [Google Scholar]
- 11.Gough GR, Miller TJ, Mantick NA. p-Nitrobenzyloxymethyl: a new fluoride-removable protecting group for ribonucleoside 2′-hydroxyls. Tetrahedron Lett. 1996;37:981–982. [Google Scholar]
- 12.Welz R, Müller S. 5-(Benzylmercapto)-1H-tetrazole as activator for 2′-O-TBDMS phosphoramidite building blocks in RNA synthesis. Tetrahedron Lett. 2002;43:795–797. [Google Scholar]
- 13.Scaringe SA, Wincott FE, Caruthers MH. Novel RNA synthesis method using 5′-O-silyl-2′-O-orthoester protecting groups. J. Am. Chem. Soc. 1998;120:11820–11821. [Google Scholar]
- 14.Pitsch S, Weiss PA, Jenny L, Stutz A, Wu X. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2′-O-[(triisopropylsilyl)oxy]methyl (2′-O-tom)-protected phosphoramidites. Helv. Chim. Acta. 2001;84:3773–3795. [Google Scholar]
- 15.Semenyuk A, Földesi A, Johansson T, Estmer-Nilsson C, Blomgren P, Brännvall M, Kirsebom LA, Kwiatkowski M. Synthesis of RNA using 2′-O-DTM protection. J. Am. Chem. Soc. 2006;128:12356–12357. doi: 10.1021/ja0636587. [DOI] [PubMed] [Google Scholar]
- 16.Micura R. Small interfering RNAs and their chemical synthesis. Angew. Chem. Int. Ed. 2002;41:2265–2269. doi: 10.1002/1521-3773(20020703)41:13<2265::AID-ANIE2265>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Helm M, Brulé H, Giegé R, Florentz C. More mistakes by T7 RNA polymerase at the 5′ ends of in vitro-transcribed RNAs. RNA. 1999;5:618–621. doi: 10.1017/s1355838299982328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maglott EJ, Glick GD. Probing structural elements in RNA using engineered disulfide cross-links. Nucleic Acids Res. 1998;26:1301–1308. doi: 10.1093/nar/26.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Persson T, Kutzke U, Busch S, Held R, Hartmann RK. Chemical synthesis and biological investigation of a 77-mer oligoribonucleotide with a sequence corresponding to E. coli tRNAAsp. Bioorg. Med. Chem. 2001;9:51–56. doi: 10.1016/s0968-0896(00)00218-2. [DOI] [PubMed] [Google Scholar]
- 20.Ogilvie KK, Usman N, Nicoghosian K, Cedergren RJ. Total chemical synthesis of a 77-nucleotide-long RNA sequence having methionine-acceptance activity. Proc. Natl Acad. Sci. USA. 1988;85:5764–5768. doi: 10.1073/pnas.85.16.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bratty J, Wu T, Nicoghosian K, Ogilvie KK, Perreault J.-P, Keith G, Cedergren RJ. Characterization of a chemically synthesized RNA having the sequence of the yeast initiator tRNAMet. FEBS Lett. 1990;269:60–64. doi: 10.1016/0014-5793(90)81118-8. [DOI] [PubMed] [Google Scholar]
- 22.Gasparutto D, Livache T, Bazin H, Duplaa AM, Guy A, Khorlin A, Molko D, Roget A, Téoule R. Chemical synthesis of a biologically active natural tRNA with its minor bases. Nucleic Acids Res. 1992;20:5159–5166. doi: 10.1093/nar/20.19.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodwin JT, Stanick WA, Glick GD. Improved solid-phase synthesis of long oligoribonucleotides: application to tRNAphe and tRNAgly. J. Org. Chem. 1994;59:7941–7943. [Google Scholar]
- 24.Ohtsuki T, Kawai G, Watanabe Y, Kita K, Nishikawa K, Watanabe K. Preparation of biologically active Ascaris suum mitochondrial tRNAMet with a TV-replacement loop by ligation of chemically synthesized RNA fragments. Nucleic Acids Res. 1996;24:662–667. doi: 10.1093/nar/24.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohgi T, Masutomi Y, Ishiyama K, Kitagawa H, Shiba Y, Yano J. A new RNA synthetic method with a 2′-O-(2-cyanoethoxymethyl) protecting group. Org. Lett. 2005;7:3477–3480. doi: 10.1021/ol051151f. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou C, Honcharenko D, Chattopadhyaya J. 2-(4-Tolylsulfonyl)ethoxymethyl (TEM)—a new 2′-OH protecting group for solid-supported RNA synthesis. Org. Biomol. Chem. 2007;5:333–343. doi: 10.1039/b614210a. [DOI] [PubMed] [Google Scholar]
- 28.Hail ME, Elliot B, Anderson A. High-throughput analysis of oligonucleotides using automated electrospray ionization mass spectrometry. Am. Biotech. Lab. 2004;22:12–14. [Google Scholar]
- 29.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2005;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 30.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNA and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 31.Yekta S, Shih I.-H, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 32.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 33.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 34.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.