Abstract
Friedreich ataxia (FRDA), the most common hereditary ataxia, is caused by mutations in the frataxin (FXN) gene. The vast majority of FRDA mutations involve expansion of a GAA•TTC-repeat tract in intron 1, which leads to an FXN mRNA deficit. Bisulfite mapping demonstrates that the region adjacent to the repeat was methylated in both unaffected and affected individuals. However, methylation was more extensive in patients. Additionally, three residues were almost completely methylation-free in unaffected individuals but almost always methylated in those with FRDA. One of these residues is located within an E-box whose deletion caused a significant drop in promoter activity in reporter assays. Elevated levels of histone H3 dimethylated on lysine 9 were seen in FRDA cells consistent with a more repressive chromatin organization. Such chromatin is known to reduce transcription elongation. This may be one way in which the expanded repeats contribute to the frataxin deficit in FRDA. Our data also suggest that repeat-mediated chromatin changes may also affect transcription initiation by blocking binding of factors that increase frataxin promoter activity. Our results also raise the possibility that the repeat-mediated increases in DNA methylation in the FXN gene in FRDA patients are secondary to the chromatin changes.
INTRODUCTION
Friedreich ataxia (FRDA) is the most common inherited ataxia (1). The ataxia, which is recessively inherited, is relentlessly progressive with patients frequently becoming wheelchair-bound in their early teens. Hypertrophic cardiomyopathy is a common co-morbid feature of FRDA, with congestive heart failure being a frequent cause of death. The most common cause of FRDA is the expansion of a GAA•TTC-repeat (henceforth referred to as the FXN repeats) in intron 1 of the frataxin gene (FXN). Unaffected individuals have at least one allele with 8–33 repeats, while most individuals with FRDA have 90 or more repeats in both alleles (1). These expanded alleles produce less mature FXN mRNA than alleles in the normal range (2,3). This results in a deficiency of frataxin, an essential protein thought to be involved in mitochondrial iron metabolism (1).
The FXN repeats form an intrinsic block to RNA polymerase in vitro (4,5), suggesting one potential mechanism for the FXN mRNA deficit. However, the FXN repeats may affect transcription in other ways. For example, the long stretches of tandem repeats present in normal human centromeres are associated with transcriptionally silent chromatin (6) as are the genes containing the expanded triplet repeat tracts responsible for Fragile X syndrome and congenital myotonic dystrophy (7). In addition, transgenes containing long GAA•TTC-tracts become associated with heterochromatin when inserted into arbitrary locations in the mouse genome (8). It is thus possible that repeat-mediated chromatin changes also contribute to the reduction in FXN transcript in FRDA. In principle, changes in chromatin modifications in non-coding sequence could affect transcription by affecting RNA polymerase II elongation or by changing the accessibility of this region to regulatory factors important for transcription initiation (9). Since the first intron of many genes contains regulatory sequences important for gene expression (10–13) and the sequence adjacent to the FXN repeat is the region of intron 1 most likely to be affected by repeat expansion, we examined the contributions of this region to FXN promoter activity. We also examined the chromatin modifications and DNA methylation status of this region in cells from unaffected individuals and individuals with FRDA. We have identified a region that is important for maximal FXN gene expression and shown that repeat expansion leads to changes in DNA methylation and chromatin organization in this region. Our data also suggests how these changes may lead to the frataxin deficit responsible for FRDA.
MATERIALS AND METHODS
In silico analysis of intron 1
Primate sequences in the region of intron 1 immediately 5′ of the FXN repeat were aligned using the online version of the Multalin algorithm [(14); http://prodes.toulouse.inra.fr/multalin/multalin.html]. The repeated DNA elements in this region were identified using the Repeat Masker track generated using the RepBase database (15). This region was also analyzed for potential transcription factor binding sites using MatInspector and Matrix Family Library Version 4.1 (https://www.genomatix.de).
Cell lines and DNA
Lymphoblasts from individuals without FRDA (GM06895, GM06865, GM06891 and GM09145), and those with FRDA (GM04079, GM15850, GM16207, GM16209 and GM16243) were obtained from the Coriell Cell Repository (Camden, NJ). Lymphoblasts were grown in RPMI 1640 medium supplemented with 10% fetal calf serum under standard conditions (Invitrogen Life Technologies Inc., Carlsbad, CA). Genomic DNA was prepared from these cell lines using standard procedures. The FXN repeat numbers in normal individuals were determined as described earlier (3,16). DNA obtained from fresh blood of affected and unaffected individuals was a kind gift of Dr Ed Grabczyk, Louisiana State University. The plasmid construct expressing the M.eco72I methylase was a kind gift of Fermentas AB (Vilnius, Lithuania). The primers and double-stranded oligonucleotides used were synthesized by Integrated DNA Technologies (Coralville, IA) and are listed in Table 1.
