Abstract
YY1 is a DNA-binding transcription factor found in both vertebrates and invertebrates. Database searches identified 62 YY1 related sequences from all the available genome sequences ranging from flying insects to human. These sequences are characterized by high levels of sequence conservation, ranging from 66% to 100% similarity, in the zinc finger DNA-binding domain of the predicted proteins. Phylogenetic analyses uncovered duplication events of YY1 in several different lineages, including flies, fish and mammals. Retroposition is responsible for generating one duplicate in flies, PHOL from PHO, and two duplicates in placental mammals, YY2 and Reduced Expression 1 (REX1) from YY1. DNA-binding motif studies have demonstrated that YY2 still binds to the same consensus sequence as YY1 but with much lower affinity. In contrast, REX1 binds to DNA motifs divergent from YY1, but the binding motifs of REX1 and YY1 share some similarity at their core regions (5′-CCAT-3′). This suggests that the two duplicates, YY2 and REX1, although generated through similar retroposition events have undergone different selection schemes to adapt to new roles in placental mammals. Overall, the conservation of YY2 and REX1 in all placental mammals predicts that each duplicate has co-evolved with some unique features of eutherian mammals.
INTRODUCTION
The transcription factor YY1 (Yin Yang 1) is a Gli–Kruppel type zinc finger protein, and can function as a repressor, activator or transcription initiator depending upon the sequence context of YY1-binding sites with respect to other regulator elements (1). The protein has a DNA-binding domain at the C-terminus and other modulating domains at the N-terminus displaying repression, activation and protein/protein interaction activities (2). YY1 interacts with several key transcription factors, including TBP, TAFs, TFIIB and Sp1, as well as histone-modifying complexes, such as p300, HDACs, PRMT1 and Polycomb complexes (2,3). Many cellular and viral genes are controlled by YY1. A recent survey estimated that ∼10% of human genes contain YY1 binding sites near their promoter regions (4). Another set of studies has revealed that some of mammalian imprinted genes contain very unusual tandem arrays of YY1 binding sites in their controlling regions, suggesting potential roles in mammalian genomic imprinting (5–7). A series of mouse mutagenesis experiments demonstrated the dosage-dependent essential roles of YY1 during mouse development as well as in cell cycle control (8,9).
YY1 is evolutionarily well conserved throughout the vertebrate and invertebrate lineages. It has been identified in several vertebrate species (1,10,11), and two genes very similar to YY1 are found even in flies, Pleiohomeotic (PHO) and Pho-like (PHOL) (12,13). PHO is one of the DNA-targeting proteins for the Polycomb complex and the phenotypes of pho-deficient mutants can be rescued by mammalian YY1 (14). In mammalian genomes, two other YY1-related genes have been identified, YY2 (Yin Yang 2) and Reduced Expression 1 (REX1). YY2 is functionally very similar to YY1 (15), and is a retroposed copy duplicated from YY1 based on its intronless structure and location in the intron of another X-chromosomal gene, Mbtps2 (16). REX1 was independently discovered, before the identification of YY1, due to its unique expression profile: dramatic decline of expression after retinoic acid-induced differentiation of F9 murine teratocarcinoma stem cells (17). Subsequently, REX1 has been mainly studied as a stem cell marker that is controlled by Oct3/4 (18,19). A recent comparative study, however, emphasized that REX1 is a member of the YY1 subfamily (20).
Despite the significant roles and evolutionary conservation of YY1-related sequences in animals, there has not been any systematic analysis of these sequences in terms of their origins, evolutionary patterns and implications for functional diversification. To address this, we have analyzed YY1-related sequences identified from genome sequences ranging from flying insects to placental mammals. We have identified two evolutionarily conserved protein domains within YY1 which were previously unrecognized. We have uncovered independent retroposition events that have been responsible for forming duplicate copies, such as PHOL from PHO in flies, and YY2 and REX1 from YY1 in placental mammals. Our analyses revealed that the zinc finger domains of YY2 and REX1 have been under different selection pressures compared to YY1. Their DNA-binding properties have evolved from YY1 by weakening DNA-binding affinity in both YY2 and REX1, and changing DNA-binding motifs in REX1. The evolution patterns of YY1 and other YY1-related genes described in the current study provide a unique paradigm for gene duplication and functional diversification.
MATERIALS AND METHODS
Database search and sequence analyses
A series of database searches were conducted using the BLAST program (http://www.ncbi.nlm.gov/BLAST) to obtain YY1-related sequences. Human YY1 (NP_003394.1) was first used as a query sequence to search sequence databases, including NCBI, the Genome Browser at University of California Santa Cruz and Ensembl. Later, human REX1 (NP_777560.2) and YY2 (NP_996806.1) were used to further characterize the identified YY1-related sequences from chordates, while Drosophila melanogaster PHO (NP_524630.1) and PHOL (NP_648317.1) were used for the identified insect sequences. The detailed information regarding all the YY1-related sequences described in this study is available as Supplementary Data 1 through the following website (http://JooKimLab.lsu.edu/JooKimLab/Data.html).
Multiple sequence alignments were performed with ClustalW using the following parameters: gap opening penalty = 10, gap extension penalty = 0.1 (0.2 for multiple alignment), Gonnet Protein Weight Matrix, residue specific penalties = ON, hydrophilic penalties = ON, gap separation distance = 4, end gap separation = OFF (21). Sequences were edited manually in Mega3 V3.1 to remove spurious introns from some sequences (22). Separate multiple alignments were performed for insects’ and chordates’ sequences. Subsequently, two phylogeny gene trees were constructed and analyzed using both the neighbor-joining and maximum parsimony methods as implemented in Mega3 V3.1 with Poisson correction and confirmed by bootstrapping 1000 iterations (23). Synonymous and non-synonymous substitution rates were estimated using two different approaches: Nei–Gojobori (24) and Yang–Nielsen methods (25).
