Abstract
Widely used nucleic acid assays are poorly suited for field deployment where access to laboratory instrumentation is limited or unavailable. The need for field deployable nucleic acid detection demands inexpensive, facile systems without sacrificing information capacity or sensitivity. Here we describe a novel microarray platform capable of rapid, sensitive nucleic acid detection without specialized instrumentation. The approach is based on a miniaturized lateral flow device that makes use of hybridization-mediated target capture. The miniaturization of lateral flow nucleic acid detection provides multiple advantages over traditional lateral flow devices. Ten-microliter sample volumes reduce reagent consumption and yield analyte detection times, excluding sample preparation and amplification, of <120 s while providing sub-femtomole sensitivity. Moreover, the use of microarray technology increases the potential information capacity of lateral flow. Coupled with a hybridization-based detection scheme, the lateral flow microarray (LFM) enables sequence-specific detection, opening the door to highly multiplexed implementations for broad-range assays well suited for point-of-care and other field applications. The LFM system is demonstrated using an isothermal amplification strategy for detection of Bacillus anthracis, the etiologic agent of anthrax. RNA from as few as two B. anthracis cells was detected without thermocycling hardware or fluorescence detection systems.
INTRODUCTION
The challenges presented by biological weapons, global health-care issues and emerging diseases of natural origin lend urgency to the development of rapid, field-deployable pathogen detection and diagnostic tools (1,2). Ideally, to be of general field utility, a diagnostic device must be capable of sensitive and specific pathogen detection while retaining simplicity of use and independence from complex laboratory instrumentation (3). Additional challenges are presented by the need to screen samples for multiple pathogenic or toxic agents, a characteristic highly desirable in cases where commonalities in early symptom presentation confound differential diagnoses. While nucleic acid-based assays for pathogen detection and identification offer sensitivity, specificity and resolution, they are relatively elaborate and often costly, limiting their utility for point-of-care diagnostics and deployment under field conditions where a supporting laboratory infrastructure is limited or absent. Reliance upon polymerase chain reaction (PCR) and fluorescent detection of amplified nucleic acids has contributed significantly to the complexity and cost of nucleic acid diagnostics (2,4–6). Retaining assay sensitivity while circumventing requirements for thermocyclers and fluorescence detection hardware remains a significant challenge.
The recent advent of DNA microarray technology has promised to increase the information capacity of nucleic acid diagnostics and enable the highly multiplexed detection of genetic signatures (7). The potential of DNA microarrays to detect, in parallel, large panels of distinct nucleic acid sequences has proven to be a powerful technique for many laboratory applications (8). Nonetheless, the reliance of this technology on costly instrumentation for high-resolution fluorescence signal transduction severely limits the utility of microarrays for field applications where a laboratory infrastructure is limited or unavailable. Additionally, the long hybridization incubations required for microarray assays increase sample-to-answer times beyond what would be acceptable for a rapid screening assay. Though microarray hybridization times as short as 300 and 500 s have been reported (9,10), such methods employ designs that remain reliant upon fluorescent detection and supporting instrumentation and do not address the need for low-cost, easily manufactured devices that can be used in the absence of laboratory infrastructures.
In contrast to DNA-based assays, immunoassays have found widespread acceptance in low-cost, easily used formats, perhaps the most notable of which is the chromatographic lateral flow immunoassay (11). Lateral flow assays, also known as hand-held assays or dipstick assays, are used for a broad range of applications where rapid antigen detection is required in an easily used, low-cost format. Expanding the domain of lateral flow chromatography to nucleic acid detection, a number of recent reports have described lateral flow detection of PCR products using a variety of capture and detection schemes (12–15). Unfortunately, the utility of lateral flow detection in the context of a PCR-based assay is severely limited by the fact that reliance on thermocycling hardware largely negates the potential benefit of the otherwise highly simplified lateral flow platform. Additionally, a PCR-based approach to lateral flow detection necessitates each PCR reaction be subjected to post-amplification manipulations required to generate single-stranded products for hybridization-based detection.
Recent work has sought to alleviate reliance on PCR through employing isothermal nucleic acid amplification schemes or direct detection of unamplified genetic material. Enabled by the use of up-converting phosphor reporters, unamplified Streptococcus pneumoniae DNA sequence has been detected using a lateral flow assay format (16). Up-converting phosphor technology, while sensitive, remains dependent upon the hardware required to detect phosphor emission (17). The use of simple colorimetric detection schemes that circumvent the requirements for complex instrumentation require an upstream amplification strategy to attain suitable sensitivity. Isothermal nucleic acid amplification coupled with lateral flow detection has been reported for assays making use of cycling probe technology [CPT, (18)], recombinase polymerase amplification [RPA, (19)] and nucleic acid sequence-based amplification [NASBA, (20–27)]. While the work by Fong et al. (23) and Piepenburg et al. (19) made use of a lateral flow immuno-assay for DNA detection, the RNA targets amplified by NASBA in the work from Baeumner's group (24–27) were detected using a lateral flow system enabled by the use of liposome encapsulated dye and a sandwich hybridization assay similar to that reported by Rule et al. (13). While shown to display nanomolar sensitivity, the reported dye encapsulating liposome-based methods require additional washing steps and the liposomes are relatively labile, must be custom synthesized, and stored under stabilizing hydrated conditions (28).