Table 1.
Oligonucleotides used in these experiments
| Primers | |
| FXNMe74-F | 5′-TTTTTCTCGAGAATAAGGTATAGAGAGGTTAATTAATTTGTTTTTTGGTTATA-3′ |
| FXNMe956-R | 5′-TTTTTGAATTCCCCTCCACCAACCTACCCTACCTTC-3′ |
| FXNMe1212-F | 5′-TTTTTCTCGAGGAGGTGAAATTTTTAGAGTTGTAGAATAGTTAGAGTAGTAG-3′ |
| FXNMe1930-R | 5′-TTTTTGAATTCACCTCCCAAAATACTAAAATTATAAACATAAACCA-3′ |
| FXNPrF | 5′-TTTTTTACGCGTGCTATGATGGCCGGGCTGGAATGCTGTGGCTATTCACAG-3′ |
| FXN-Luc-F1 | 5′-ACCGACATCGATGCGACCT-3′ |
| FXN-Luc-R2 | 5′-TTGCTCTCCAGCGGTTCCATCTT-3′ |
| FXNchip-QF1 | 5′-GCAAAGGCCAGGAAGGCGGA-3′ |
| FXNchip-QR1 | 5′-ATGGCTTGGACGTGGCCTGC-3′ |
| Double-stranded oligonucleotides (for EMSAs) | |
| E-boxcon | 5′-ATCTGTATCACGTGTGTAGTCGTGATG-3′ |
| FXN E-box | 5′-GAACTTCCCACACGTGTTATTTGGCCCACA-3′ |
| FXN E-boxΔ | 5′-ACACGTGTTATTTGGCCCAC-3′ |
DNA methylation analysis
Genomic DNA from cell lines of four unaffected and four affected individuals was bisulphite modified according to standard procedures except that the bisulphite treatment was carried out overnight at 55°C. The methylation status of the promoter and intron 1 were examined. The promoter was analyzed using the primers FXNMe74-F and FXNMe956-R. The region 5′ of the GAA repeat in the FXN gene was amplified by PCR using the primers FXNMe1212-F and FXNMe1930-R. The PCR fragments were gel purified, digested with XhoI and EcoRI, cloned into pBS SK+ (Stratagene, La Jolla, CA, USA) and sequenced. Five clones from each individual were sequenced. The data for each population was pooled and the frequency with which each residue was methylated was then plotted along with the standard deviation.
Reporter assays
The primer FXNPrF was used as the forward primer for all constructs. This primer has an MluI site at its 5′ end and primes 1118 bp upstream of the start of FXN translation. Each of the reverse primers contained the splice acceptor site from the 3′ end of intron 1 and an NcoI site and were designed so that the FXN coding sequence in exon 1 would be translated in frame with the luciferase open reading frame from pGL3-Basic (Promega, Madison, WI, USA). After PCR amplification, the gel-purified fragment was digested with MluI and NcoI and cloned into pGL3-Basic. The numbers in the construct names are based on the location of the reverse primer in the chromosome 9 sequence from the May 2004 freeze of the human genome in the UCSC database with the numbering starting at 68 800 000. Thus the 3′ end of the FXN sequence in construct FXN80940 corresponds to base 68 880 940 in the chromosome 9 sequence. FXN81423 contains 1006 bases from the 5′ end of intron 1 terminating at position 68 881 423, and FXN81658 contains the first 1241 bases of intron 1 and terminates at position 68 881 658. FXN E-box/Mt+ and FXN E-box/Mt− clones were generated using reverse primers that contain either the FXN intron E-box/Mt sequence or the sequence immediately downstream of this region. The resultant clones differ only by eight bases. The second C in the E-box core sequence 5′-CACGTG-3′ can be methylated by M.eco72I, a DNA methyltransferase from Escherichia coli RFL72 (17). Methylation was carried out as previously described (18), and the extent of methylation checked by digestion with PmlI, a restriction enzyme whose cleavage is blocked by M.eco72I methylation. Reporter assays were carried out using mouse C2C12 myoblasts as previously described (19). To assess any differences in the RNA stability of these constructs, cells were treated 24 h post-transfection with 5 µg/ml actinomycin D. RNA and protein samples were collected after 8 h. The luciferase protein levels were determined as previously described (19). The luciferase mRNA levels were determined by real-time PCR as described below. The RNA and protein values after 8 h of actinomycin treatment were calculated as the percentage of the values obtained at 0 h.