Expression of fusion proteins and DNA-binding motif study
The zinc finger regions of YY1 (NM_009537.2), YY2 (NM_178266) and REX1 (NM_009556.2) were amplified from either mouse brain cDNAs or genomic DNAs by the following primer sets: YY1 (mYY1Zn5, 5′-CCAAGAACAATAGCTTGCCCTC-3′ and mYY1Zn3, 5′-TCACTGGTTGTTTTTGGCTTTAGCG-3′), YY2 (mYY2Zn5, 5′-CCAAGACCTATAGCATGCTCTC-3′ and mYY2Zn3, 5′-TTACTGGTCATTCTT GTTCTTAACATGGG-3′) and REX1 (mRexZn5, 5′-TTATCGATGCTGGAGTGTCCTCAAGC-3′ and mRexZn3, 5′-TCAGCATTTCTTCCCTGCCTTTGC-3′). The amplified products were first cloned into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA, USA), and later transferred to the EcoRI site of the pGEX-4T-2 vector (Amersham Biosciences, Piscataway, NJ, USA) after sequence confirmation. The constructed vectors were transformed into BL21 (DE3) competent cells for bacterial expression (Strategen, La Jolla, CA, USA). The optimum induction of the constructs by IPTG was monitored through SDS-PAGE (Supplementary Data 4 from http://JooKimLab.lsu.edu/JooKimLab/Data.html).
DNA-binding motif studies were conducted as described in the previous studies (26,27) with slight modifications. Briefly, the transformed cells were grown at 37°C in LB media (100 ml) to an optical density of 0.6 at 600 nm, and protein expression was induced with 0.4 mM IPTG for additional 3.5 h. Cells were harvested by centrifugation at 4000g for 10 min at 4°C. Lysates were prepared from the cell pellets by sonication in 6 ml of ice-cold NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris–HCl, pH 8.0, 0.5% NP-40). Protein concentration in cell lysates was determined using the Bradford assay (Pierce, Rockford, IL, USA). Aliquots of 500 μg/100 μl were frozen at −80°C.
Immobilized glutathione agarose (Pierce) was washed three times with 1 ml ice-cold NETN buffer and used to isolate fusion proteins by incubating 500 μg lysate with 50 μl washed agarose beads at 4°C for 30 min while rotating. The agarose beads were precipitated by centrifugation, and washed twice first with 1 ml ice-cold NTEN buffer and later with 1 ml 1× binding buffer (12 mM HEPES, pH 7.9, 60 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, 0.05% NP-40, 50 μg/ml bovine serum albumin, 10% glycerol). The final pellet was resuspended in 100 μl 1× binding buffer. Randomized duplex DNAs were prepared with PCR using following oligonucleotides (10 ng of NT55, 5′-CTGTCGGAATTCGCTGACGT(N)15CGTCTTATCGGATCCTACGT-3′, 0.1 μg of UpNt, 5′-CTGTCGGAATTCGCTGACGT-3′ and 0.1 μg of DwNt, 5′-ACGTAGGAT CCGATAAGACG-3′ as a template and primers for extension reaction, respectively). Duplex DNAs were labeled 10 μCi [α-32P] dATP for the easy chase of the bound DNAs with the PCR reaction containing 5 U of i-StarTaq DNA polymerase (Intron Biotech), 0.2 mM each of dGTP, dTTP and dCTP and 10 μM dATP. PCR was performed for 25 cycles (95°C 30 s; 65°C 1 min; 72°C 1 min). The labeled DNAs were allowed to bind to the fusion protein immobilized on the agarose beads at room temperature for 30 min with rotation. The bound DNAs were washed three times with 1 ml of 1× binding buffer, eluted by phenol: chloroform extraction, and finally precipitated ethanol. The eluted DNAs were amplified again with the same conditions described earlier for another round of DNA-binding. The following PCRs were performed only for 10 cycles. After five rounds of DNA-binding and amplification (Supplementary Data 4), the DNAs were subcloned into pCR4-TOPO vector (Invitrogen). For each fusion protein, 40–60 clones were purified and sequenced.
Gel shift assay of DNA-binding motifs
The identified DNA motifs for each fusion protein were further analyzed with gel shift assays (Gel shift Assay System, Promega, Madison, WI, USA). About 10–20 μg of each fusion protein was used for each experiment with the [γ-32P] ATP-labeled duplex probes prepared from the following oligonucleotides: CSE2-A, 5′-CCCACCCACCTGGGCGCCATCTTTAATGAAAG-3′, and CSE2-B, 5′-CTTTCATTAAAGATGGCGCCCAGGTGGGTGGG-3′; 2a-A, 5′-CCCACCCACCTGGGTGCCATCTTTAATGAAAG-3′, and 2a-B, 5′-CTTTCATTAAAGATGGCACCCAGGTGGGTGGG-3′; 2b-A, 5′-CCCACCCACCTGGGCACCATCTTTAATGAAAG-3′, and 2b-B, 5′-CTTTCATTAAAGATGGTGCCCAGGTGGGTGGG-3′; Probe1-A, 5′-GATAAGACGCGGCAGCCATTTGGAACGTCAGCG-3′, and Probe1-B, 5′-CGCTGACGTTCCAAATGGCTGCCGCGTCTTATC-3′; Probe2-A, 5′-GATAAGACGGCAGCCATTTTGAGGCCCACGTCAGCG-3′, and Probe2-B, 5′-CGCTGACGTGGGCCTCAAAATGGCTGCCGTCTTATC-3′; Probe3-A, 5′-GATAAGACGCGGCAGCCATTAGGAACGTCAGCG-3′, and Probe3-B, 5′-CGCTGACGTTCCTAATGGCTGCCGCGTCTTATC-3′; Probe4-A, 5′-GATAAGACGGCCATTATGAGGCCCACGTCAGCG-3′, and Probe4-B, 5′-CGCTGACGTGGGCCTCATAATGGCCGTCTTATC-3′; Probe5-A, 5′-GATAAGACGGCCATTTTGAGGCCCACGTCAGCG-3′, and Probe5-B, 5′-CGCTGACGTGGGCCTCAAAATGGCCGTCTTATC-3′; Probe6-A, 5′-GATAAGACAGCCATTTTGAGGCCCACGTCAGCG-3′, and Probe6-B, 5′-CGCTGACGTGGGCCTCAAAATGGCTGTCTTATC-3′; Probe7-A, 5′-GATAAGACCGCCATTTTGAGGCCCACGTCAGCG-3′, and Probe7-B, 5′-CGCTGACGTGGGCCTCAAAATGGCGGTCTTATC-3′. To monitor our gel shift assays, we also performed a set of control experiments using endogenous YY1 from HeLa nuclear extracts (Promega).