To develop more capable nucleic acid detection methods that offer many of the advantages of microarray technology yet retain the simplicity of lateral flow-based platforms, we have developed a microarray-based lateral flow technology. Using an assay based on the nonsense mutation in the plcR gene of Bacillus anthracis, that is absent in the near phylogenetic neighbors B. thuringiensis and B. cereus (29,30), we illustrate the utility of the lateral flow microarray (LFM) approach for sensitive detection and discrimination of closely related microbial signatures when present as minority sequences in complex nucleic acid mixtures. The results demonstrate that LFMs, making use of stable detection reagents suitable for dry storage, can be used to detect as little as 250 amol analyte within 2 min of sample addition. The miniaturization of lateral flow detection decreases reagent consumption and sample-to-answer times while increasing the potential information capacity of the platform to enable the development of highly multiplexed nucleic acid detection assays.
MATERIALS AND METHODS
RNA isolation
Total RNA was isolated from B. anthracis strain Sterne 7702 and B. thuringiensis strain HD 621 (31) using a previously reported protocol (32). Purified RNA was quantified by measuring OD260 and evaluated by gel electrophoresis. 3 × 108 cells were used for RNA isolation typically yielding 50–75 μg of total RNA.
Amplification primer design
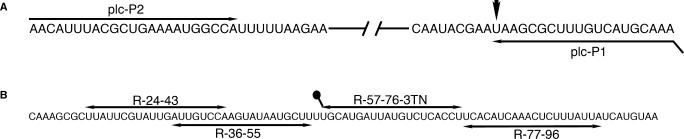
NASBA (22) primers, plc-P1 and plc-P2, were designed to amplify a fragment of the plcR locus from B. anthracis. Primer sequences used for NASBA reactions are provided in Table 1, the T7 promoter sequence is italicized in plc-P1. Plc-P1 hybridizes to the plcR transcript such that the 3′-end of the primer forms a base pair with the previously reported polymorphism strictly associated with B. anthracis (29,30). The NASBA P2 primer, plc-P2, is located such that the amplified RNA resulting from NASBA is 179 bases in length (Figure 1A). Previously reported plcR-based B. anthracis real-time PCR assays (29,30) have made use of an alternate upstream primer that generates a 83 bp product but may be poorly suited for NASBA given the optimal NASBA product size of 120–250 bases (33).
Table 1.
| Function | Name | Sequence |
|---|---|---|
| Bead Conjugation/Detection Probe | R-57-76-3TN | 5′-AGGTGAGACATAATCATGCA TTTTTTTTTTTTTTTTTT-NH2-3′ |
| Detection Probe/Negative Control | R-57-76-3N | 5′-AGGTGAGACATAATCATGCA-NH2-3′ |
| LFM Immobilized Capture Probe | R-77-96 | 5′-TAATAAAGAGTTTGATGTGA-3′ |
| LFM Immobilized Capture Probe | R-36-55 | 5′-AAGCATTATACTTGGACAAT-3′ |
| LFM Immobilized Capture Probe | R-24-43 | 5′-TGGACAATCAATACGAATAA-3′ |
| Synthetic target/Positive Hyb Control | dnaR89 | 5′-CAAAGCGCTTATTCGTATTGATTGTCCAAGTATAATGCTTTTGC ATGATTATGTCTCACCTTCACATCAAACTCTTTATTATCATGTAA-3′ |
| NASBA/In vitro transcription product | plcRivt | 5′-GGGAGAUUUGCAUGACAAAGCGCUUAUUCGUAUUGAUUGU CCAAGUAUAAUGCUUUUGCAUGAUUAUGUCUCACCUUCACAU CAAACUCUUUAUUAUCAUGUAAUACUUCUAAUUGCUUUAAUA UAUUUUCAUAUAACUCAAUACUCUUCUUAAAAUGGCCAUUUU CAGCGUAAAUGUU-3′ |
| Negative Hyb Control | FT-S18 | 5′-GCGGTCCCAAAAGGGTCAGTCGTAGCACACCACTTTCA-3′ |
| Negative Hyb Control | F-24-43 | 5′-TTATTCGTATTGATTGTCCA-3′ |
| NASBA-P1/Allele Discrimination | plc-P1 | 5′-TTCTAATACGACTCACTATAGGGAGATTTGCATGACAAAGCGCTTA-3′ |
| NASBA-P2 | plc-P2 | 5′-AACATTTACGCTGAAAATGGCCA-3′ |
Figure 1.