Electrophoretic mobility shift assays (EMSAs)
A double-stranded oligonucleotide FXN E-box was radiolabeled using [γ-P32] ATP (MP Biomedicals, Solon, OH) and T4 polynucleotide kinase (New England Biolabs, Beverly, MA, USA) according to standard procedures. Nuclear extracts were prepared from C2C12 cells using NE-PER Nuclear Extraction Reagents (Pierce, Rockford, IL, USA) according to the supplier's instructions. Binding was carried out at 20°C for 30 min in 30 μl of reaction buffer containing 5 µg of nuclear extract, 25 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM EDTA and 4 mM DTT with 0.005 pmol of labeled probe. A 1000-fold molar excess of oligonucleotides containing a consensus E-box, E-boxcon, the FXN E-box and FXN E-boxΔ were used as competitors in this assay. The samples were subjected to electrophoresis on 5% native polyacrylamide (60:1, acrylamide:bisacrylamide) gels.
Real-time RT–PCR
Total RNA was isolated from the cell lines using Trizol (Invitrogen Life Technologies Inc.) and reverse transcribed using SuperScript™ III RT First strand synthesis system for RT–PCR (Invitrogen Life Technologies Inc.), as per the manufacturer's instructions. Real-time PCRs for measuring the endogenous FXN, GUS and 18S RNA were carried out using an ABI 7500 FAST PCR system (Applied Biosystems, Foster City, CA, USA) and appropriate Taqman probe primer mixes (Applied Biosystems). Quantitation of luciferase mRNA was carried out using Power SYBR® Green PCR mix (Applied Biosystems) and the primer pair FXN-Luc-F1 and FXN-Luc-R2 which amplifies across the intron between the Frataxin exon 1 and the luciferase coding sequence.
Chromatin immunoprecipitation assays
The anti-dimethyl-Histone H3 (Lys9) (H3K9me2) antibody (Cat. no. 07-441) was purchased from Upstate (Charlottesville, VA, USA). The rabbit preimmune serum was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). The ChIP assay kit from Upstate was used according to the manufacturer's instructions with slight modifications. Briefly, cells were cross-linked in RPMI medium 1640 with 1% formaldehyde at 23°C for 5 min. The cells were washed in PBS and lysed in a buffer containing 1% SDS, 10 mM EDTA and 50 mM Tris–HCl pH 8.0. The chromatin from lysed cells was sonicated to lengths between 200 and 1000 bp using a Bioruptor sonicator (Diagenode, Philadelphia, PA, USA) and the cell debris was removed by centrifugation. The sonicated lysates were precleared and incubated with either preimmune serum or the anti-H3K9Me2 antibody at 4°C overnight. The immunocomplexes were recovered by Protein A agarose, washed and eluted in 1% SDS, 0.1 M NaHCO3 and the crosslinking reversed by incubation at 65°C for 4 h in 200 mM NaCl. The samples were phenol: chloroform extracted and ethanol precipitated in the presence of Pelletpaint (EMD Biosciences, San Diego, CA, USA). The amount of FXN DNA immunoprecipitated was determined using quantitative real-time PCR using Power SYBR® green PCR master mix (Applied Biosystems). The PCR primers used were FXNchip-QF1 and FXNchip-QR1. The ChIP experiments were performed in triplicate and each PCR reaction was done in duplicate. The immunoprecipitated DNA was normalized to the amount of input DNA and plotted as the fold enrichment over the preimmune serum (IgG).