RESULTS
Identification of YY1-related sequences from invertebrates and vertebrates
The protein sequence of human YY1 (GenBank accession no. NP_0034941, 414 amino acid long) was used to search databases to identify YY1-related sequences from all available genome sequences. One YY1 homolog, known as PHO, was identified from each of the flying insects, including mosquitoes, honeybees, beetles and 10 different species of flies. In flies, a similar sequence, known as PHOL, was identified from each of the 10 different fly species. This totals to 23 different YY1-related sequences from insects. Database searches identified 39 different YY1-related sequences in chordates, ranging from urochordates (sea squirts) to placental mammals: one each from sea squirts and purple sea urchins, six from fish, one from frog, one from chicken, 29 from mammals. In fish, two copies of YY1 sequences were identified from each of three sequenced genomes, zebrafish, pufferfish and spotted pufferfish whereas three copies of YY1-related sequences were identified from each placental mammal. Database searches have identified a total of 62 YY1-related sequences. Based on sequence similarity, these are categorized into five groups: the PHO and PHOL groups from flying insects, the YY1 group from vertebrates, and the YY2 and REX1 groups from placental mammals (Figure 1). Individual sequences and other related information are available through the following website (http://JooKimLab.lsu.edu/JooKimLab/Data.html).
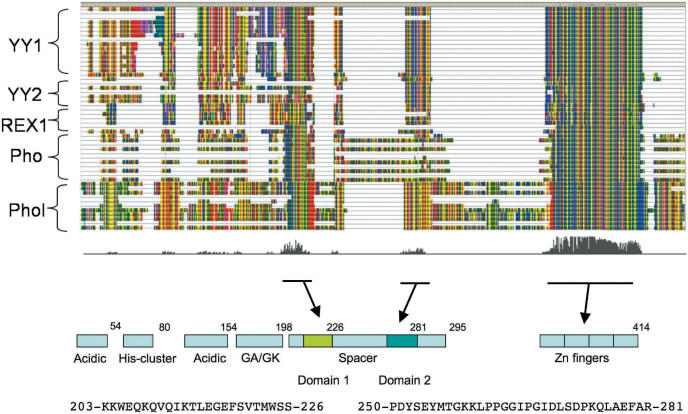
Figure 1.
Global alignment of YY1, YY2, REX1, PHO and PHOL. Protein sequences derived from 62 YY1-related sequences are aligned using the ClustalW program. The zoom-out view of this result is shown for global representation. The actual sequence alignment is available as Supplementary Data 2 through the following website (http://JooKimLab.lsu.edu/JooKimLab/Data.html). Each row represents one individual sequence and these sequences are categorized into different groups indicated by parentheses on the left. Different amino acids are represented by different background colors, and thus a vertical line with the same color indicates the conservation (or identical amino acid residue) at that position amongst all the sequences analyzed. The different levels of evolutionary conservation throughout the entire region of YY1-related sequences are represented by a graph underneath the alignment. Three regions are evolutionarily conserved, and thus highlighted by underlines with arrows. These include Domain I and II that are located within the previously defined Spacer region, and the DNA-binding zinc finger domain at the C-terminus. The protein domain structure of human YY1 is shown as a reference at the bottom (2).
Comparison of the amino acid sequences derived from the YY1-related sequences identified three evolutionarily conserved protein domains (Figure 1). These include two domains in the middle of the protein (amino acid position 203–226 and 250–281 in the human YY1, respectively), and one DNA-binding zinc finger domain at the C-terminus (aa 298–414). The two domains in the middle, Domains I and II, are located within the region previously known as the Spacer between several N-terminal domains and the C-terminal DNA-binding domain (2). These two domains are found in the YY1-related sequences of most, but not all, vertebrates and insects. In flies, only Domain I is found in both PHO and PHOL sequences. In placental mammals, two domains are found in YY2, but only Domain II is found in the REX1 sequences. However, the zinc finger domain is found in all the YY1-related sequences with high levels of sequence conservation, ranging from 66 to 100% similarity. The relative positions of these three protein domains are also conserved among all the identified sequences. The conservation of these three domains in the YY1-related sequences suggests that these three domains constitute the original domain structure of the YY1 protein.
Retroposition-mediated YY1 duplications in flies and placental mammals
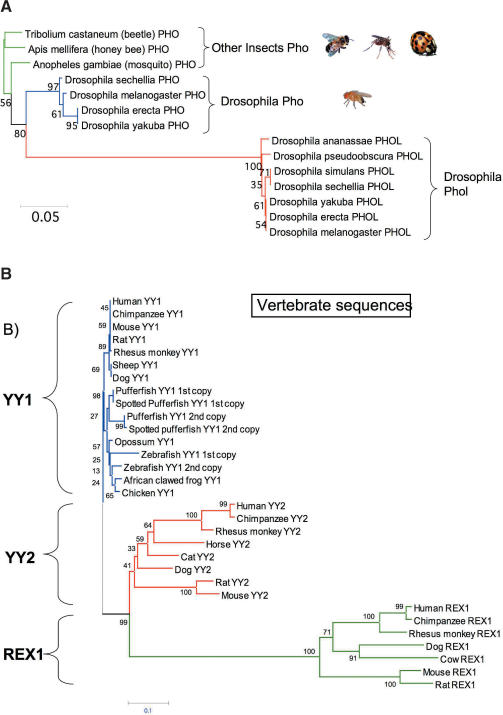
Several lineages have more than one copy of YY1-related sequences, including flies, fish and placental mammals. Two copies of YY1-related sequences, PHO and PHOL, are found in all the fly species examined to date while only one copy, PHO, is found in the other flying insects. This suggests a gene duplication unique to the fly lineage. According to the results of phylogenetic analyses (Figure 2A), the topology of the two gene trees corresponding to the PHO and PHOL groups in flies is very similar to that of the known species tree of the fruitfly genus Drosophila, indicating that this gene duplication predates the radiation of all fly species. The PHO sequences of the other flying insects show slightly greater levels of sequence similarity to the PHO rather than PHOL sequences in flies, suggesting that PHO is the original sequence that gave rise to the duplicated copy PHOL. This is further confirmed by the different exon structures of PHO and PHOL (Figure 3A). The coding region of PHO is split into five exons, and a similar split exon structure is also found in the PHO of other insects, such as beetles and honeybees. In contrast, the entire coding region of PHOL is located within one exon, an intronless structure of its coding region. This intronless genomic structure is usually observed in the sequences that have been duplicated through an RNA-mediated mechanism, retroposition, by which processed mRNAs are reverse-transcribed and transposed to other genomic loci without introns in germ cells (28). These data therefore indicate that PHOL has been duplicated from PHO through retroposition.