(A) NASBA primer-binding sites are shown in the relevant region of the predicted B. anthracis plcR mRNA sequence based on GenBank accession number AY265698. The terminal 3′ base of plc-P1 is complementary to the U of the ochre stop codon, indicated with an arrowhead, diagnostic for B. anthracis. (B) The predicted nucleotide sequence plcR mRNA in the region represented by synthetic target dnaR89. The binding sites of detection probe R-57-76-3TN, as well as capture probes R-77-96, R36-55 and R-24-43 are indicated.
Nucleic acid sequence-based amplification (NASBA)
NASBA reactions were prepared according to the manufacturer's instructions using the NucliSens Basic kit (Biomerieux) and primers plc-P1 and plc-P2 at 0.4 μM each. Amounts of total cellular bacterial RNA were varied, as indicated, between 0 and 2 ng. Bacillus anthracis Sterne 7702 was used as a test strain and B. thuringiensis strain HD 621 was employed as a negative control. One microgram of human total cellular RNA isolated from HeLa S3 cells (Stratagene) was included in all NASBA reactions to provide a complex RNA background consistent with the composition of human diagnostic samples. Following a 60-min incubation at 41°C, NASBA reaction products were detected by using a LFM.
LFM fabrication
LFMs were printed using a NanoPlotter 2.0 (GeSim, mbH, Dresden, Germany) non-contact picoliter deposition system equipped with NanoTips (GeSim). Unless otherwise indicated, LFMs were patterned with 400 μM solutions of oligonucleotide in H2O containing a 1:50 dilution of Ponceau S (P7767, Sigma) as a tracking dye. A lateral flow compatible nitrocellulose membrane (HiFlow 135, Millipore) was used as the LFM substrate. Following oligonucleotide deposition, nitrocellulose membranes were air dried and exposed to 5000 μJ UV in a StrataLinker (Stratagene). The resulting membrane sheets were cut into 3-mm wide, 30-mm long strips, which were either used directly with buffer-suspended dyed microspheres or assembled with conjugate release pads into a custom plastic housing. Housings were fabricated from polycarbonate sheet cut using a CO2 laser (VersaLaser VL-300, Universal Laser Systems, Inc., Scottsdale, AZ, USA). A gasket of 500-μm thickness was used to generate an internal chamber of sufficient size to accommodate the LFM substrate and the conjugate release pad. The small sample volumes used obviated the need for sample and downstream absorbent pads, the function of which was supplied by the conjugate release pad and unpatterned regions of the LFM substrate, respectively. Conjugate release pads were made by impregnating glass fiber conjugate pad (GFCP203000, Millipore) with dyed microspheres covalently conjugated to R-57-76-3TN in 1% SDS. Conjugate release pads measuring 3.5 mm × 4.5 mm were doped with ∼8 × 109 oligonucleotide conjugated dyed microspheres. Microsphere saturated release pads were allowed to air dry under ambient conditions prior to assembly with LFM membranes.
Colorimetric hybridization sandwich assay reagents
Table 1 provides capture and detection oligonucleotide sequences, their binding sites within the plcR amplicon are depicted in Figure 1B. Amine modification and a T18 spacer sequence were included on the 3′-end of detection oligonucleotide R-57-76-3TN to allow covalent cross-linking to dyed microspheres and to facilitate hybridization in lateral flow sandwich assays, respectively.
SPHERO™ carboxyl-polystyrene 0.35-μm blue microspheres (Spherotech) were covalently conjugated to amino modified oligonucleotide R-57-76-3TN using the coupling agent 1-etyl-3-(3-dimethylaminopropyl-diimide HCl) (EDAC, Pierce) under conditions adapted from Spiro et al. (34). Briefly, 1.1 × 1012 microspheres were suspended in 100 mM 2-(N-morpholino)ethanesulfonic acid pH 4.5 (MES, Sigma). Indicated amounts of oligonucleotide were introduced to MES suspended microspheres, vortexed and incubated in the presence of 0.5 mg/ml EDAC. Reactions were protected from light in aluminum foil wrapped tubes and incubated at room temperature for 30 min followed by the introduction of additional EDAC to bring the final EDAC concentration to 1 mg/ml. Incubation was continued for an additional 30 min after which beads were washed once with 1 ml 0.02% tween-20 (Sigma) and twice with 0.5 ml 0.1% SDS (Fisher Scientific). Beads were re-suspended in 0.5 ml DNAase/RNAase free H2O. Bead suspensions were assessed for aggregation by phase-contrast light microscopy using a Zeiss IM135 inverted microscope.