RESULTS AND DISCUSSION
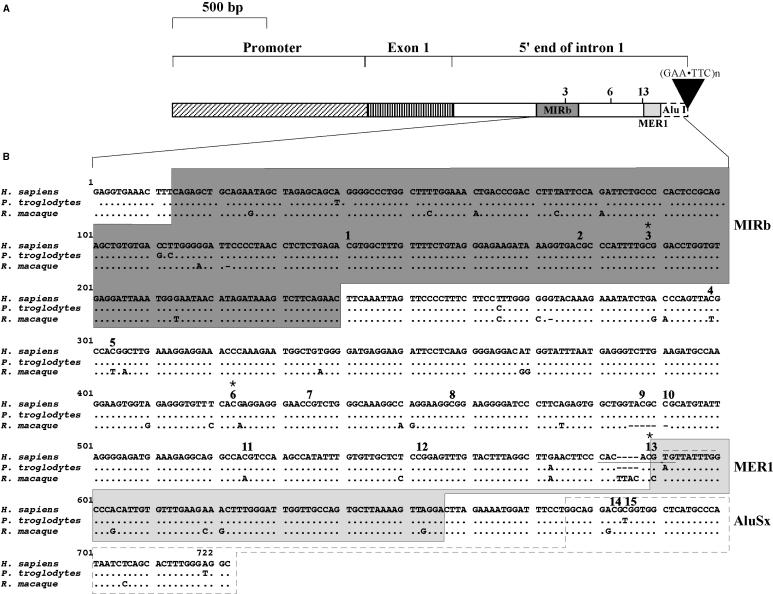
We have previously analyzed the region upstream of the transcription start site of the FXN gene and identified the regulatory elements important for FXN promoter activity (19). However, almost nothing was known about the region of intron 1 containing the FXN repeat. Since sequences important for promoter activity can be located in the first intron (10–13) and the sequences closest to the repeat are the ones most likely to be affected by repeat expansion, we set out to examine this region in some detail. The organization of the 5′ end of the FXN gene is shown in Figure 1A. Intron 1 is 10 436 bp long. The FXN repeat is located 1340 bp from the beginning of the intron. The region of intron 1 between exon 1 and the FXN repeat contains three interspersed repeated DNA elements. This includes a member of the ancient MIRb family of repeated DNA elements that is thought to have been active in transposition before the mammalian radiation, Charlie8, a member of the primate-specific MER1 (medium reiteration 1) family (20) and the first half of the Alu element from which the FXN repeat is thought to have been derived (21).
Figure 1.
The 5′ end of the frataxin gene. (A) Schematic representation of the 5′ end of the human FXN gene showing the location of the FXN repeat relative to the other repeated DNA elements and to the FXN promoter. The 5′ end of the region depicted here corresponds to base 68 878 851 of chromosome 9 in the 2004 build of the UCSC human DNA sequence database. The beginning of the FXN repeat corresponds to base 68 881 758 of this sequence. The numbers 3, 6 and 13 refer to those CpG residues that are protected from methylation in cells from unaffected individuals but that are methylated in FRDA cells. These residues correspond to the Cs marked with asterisks in panel B. (B) The sequence of region of intron 1 analyzed by bisulfite mapping showing an alignment of the human with chimpanzee and macaque FXN genes. The bold numbers 1–15 indicate the 15 methylatable C residues that were analyzed. The boxed sequences indicate interspersed repeated DNA elements. The dotted gray line above the sequence indicates the core muscle-specific Mt-binding site. The solid gray line below the sequence marks the core E-box site.
Intron 1 in affected and unaffected individuals show differences in DNA methylation
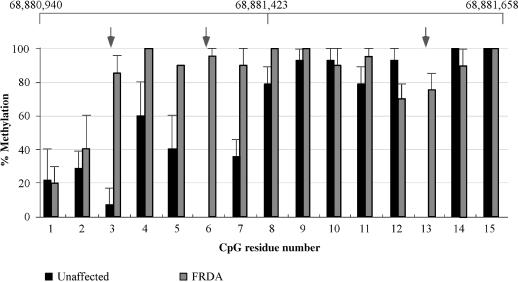
Since repeat-mediated epigenetic changes have been suggested to occur in regions containing expanded FXN repeats (8), we examined the methylation changes in the sequence immediately upstream of the FXN repeat. Bisulfite analysis demonstrates that a number of CpG residues were methylated in cell lines from both individuals with FRDA and unaffected individuals (Figure 2). However, the average methylation density was ∼50% higher in cells from affected individuals (compare the heights of the black and gray bars in Figure 2). This reflects, in part, the fact that the methylation density decreased more slowly 5′ of the repeat tract in these cells. The basal level of methylation seen in unaffected individuals may be due to methylation spreading from the downstream Alu element since these elements are themselves frequently methylated (22). In addition to an overall increase in methylation in FRDA cells, certain residues were much more likely to be methylated in these cells than in those without the disease. For example residues 3, 6, and 13 are methylated in 75–100% of alleles from patients, while they were completely free from methylation in unaffected individuals (Figure 2). DNA from fresh blood of four unrelated individuals with FRDA showed a similar methylation of residues 3, 6 and 13 to DNA (data not shown), suggesting that this methylation is not an artifact of the transformation process. Thus normal alleles seem to have a naturally occurring ‘methylation footprint’ that is absent from FRDA alleles. The elevated level of methylation seen in FRDA cells disappeared 5′ of residue 3 with overall levels of methylation in both populations falling to 20% at residue 1. Thus the repeat-mediated increase in DNA methylation is limited to the region immediately adjacent to the repeat. This conclusion is consistent with the lack of any DNA methylation in the promoter of either population group (data not shown).