Figure 2.
Gene trees connecting (A) PHO and PHOL and (B) YY1, YY2 and REX1. Alignments were first created using a subset of sequences, the protein sequences of which are available or can be predicted with certainty. Later, the trees were constructed with the neighbor-joining method using the Mega3 program. In each tree, the bootstrap values calculated from 1000 replicates are indicated above each branch. The trees constructed with the maximum parsimony method are also available as Supplementary Data 6.
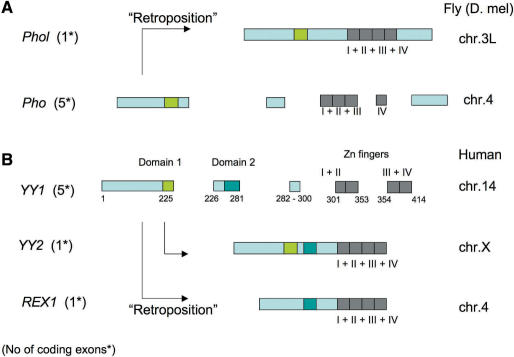
Figure 3.
Exon structures of (A) PHO and PHOL and (B) YY1, YY2 and REX1. The protein coding regions of PHO and YY1 both are split into five different exons depicted by boxes. Three conserved domains are marked by different colors: green for Domain I, blue for Domain II and gray for the zinc finger domain. This multi-exonic structure of both PHO and YY1 is conserved throughout all studied lineages. In contrast, the entire coding region of each PHOL, YY2 and REX1 is localized within one exon, suggesting the retroposition-driven formation of these duplicates in both the fly and placental mammal lineages. This retroposition-mediated duplication also resulted in the different chromosomal positions among these duplicates as shown in the right column.
The two copies of YY1 sequences found in the fish lineage show an almost identical sequence and exon structure to each other (data not shown). Chromosome-wide duplications are known to have been prevalent at the early stage of the fish genome evolution (29,30). Therefore, the two copies of YY1 present in each fish genome are thought to be another outcome of this chromosome-wide duplication event. In contrast, the two additional copies in placental mammals, YY2 and REX1, show quite different evolution patterns. First, like PHOL, the coding regions of both YY2 and REX1 are also located within one exon while the YY1 genes of all vertebrates show a very similar split exon structure with five coding exons (Figure 3B). This suggests that both YY2 and REX1 were also duplicated from YY1 by retroposition. The detection of YY2 and REX1 exclusively in placental mammals further suggests relatively recent formation of these two copies during mammalian evolution with the estimated time being about 60–100 million years ago. In mammals, both YY2 and REX1 are transcribed and maintain their Open Reading Frames (ORFs), confirming the functionality of these two retroposed copies. Second, despite this recent origin, inter-species sequence divergence levels of YY2 and REX1 are much greater than those of YY1, as reflected on the phylogenetic tree shown in Figure 2B. Very low levels of sequence divergence are observed between all the YY1 sequences of different vertebrates whereas each sequence from the YY2 and REX1 groups exhibits average 20% divergence between different species. This indicates relaxation of evolutionary constraints on both the YY2 and REX1 genes. As compared to REX1, YY2 displays greater levels of similarity to YY1 in terms of its overall sequence and protein domain structure, suggesting that the retroposition of YY2 may have occurred in more recent times than that of REX1. Pairwise sequence comparison also revealed that both YY2 and REX1 share higher sequence identity with YY1 than each other (Supplementary Data 3), suggesting that both REX1 and YY2 have been independently derived from YY1. The presence of two conserved domains, Domains I and II, in YY2 also supports the idea that YY2 has been derived from YY1, not from REX1, since REX1 has only Domain I. Overall, exon structure and sequence conservation levels suggest that the two retroposed copies, YY2 and REX1, have been under different levels of functional constraints than the original gene, YY1.
Different selection pressures on the DNA-binding domains of YY1, YY2 and REX1
All the YY1-related sequences show very unusual levels of sequence conservation in the DNA-binding domain of the predicted proteins (Figure 4). The zinc finger domains of PHO and PHOL from all of the different fly species share 5 and 18 amino acid differences, respectively, as compared to those of vertebrate YY1. The zinc finger domains of the other flying insects, however, show an almost identical sequence to those of vertebrate YY1. Thus, the observed amino acid differences in flies represent the substitutions that had occurred in the fly lineage. Apparently, the overall consensus sequence of flying insects’ PHO is still identical to that of vertebrate YY1. Similarly, the zinc finger domains of vertebrates’ YY1 also do not show any shared substitution except for one or two species-specific amino acid changes. Thus, YY1 is believed to have maintained its DNA-binding domain without any amino acid changes in the past 600 million year period, representing one of the most extreme cases for functional selection imposed on an eukaryote gene.