Figure 2.
Methylation status of 15 CpG residues in the region 5′ of the FXN repeat in cells from unaffected and affected individuals. DNA from lymphoblastoid cell lines from four unaffected individuals and four individuals with FRDA was bisulphite modified and cloned as described in the Materials and Methods section. The numbers below the bars indicate the residue number using the same numbering convention used in Figure 1. The gray arrows indicate the three residues that are frequently methylated in affected individuals but that are very infrequently methylated, if at all, in the unaffected population.
Intron 1 contributes to FXN promoter activity
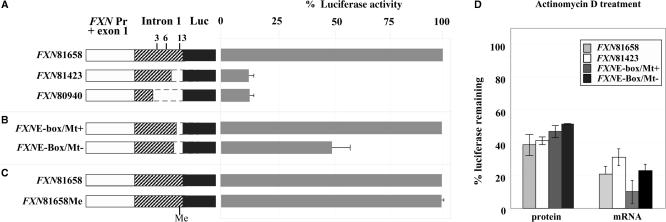
The methylation ‘footprints’ at residues 3, 6 and 13 in unaffected individuals could reflect either the existence of positioned nucleosomes or binding of sequence-specific DNA-binding proteins. If the latter were true, we might expect that deletion of these regions could affect FXN promoter activity in transient transfection assays. We therefore generated reporter constructs containing the promoter, exon 1, and various lengths of intron 1. Downstream of the intron 1 sequence in each construct was the branch and splice acceptor sites from the end of intron 1, and the luciferase coding sequence. The results of transfections of these constructs into mouse C2C12 myoblasts are shown in Figure 3A. The longest construct tested, FXN81658 contained the region of intron 1 from the end of exon 1 to just 5′ of the FXN repeat that encompasses all three protected residues. This construct gave the highest luciferase activities of all the constructs tested with an activity that was ∼45-fold above that of the empty vector. A construct FXN81423 that terminated 235 bases 5′ of the sequence in FXN81658, and therefore that lacked residue 13, had a luciferase activity that was only ∼10% that of the full length construct FXN81658 (Figure 3A). A construct FXN80940 that terminated 718 bases 5′ of FXN81423, and thus lacked residue 3 and 6 had an activity very similar to that of FXN81423. As can be seen from Figure 3D, treatment of FXN81658 and FXN81423 transfected cells with the transcription inhibitor Actinomycin D resulted in very similar levels of luciferase protein and mRNA after 8 h of treatment. This suggests that the absolute difference in luciferase activity between the two constructs reflects differences in transcription rates rather than differences in RNA stability. Thus, it is likely that the 235 bp region of intron 1 containing residue 13 has a binding site for one or more transcription factors that make a significant positive contribution to the FXN promoter activity.
Figure 3.
The effect of different lengths of intron 1 sequence on the activity of the FXN promoter. (A–C). Luciferase activities of constructs containing different amounts of intron 1. Constructs containing the promoter, exon 1 and different lengths of intron 1 sequence were transfected into mouse myoblasts as described in the Materials and Methods section. The dotted lines demarcate deleted regions. The suffix ‘Me’ in the construct FXN81658Me indicates that the FXN81658 construct was specifically methylated at the E-box/Mt-binding site. The luciferase levels from the FXN reporter constructs were normalized to luciferase levels from the pRLNull vector and expressed as a percentage of the normalized luciferase activity of the full-length construct. The data are an average of three independent transfection experiments. (D) Actinomycin D treatment of cells transfected with the FXN81658, FXN81423, FXNE-box/Mt+ and FXNE-box/Mt- constructs. The values for the luciferase protein and mRNA after exposure to Actinomycin D for 8 h are shown expressed as a percentage of the values at 0 h. The data shown represent the average of three independent experiments.
Residue 13 is located within a putative E-box. This site overlaps with a binding site for the muscle specific Mt transcription factor. We generated two constructs differing only in eight bases. The first construct included the E-box/Mt-binding site and the second terminated just 5′ of the E-box and was thus missing both sites. We measured the activity of these constructs in C2C12 cells. The construct lacking the E-box/Mt-binding site had a significantly lower luciferase activity than the construct containing these regions (Figure 3B). As can be seen from Figure 3D, Actinomycin D treatment had a similar effect on the yield of luciferase RNA and protein of both of these constructs. These findings support the idea that the original differences in luciferase activity we observe without Actinomycin D reflects a higher promoter activity for the construct with the E-box/Mt-binding site rather than enhanced mRNA stability.
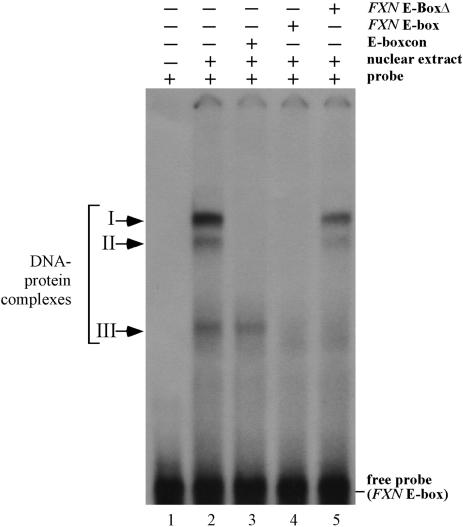
The E-box/Mt region binds transcription factors in extracts from muscle cells
Three protein–DNA complexes were found in electrophoretic mobility shift assays with C2C12 cells using the E-box/Mt region as a probe (complexes I, II and III in Figure 4, lane 2). Complexes I and II could be competed out by addition of E-boxcon, an oligonucleotide that contains a consensus E-box-binding site. Thus members of a large family of basic Helix-Loop-Helix E-box-binding proteins that includes MyoD and c-myc have the potential to bind this region. Complex III could be competed out by an oligonucleotide that contains a truncated E-box and the intact Mt-binding site (FXN E-boxΔ, F Figure 4, lane 5). Complex III thus probably represents the complex formed by a protein bound at the Mt-binding site. Thus, a number of proteins have the potential to bind to this region in vivo.
Figure 4.
Electrophoretic gel mobility shift analysis (EMSA) of proteins binding to FXN intron 1. EMSA was carried in the presence of molar excesses of E-boxcon, a canonical E-box sequence, FXN E-box which contains the FXN E-box together with the muscle-specific Mt-binding site, and FXN E-BoxΔ which contains the 3′ end of the E-box together with the full Mt-binding site.
Specific methylation of residue 13 does not affect FXN promoter activity
The methylation of the E-box in FRDA patients could lead to a FXN deficit via a direct effect on the binding of one or more DNA-binding proteins. M.eco72I from E. coli RFL72 is a DNA methyltransferase that specifically methylates the CpG residue in the sequence 5′-CACGTG-3′ (17). Thus the FXN E-box is a substrate for this enzyme and the site it methylates corresponds to residue 13. Since there are no other sites that can be methylated by this enzyme in the reporter construct, this methylase can be used to examine the effect of methylation of this residue specifically. Methylation had no effect on the activity of a reporter construct transfected into C2C12 cells (Figure 3C). Thus methylation of residue 13 per se is unlikely to be the primary cause of the FXN deficiency.