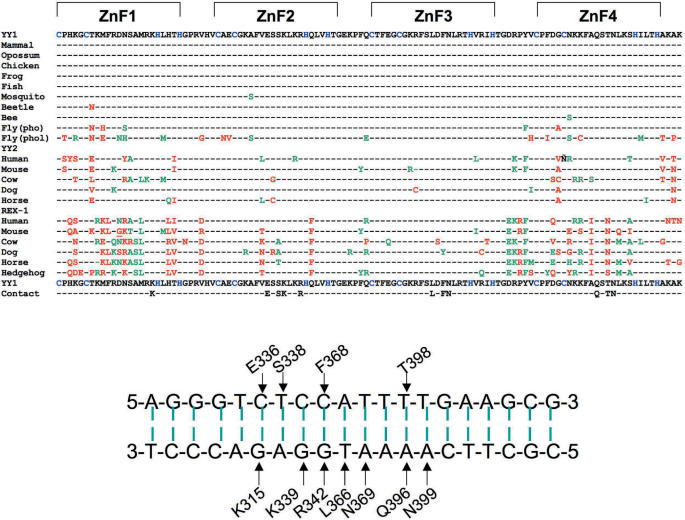
Figure 4.
Sequence alignment of the zinc finger domains of YY1-related sequences. The zinc finger domains of YY1-related sequences are compared with that of human YY1 (aa 298–411). The amino acid residues identical to human YY1 are indicated by dashes (–). The residues that differ from human YY1 are indicated by the single letter amino acid code with colors: green for conservative substitution and red for non-conservative substitution. Several entries underneath the YY1 sequence correspond to the sequences from different lineages. The entries in the middle and bottom of the alignment represent the YY2 and REX1 sequences, respectively. The amino acid residues known to contact directly with the bases of target DNAs are indicated on the last row. The duplex sequence of a known YY1 target DNA from the Adeno-Associated Virus (AAV) P5 promoter is shown along with contacting amino acid residues, which are indicated by the single letter code with the amino acid position information based on human YY1 sequence.
As described earlier, YY2 and REX1 have been under different levels of evolutionary constraints since their formation in placental mammals. This is in stark contrast to the extreme conservation of YY1. The zinc finger domains of different species’ YY2 protein show an average of 6–11 amino acid differences as compared to that of YY1 (Figure 4). None of these changes are shared among different mammals, indicating that these changes represent independent substitutions that occurred in each species. Similarly, the zinc finger domains of different species’ REX1 proteins also show an average of 11–20 amino acid differences between each other, implying a slightly higher level of relaxation of evolutionary constraint on REX1. As compared to vertebrate YY1, however, the zinc finger domains of all REX1 sequences share 8 amino acid substitutions (Figure 4). These substitutions represent the changes that occurred and were fixed before the radiation of eutherian mammals. The sudden fixation of these substitutions might be an evolutionary remnant suggesting positive selection that might have occurred in the early stages of REX1 evolution, although our analyses point toward purifying selection with relaxed constraints for the REX1 evolution (Supplementary Data 5). Interestingly, most of these changes are localized within Fingers 1 and 4, and are also non-conservative amino acid substitutions from the original amino acid residues of YY1. In particular, the amino acid change T398N in Finger 4 is localized within the region known to contact directly with the bases of target DNAs (31). Therefore, this change along with other amino acid substitutions in REX1 may have a functional outcome possibly allowing REX1 to bind to DNA motifs divergent from the YY1 DNA-binding motif. Similarly, the amino acid substitutions within YY2 also appear to be slightly more frequent in Fingers 1 and 4, suggesting the presence of different selection pressures on each zinc finger. However, none of YY2 changes appear to be located within critical regions for its DNA binding, predicting no major difference between the DNA binding motifs between YY1 and YY2.
DNA-binding motifs of YY1, YY2 and REX1
We have further investigated the functional consequences of different selection pressures imposed on the zinc finger domains of YY1, YY2 and REX1 by characterizing their DNA-binding motifs. For this experiment, the zinc finger domain of each protein was subcloned into the downstream region of the GST protein, expressed as part of a fusion protein in bacteria, fixed on agarose beads, and finally we allowed them to bind to duplex DNAs derived from randomized oligonucleotide sequences (4n=15). After five rounds of selection, the bound DNAs were subcloned and sequenced (Figure 5). In the case of YY1, 20 of 34 bound DNAs contain DNA motifs that have either a perfect match or 1 base difference from the known YY1 consensus sequence. All of the remaining 14 bound DNAs still show an almost identical sequence as YY1 but have an average of two base differences from YY1. Our approach used only the zinc finger domain of YY1, but most of the bound DNAs are identical to the known consensus sequence of YY1. This confirms the modular nature of the zinc finger domain of YY1, and subsequently the feasibility of this approach.
Figure 5.
DNA-binding motifs of YY1, YY2 and REX1. The sequences of DNAs bound by YY1 (left), YY2 (middle) and REX1 (right) are shown with the clone numbers on the right. The uppercase sequences are derived from the randomized portion of the input DNAs for binding whereas the lowercase dinucleotides represent the surrounding, fixed portion of the input DNAs. The majority of the DNAs bound by both YY1 and YY2 contain the known YY1 consensus motif (CGCCAT.TT), which is marked blue in the forward direction and by red in the reverse direction. The DNAs bound by REX1 are divided into two groups: one group indicated by blue and the other by bold-type. The total number of analyzed DNA molecules for each individual protein is indicated inside the parenthesis. For YY1 and YY2, the first number corresponds to the number of bound DNAs with either a perfect match or one base difference, while the second number to bound DNAs with more than two base differences.