Affected individuals have elevated levels of H3K9Me2
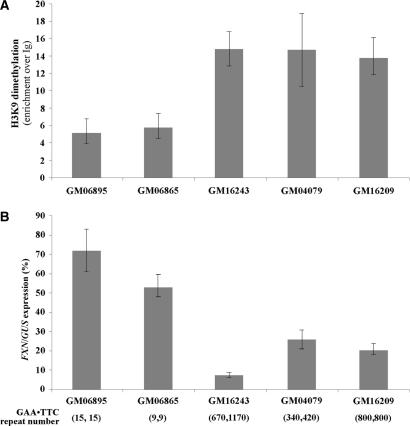
Increased methylation of residue 13 may be a secondary consequence of a more compact chromatin organization which blocks factor binding. Since chromatin compaction is associated with elevated levels of H3K9Me2, we analyzed intron 1 in four FRDA patients for the presence of this histone modification. Elevated levels of H3K9Me2 were seen in these individuals (Figure 5A), all of whom had FXN mRNA levels 10–30% that of unaffected individuals (Figure 5B). While this manuscript was in preparation a similar observation was reported for another lymphoblastoid cell line from a FRDA patient (23). The increase in H3K9Me2 is consistent with the fact that treatment of FRDA lymphocytes with histone deacetylase inhibitors increases FXN transcription significantly (23) since H3K9 acetylation blocks K9 methylation. Our data thus support the idea that a chromatin-based mechanism is responsible for the FXN mRNA deficit in FRDA (23).
Figure 5.
FXN histone H3 K9 dimethylation and mRNA levels in unaffected and affected individuals. (A) The amount of FXN DNA immunoprecipitated by antibodies to histone H3-K9Me2 in each lymphoblastoid cell line was determined by real-time PCR as described in the Materials and Methods section. The data shown is an average of three independent ChIP experiments. In all instances P < 0.002. When the data from all the patient samples was combined and compared to the combined data from the unaffected individuals P = 0.0001. (B) FXN RNA levels were determined in the indicated cell lines using real-time PCR and expressed relative to the amount of GUS mRNA in the same cells.
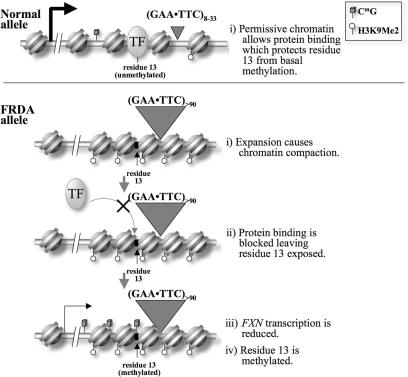
The altered chromatin conformation may decrease transcription by impeding transcription elongation (9). Our data also suggests another way in which the chromatin modification at the FXN gene can lead to the mRNA deficit. In the absence of an expanded repeat tract, intron 1 binds proteins that positively affect FXN transcription. A secondary effect of this binding is that it creates natural ‘methylation footprints’ by protecting the binding sites from DNA methylation (Figure 6). The more compact chromatin formed on alleles with large FXN repeats blocks access of these proteins to the DNA as evidenced by disappearance of the ‘methylation footprints’. Absence of binding of these transcription factors may lead to a reduction in transcription initiation. One implication of this model is that DNA methylation may be a secondary consequence of the histone modifications. This may explain why histone deacetylase inhibitors are so effective at increasing FXN gene expression in FRDA cells (23).
Figure 6.
Model for chromatin organization in the region containing residue 13 in normal and FRDA alleles.
How could the long FXN-repeat tracts in patient cells generate transcriptionally silent chromatin? One possibility is that this process involves small double-stranded RNAs (dsRNAs) that target the transcriptional silencing machinery to homologous regions of the genome (24). Potential sources of these dsRNAs are regions of transcripts that can form hairpins (25) or overlapping sense and antisense transcripts (26). Long GAA-repeat tracts form DNA hairpins (27). This raises the possibility that RNA containing these repeats may also do so. However, since a low level of modified chromatin exists at the frataxin locus even when the repeat number is too small to form stable hairpins, it is tempting to speculate that an antisense mechanism is involved at least when the repeat number is low.
Repeat-mediated heterochromatin formation is seen in two other Repeat Expansion diseases: Fragile X syndrome and the congenital form of Myotonic Dystrophy type 1 (7). Thus repeat-mediated transcriptional silencing may provide a common thread linking these diseases where the repeat is transcribed but not translated.
ACKNOWLEDGEMENTS
The authors thank Ann Dean (NIDDK) for her thoughtful comments and advice. This work was supported by a grant from the Intramural Program of the NIDDK (NIH). Funding to pay the Open Access publication charges for this article was provided by the Intramural Program of the NIDDK (NIH).
Conflict of interest. None declared.