In the case of YY2, 16 of 46 sequences contain DNA motifs similar to the YY1 consensus sequence. As with the YY1 fusion protein, the remaining sequences also contain a motif similar to YY1 with two base differences, confirming our initial prediction: there is no major difference between YY1 and YY2 motifs. Interestingly, however, most of the YY2-bound sequences have more than two binding motifs within the randomized portion of each sequence. About half of the bound sequences show two motifs in an opposite orientation, with the other half in the same orientation. In contrast to YY2, the DNAs bound by REX1 seem to be slightly different from those bound by either YY1 or YY2. The sequences bound by REX1 can be divided into two groups. These two groups can be represented by two slightly different consensus sequences (Figure 5): Type 1 (5′-GGCAGCCATTA-3′) and Type 2 (5′-GGCCATTA-3′). The consensus sequences of these two groups differ by the presence (or absence) of three bases (GGC) at the 5′-side. These two consensus sequences also show one unique difference at their 3′-side final position: all the DNAs bound by REX1 contain A instead of T. This is consistent with the amino acid change detected in the critical DNA binding region of REX1, T388N in Figure 4. Despite these changes, the core sequences of the YY2 and REX1 binding motifs are still the same as that of YY1 (5′-CCAT-3′), suggesting that the conservation of the two fingers, Fingers 2 and 3, may be responsible for maintaining a similar core target motif among these three genes.
Gel shift assays of DNA-binding motifs of YY1, YY2 and REX1
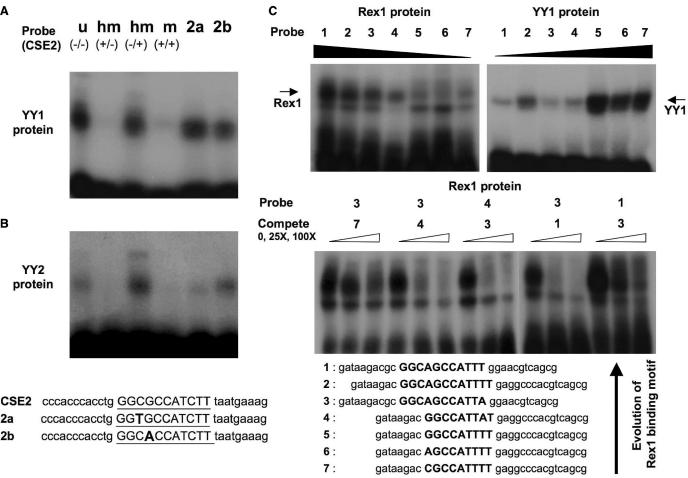
The DNA-binding motifs of YY1, YY2 and REX1 were further analyzed using gel shift assays (Figure 6). In the case of YY1, we have used the same set of duplex oligonucleotides used in a previous study to demonstrate the subtle but unique property of YY1, methylation-sensitive DNA-binding (5). As expected, the DNA-binding domain, as part of the GST-YY1 fusion protein, showed an almost identical pattern of DNA binding as endogenous YY1 protein (Figure 6A). The GST-YY1 protein is methylation-sensitive: methylation on the upper strand is inhibitory to the binding (Figure 6A, Lanes 1–4). One base change in this CpG site, either CpA or TpG, somewhat reduced the affinity of the YY1 binding, but still allowed YY1 binding to these probes (Figure 6A, Lanes 5–6). The DNA-binding domain of YY2 also showed a similar pattern of DNA binding: methylation-sensitive binding and subtle effects by single base changes caused by the CpG site (Figure 6B). However, the DNA-binding affinity of YY2 is much weaker than YY1 based on the results derived from our control experiments for gel shift assays (Supplementary Data 4). We have also tested some of the DNAs that contain two motifs within the randomized portion of the target DNAs (Figure 5B). We did not observe any difference in binding between the duplex DNAs with two binding motifs versus single binding motif (data not shown). Overall, the DNA-binding patterns of YY1 and YY2 appear to be similar except for the fact that the binding affinity of YY2 is much weaker than YY1, consistent with the observed relaxation of evolutionary constraint on the DNA-binding domain of YY2.
Figure 6.
Gel shift assays of DNA-binding motifs of (A) YY1, (B) YY2 and (C) REX1. Identified DNA-binding motifs were further confirmed with gel shift assays using three fusion proteins. For the gel shift assays of the YY1 and YY2 fusion proteins, we have used a set of the six different duplex probes that have been previously used for testing the methylation-sensitive DNA-binding activity of endogenous YY1 (5). Four different probes have an identical sequence as the CSE2 probe containing one YY1 binding site indicated by an underline. However, their methylation status at the CpG site is different: u (−/−), unmethylated on both strands; hm (+/−), methylated on the upper strand; hm (−/+), methylated on the bottom strand; and m (+/+), methylated on both strands. For the DNA-binding motif studies of (C) REX1, we have used seven probes: the YY1 consensus motif probe (Probe 7), and two YY1-related probes with one base difference (Probe 6&5), the Type 2 and 1 motifs of REX1 (Probe 4&3), and two variants of the Type 2 motif (Probe 2&1). The REX1 and YY1 proteins were individually used for the left and right sets of gel shift assays, respectively (Upper panel). Three representative probes were also used for competition assays (Lower panel). One minor band below the REX1 protein is from non-specific binding by an unidentified Escherichia coli protein in crude extracts. The sequences of these probes are shown on the bottom, and the relevant binding motifs within these sequences are bold-typed.
Several sets of gel shift assays were performed for the identified DNA-binding motifs of REX1 (Upper panel in Figure 6C). The REX1 and YY1 fusion proteins were individually allowed to bind to seven duplex probes. These include three consensus motifs, the consensus of YY1 (Probe 7), the consensus of REX1 Type 1 (Probe 3) and Type 2 (Probe 4). We have also included four other probes containing one or two base variations from the three consensus motifs to further dissect the binding specificity of REX1 and YY1. The REX1 protein bound to the four probes containing REX1 motifs (Probes 1–4), but not to the YY1 or related probes (Probes 5–7). On the other hand, the binding of the YY1 protein to the REX1 probes was detected but very marginal compared to its binding to the YY1 or related probes (Probes 5–7). This indicates the different binding specificity between the YY1 and REX1 proteins. This different binding specificity is originated from three key differences found in the REX1 binding motifs as compared to the YY1 binding motifs. First, the REX1 motifs have A instead of T at the 8th position of the YY1 consensus (CGCCATNTT). This change reduced dramatically the binding affinity of the YY1 protein, but increased the binding affinity of the REX1 protein (Probe 4 versus Probe 5). Second, the REX1 motifs do not show any base preference at the 9th position of the YY1 consensus (CGCCATNTT). Interestingly, the T base at this position reduced slightly the binding affinity of the REX1 protein, but is required for the binding of the YY1 protein (Probe 1 versus Probe 2). Third, one of the REX1 motifs contains additional three bases (5′-GGC-3′) at the 5′-side of its sequence. The addition of these three bases reduced the binding affinity of the YY1 protein, but increased the affinity of the REX1 protein (Probe 2 versus Probe 6). The significance of these key differences was further demonstrated by competition assays using three representative probes (Lower panel in Figure 6C). Overall, these data clearly demonstrate the different binding specificity between the YY1 and REX1 proteins, and also prove that the two identified motifs, Types 1 and 2, represent bona fide DNA-binding motifs for REX1. The positions of these three critical base differences in the surrounding regions of the core motif (5′-CCAT-3′) are consistent with an observed evolution pattern (Figure 4), differential selection pressures on each of the four zinc finger units of the REX1 protein.
DISCUSSION
In the current study, we have analyzed all the YY1-related sequences identified from genome sequences of invertebrates and vertebrates. We have identified two other protein domains, besides the zinc finger domain, that are conserved throughout all the YY1 and YY1-related sequences. Our analyses also confirmed that independent retroposition events have been responsible for forming duplicated copies, such as PHOL from PHO in flies, and YY2 and REX1 from YY1 in placental mammals. The zinc finger domains of YY2 and REX1 have been under different selection pressures than YY1, and consequently their DNA-binding properties have evolved from those of YY1 by weakening DNA-binding affinity in YY2 and REX1, and changing DNA-binding motifs in REX1. The evolution patterns of YY1 and other YY1-related proteins appear to be unique in several regards, as discussed subsequently.
Besides the zinc finger domain, two other protein domains, Domains I and II, are evolutionarily well conserved throughout all the YY1-related sequences ranging from flying insects to mammals (Figure 1). The conservation of these two domains is somewhat less obvious within the sequences of flies, but the detection of these domains within the PHO sequences of honeybees and beetles undoubtedly indicates that these two domains are part of the original domains of YY1. Database searches with these two domains did not find any proteins other than YY1 or YY1-related sequences, suggesting that these two domains are unique to YY1-related sequences (data not shown). According to previous studies analyzing protein–protein interactions, the Spacer region, a relatively large region of YY1 (aa 201–298 in human YY1) encompassing these two domains, is responsible for the interaction with the viral oncoprotein E1A and the p53-interacting partner Hdm2 (11,32). It should be interesting to test whether YY2 and REX1 also interact with the above two proteins. Nevertheless, the functional roles played by these two domains are predicted to be essential for YY1 functions based on their conservation in most of the YY1-related sequences.
There are several key transcription factors with similar evolutionary ages as YY1, such as Sp1 and the E2F family of proteins. These transcription factors have increased their gene copy numbers along with the increase of complexity and genome size of animals (33,34), but the duplication of these genes has been mainly driven by DNA-mediated mechanisms involving the entire genomic fragments surrounding individual genes (35,36). That is, in the Sp1 and E2F families, the whole gene structure has been duplicated with exons, introns and promoters intact. In the case of YY1, however, retroposition has been the primary mechanism for its duplication: PHOL duplication from PHO, and YY2 and REX1 duplications from YY1 (Figure 3), which is quite different from the general duplication mode observed in other key transcription factors. A gene copy duplicated through retroposition is subject to transcriptional controls different from those of its original gene due to its random insertions at other genomic regions. As an outcome, the duplicate copy tends to show different expression patterns compared with its original gene. Consistently, both YY2 and REX1 also display expression patterns quite different from that of YY1. As compared to the ubiquitous expression patterns of YY1, YY2 shows more germ cell-specific expression patterns (16), and REX1 exhibits stem cell-specific expression (20). It is still puzzling why YY1 duplication has been driven by retroposition, but different expression patterns resulting from this duplication mode may have been one major factor contributing to the success of YY1 duplications in placental mammals.
The evolutionary patterns observed with YY2 and REX1 are quite different from that of YY1 (Figure 4). YY1 shows high levels of sequence conservation throughout its coding region. In particular, the zinc finger domain of YY1 has maintained its amino acid sequence without any changes in the past 600-million year period, implying that the YY1 homologs, insect PHO and vertebrate YY1, may still bind to similar DNA motifs. This turns out to be the case based on DNA-binding motif studies (12,13). In contrast, the zinc finger domains of YY2 and REX1 show much higher levels of inter-species sequence divergence, suggesting relaxed constraints on their DNA-binding domains. Consequently, both YY2 and REX1 display much weaker DNA-binding affinity than YY1 (Figure 6 and Supplementary Data 4). The loosened DNA-binding affinities of YY2 and REX1 may have allowed these duplicates to bind to slightly different binding motifs, as seen in REX1 (Figure 5), and subsequently to bind to new sets of downstream genes. Together different expression patterns, loosened affinities and different DNA-binding motifs may have contributed to the functional diversification of the two duplicates, YY2 and REX1, in the mammalian lineage.