REFERENCES
- 1.Pandolfo M. The molecular basis of Friedreich ataxia. Adv. Exp. Med. Biol. 2002;516:99–118. doi: 10.1007/978-1-4615-0117-6_5. [DOI] [PubMed] [Google Scholar]
- 2.Pianese L, Turano M, Lo Casale MS, De Biase I, Giacchetti M, Monticelli A, Criscuolo C, Filla A, Cocozza S. Real time PCR quantification of frataxin mRNA in the peripheral blood leucocytes of Friedreich ataxia patients and carriers. J. Neurol. Neurosurg. Psychiatry. 2004;75:1061–1063. doi: 10.1136/jnnp.2003.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 4.Grabczyk E, Usdin K. The GAA*TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grabczyk E, Usdin K. Alleviating transcript insufficiency caused by Friedreich's ataxia triplet repeats. Nucleic Acids Res. 2000;28:4930–4937. doi: 10.1093/nar/28.24.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fry M, Usdin K. Human Nucleotide Expansion Disorders. Heidelberg: Springer; 2006. [Google Scholar]
- 8.Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 9.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 10.De Jaco A, Camp S, Taylor P. Influence of the 5′ intron in the control of acetylcholinesterase gene expression during myogenesis. Chem. Biol. Interact. 2005;157, 158:372–373. doi: 10.1016/j.cbi.2005.10.058. [DOI] [PubMed] [Google Scholar]
- 11.LeBlanc SE, Jang SW, Ward RM, Wrabetz L, Svaren J. Direct regulation of myelin protein zero expression by the Egr2 transactivator. J. Biol. Chem. 2006;281:5453–5460. doi: 10.1074/jbc.M512159200. [DOI] [PubMed] [Google Scholar]
- 12.Zhao P, Caretti G, Mitchell S, McKeehan WL, Boskey AL, Pachman LM, Sartorelli V, Hoffman EP. Fgfr4 is required for effective muscle regeneration in vivo. Delineation of a MyoD-Tead2-Fgfr4 transcriptional pathway. J. Biol. Chem. 2006;281:429–438. doi: 10.1074/jbc.M507440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JG, Dahi S, Mahimkar R, Tulloch NL, Alfonso-Jaume MA, Lovett DH, Sarkar R. Intronic regulation of matrix metalloproteinase-2 revealed by in vivo transcriptional analysis in ischemia. Proc. Natl Acad. Sci. USA. 2005;102:16345–16350. doi: 10.1073/pnas.0508085102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 16.Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina GA, et al. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum. Mol. Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- 17.Rimseliene R, Vaisvila R, Janulaitis A. The eco72IC gene specifies a trans-acting factor which influences expression of both DNA methyltransferase and endonuclease from the Eco72I restriction-modification system. Gene. 1995;157:217–219. doi: 10.1016/0378-1119(94)00794-s. [DOI] [PubMed] [Google Scholar]
- 18.Kumari D, Usdin K. Interaction of the transcription factors USF1, USF2, and alpha -Pal/Nrf-1 with the FMR1 promoter. Implications for Fragile X mental retardation syndrome. J. Biol. Chem. 2001;276:4357–4364. doi: 10.1074/jbc.M009629200. [DOI] [PubMed] [Google Scholar]
- 19.Greene E, Entezam A, Kumari D, Usdin K. Ancient repeated DNA elements and the regulation of the human frataxin promoter. Genomics. 2005;85:221–230. doi: 10.1016/j.ygeno.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Kawashima I, Mita-Honjo K, Takiguchi Y. Characterization of the primate-specific repetitive DNA element MER1. DNA Seq. 1992;2:313–318. doi: 10.3109/10425179209030964. [DOI] [PubMed] [Google Scholar]
- 21.Clark RM, Dalgliesh GL, Endres D, Gomez M, Taylor J, Bidichandani SI. Expansion of GAA triplet repeats in the human genome: unique origin of the FRDA mutation at the center of an Alu. Genomics. 2004;83:373–383. doi: 10.1016/j.ygeno.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Liu WM, Schmid CW. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 1993;21:1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 24.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 25.Matzke MA, Aufsatz W, Kanno T, Mette MF, Matzke AJ. Homology-dependent gene silencing and host defense in plants. Adv. Genet. 2002;46:235–275. doi: 10.1016/s0065-2660(02)46009-9. [DOI] [PubMed] [Google Scholar]
- 26.Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Heidenfelder BL, Makhov AM, Topal MD. Hairpin formation in Friedreich's ataxia triplet repeat expansion. J. Biol. Chem. 2003;278:2425–2431. doi: 10.1074/jbc.M210643200. [DOI] [PubMed] [Google Scholar]