Successful gene duplication is still regarded as a rare evolutionary event (37), which is further supported by the single-copy status of YY1 in the majority of animal lineages. Then, what could be the main reason(s) underlying the sudden formation of two YY1 duplicates in placental mammals? This may be indirectly answered by observations drawn from other gene duplicates in mammals. For instance, DNMT3L is a member of the DNA methyltransferase family, which is found only in mammals (38). Yet, DNMT3L has been found to be involved in genomic imprinting (39), a gene dosage control mechanism unique to placental mammals (40). CTCFL (or BORIS), a mammal-specific duplicate of the vertebrate insulator protein CTCF (41), might be involved in establishing the gametic imprinting mark of DNA methylation for H19 during germ cell development (42). In both cases, gene duplicates appear to play specific roles in mammal lineage-specific novelties, such as genomic imprinting and epigenetic modification. These two duplicates, interestingly, share some similarities with the YY1 duplicates, YY2 and REX1, such as recent formation, rapid evolution, lineage-specific conservation in mammals and germ cell-specific expression (43). Furthermore, recent studies suggest that several imprinted domains may be controlled by YY1 or related transcription factors (6,7). This entices the speculation that both YY2 and REX1 may be also involved in novel placental mammal-specific functions, such as genomic imprinting. This idea needs to be tested, but the evolutionary patterns presented in this study clearly indicate the tight linkage of both YY2 and REX1 to the biology of placental mammals.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Lisa Stubbs for helpful discussions and Jennifer M. Huang for providing help in cloning and sequencing. This study was supported by NIH grant GM66225 (to J.K.). Funding to pay the Open Access publication charges for this article was provided by NIH grant GM66225.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shi Y, Lee JS, Galvin KM. Everything you ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta. 1997;1334:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 2.Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 3.Gordon S, Akoryan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 4.Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ., Jr Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J, Kollhoff A, Bergmann A, Stubbs L. Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene. Peg3. Hum. Mol. Genet. 2003;12:233–245. doi: 10.1093/hmg/ddg028. [DOI] [PubMed] [Google Scholar]
- 6.Kim JD, Hinz AK, Bergmann A, Thompson JM, Ovcharenko I, Stubbs L, Kim J. Identification of clustered YY1 binding sites in imprinting control regions. Genome Res. 2006;16:901–911. doi: 10.1101/gr.5091406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JD, Hinz AK, Choo JH, Stubbs L, Kim J. YY1 as a controlling factor for the Peg3 and Gnas imprinted domains. Genomics. 2007;89:262–269. doi: 10.1016/j.ygeno.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Affar EB, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satijn DPE, Hamer KM, Blaawen JD, Otte AP. The polycomb group protein EED interact with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cel. Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sui G, Affar EB, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 13.Brown JL, Fritsch C, Mueller J, Kassis JA. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;30:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan L, Pan X, Atchison ML. Transient requirements of YY1 expression for PcG transcriptional repression and phenotypic rescue. J. Cell. Biochem. 2005;96:689–699. doi: 10.1002/jcb.20562. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen N, Zhang X, Olashaw N, Seto E. Molecular cloning and functional characterization of the transcription factor YY2. J. Biol. Chem. 2004;279:25927–25934. doi: 10.1074/jbc.M402525200. [DOI] [PubMed] [Google Scholar]
- 16.Luo C, Lu X, Stubbs L, Kim J. Rapid evolution of a recently retroposed transcription factor YY2 in mammalian genomes. Genomics. 2006;87:348–355. doi: 10.1016/j.ygeno.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Hosler BA, LaRosa GJ, Grippo JF, Gudas LJ. Expression of REX-1, a gene containing zinc finger motifs, is rapidly reduced by retinoic acid in F9 teratocarcinoma cells. Mol. Cell. Biol. 1989;9:5623–5629. doi: 10.1128/mcb.9.12.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosler BA, Rogers MB, Kozak CA, Gudas LJ. An octamer motif contributes to the expression of the retinoic acid-regulated zinc finger gene Rex-1 (Zfp-42) in F9 teratocarcinoma cells. Mol. Cell. Biol. 1993;13:2919–2928. doi: 10.1128/mcb.13.5.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Shushan E, Thompson JR, Gudas LJ, Bergman Y. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol. 1998;18:1866–1878. doi: 10.1128/mcb.18.4.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mongan NP, Martin KM, Gudas LJ. The putative human stem cell marker, Rex-1 (Zfp42): structural classification and expression in normal human epithelial and carcinoma cell cultures. Mol. Carcinog. 2006;45:887–900. doi: 10.1002/mc.20186. [DOI] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DJ, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 26.Hyde-DeRuyscher RP, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yant SR, Zhu W, Millinoff D, Slightom JL, Goodman M, Gumucio DL. High affinity YY1 binding motifs: identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brosius J. The contribution of RNAs and retroposition to evolutionary novelties. Genetica. 2003;118:99–116. [PubMed] [Google Scholar]
- 29.Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christoffels A, Koh EG, Chia JM, Brenner S, Aparicio S, Venkatesh B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol. 2004;21:1146–1151. doi: 10.1093/molbev/msh114. [DOI] [PubMed] [Google Scholar]
- 31.Houbaviy HB, Usheva A, Shenk T, Burley SK. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl Acad. Sci. USA. 1996;93:13577–13582. doi: 10.1073/pnas.93.24.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 33.Carroll SB, Grenier JK, Weatherbee SD. Malden, MA: Blackwell Science; 2001. From DNA to diversity-Molecular genetics and the evolution of animal design. [Google Scholar]
- 34.Davidson EH. San Diego, CA: Academic Press; 2001. Genomic regulatory systems: development and evolution. [Google Scholar]
- 35.Dynlacht BD, Brook A, Dembski M, Yenush L, Dyson N. DNA-binding and trans-activation properties of Drosophila E2F and DP proteins. Proc. Natl Acad. Sci. USA. 1994;91:6359–6363. doi: 10.1073/pnas.91.14.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondrashov FA, Kondrashov AS. Role of selection in fixation of gene duplications. J. Theor. Biol. 2006;239:141–151. doi: 10.1016/j.jtbi.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 38.Yokomine T, Hata K, Tsudzuki M, Sasaki H. Evolution of the vertebrate DNMT3 gene family: a possible link between existence of DNMT3L and genomic imprinting. Cytogenet. Genome Res. 2006;113:75–80. doi: 10.1159/000090817. [DOI] [PubMed] [Google Scholar]
- 39.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 40.Reik W, Lewis A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat. Rev. Genet. 2005;6:403–410. doi: 10.1038/nrg1602. [DOI] [PubMed] [Google Scholar]
- 41.Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc. Natl Acad. Sci. USA. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bestor TH, Bourc'his D. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb. Symp. Quant. Biol. 2004;69:381–387. doi: 10.1101/sqb.2004.69.381. [DOI] [PubMed] [Google Scholar]