LFM assays
A DNA oligonucleotide, dnaR89, composed of sequence derived from a region of the plcR gene of B. anthracis, as shown in Figure 1B, was used to provide a readily available and quantifiable target for LFM assay development and optimization. The sequence of this synthetic target is provided in Table 1. Additionally, a full-length synthetic target RNA was generated. This RNA, referred to here as plcRivt, was used to confirm that reaction conditions established with dnaR89 were also suitable for the detection of NASBA reaction products. Synthesis of plcRivt was accomplished by using plc-P1 and plc-P2 primers in PCR reactions containing 20 ng of B. anthracis Stern strain 7702 genomic DNA. PCR reactions using Platinum PCR Supermix (Invitrogen) were conducted for 40 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 1 min following an initial 2-min incubation at 94°C. The resulting amplicon was subjected to purification using QIAquick PCR clean-up spin-columns (QIAGEN) and subsequently used to program an in vitro transcription reaction using the T7 AmpliScribe kit (EpiCentre). The in vitro transcription reaction product was subjected to treatment with RNase free DNase I (Ambion) and purified using a RNeasy column (QIAGEN). The resulting RNA was quantified by measuring the OD260. plcRivt is predicted to be identical in sequence to the NASBA product generated from B. anthracis total cellular RNA using plc-P1 and plc-P2.
Detection of NASBA reaction products by LFM was accomplished by introducing a 2-μl aliquot of a 20-μl NASBA reaction into 8 μl of LFM running buffer. Lateral flow running buffer was based on the widely used standard sodium citrate buffer (SSC; 1 × SSC = 150 mM NaCl, 15 mM sodium citrate, pH 7.0) supplemented with 1.4% Triton X-100 and 0.1% SDS to reduce microsphere aggregation and 5% formamide to increase hybridization stringency and destabilize target secondary structure (final buffer composition: 4 × SSC, 0.1% SDS, 1.4% Triton X-100, 5% deionized formamide). The final volume of solution applied to LFMs was 10 μl. Following completion of sample flow, LFM membranes were removed from plastic housings and allowed to air dry prior to scanning with a standard flatbed PC scanner (CanoScan 9950F, Canon, Inc.). Scans were performed at 2400 dpi resolution using 48-bit color. The resulting image files were converted to grayscale, inverted and saved as 16-bit TIFF files using Photoshop CS2 (Adobe). Image files were analyzed using GenePix Pro 6.0 (Molecular Devices) to quantify microarray spot intensities for NASBA product detection and for dnaR89 dilution series experiments.
For time course studies, LFM assays were conducted with sample buffer containing 0.1% w/v suspended dyed microspheres (∼4 × 109 particles) in running buffer. Lateral flow was recorded using a digital video recorder (DCR-PC1, Sony). Video frames were collected for quantification using iMovie (Apple Computer). Feature intensity was quantified for time course studies and some optimization experiments using uncalibrated optical density in ImageJ (http://rsb.info.nih.gov/ij/). For better reproduction contrast, LFM images used for figures were cropped and modified by applying the Auto Contrast function in Photoshop CS2. No other modifications were applied.
RESULTS
LFM prototype
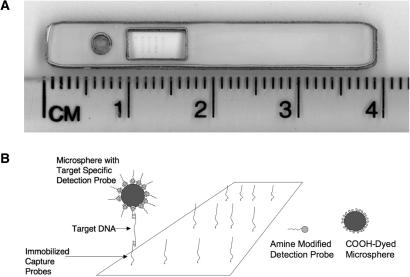
Oligonucleotides for hybridization sandwich assays were designed to detect NASBA amplified B. anthracis plcR mRNA or synthetic targets based on relevant subregions of the plcR sequence. Oligonucleotides immobilized on the lateral flow substrate are referred to here as capture oligonucleotides while those conjugated to dyed microspheres for signal generation are referred to here as detection oligonucleotides. Supported large-pore nitrocellulose membranes were patterned with varying concentrations of capture oligonucleotides using a NanoPlotter 2.0 robotic positioning system and NanoTip piezoelectronically actuated micropipets. Oligonucleotide dnaR89 was printed on LFM substrates as a positive hybridization control as this oligonucleotide carries sequence complementary to bead coupled detection oligonucleotide R-57-76-3TN. Negative hybridization control oligonucleotides included the reverse complement of capture oligonucleotide R-24-43 (F-24-43), the detection probe sequence sans T18 spacer (R-57-76-3N) and a plcR unrelated sequence complementary to a region of the Francisella tularensis sdhA locus (FT-S18). By ejecting droplets from the micropipet at a distance of 500 μm from the nitrocellulose substrate, microarray feature sizes of ∼200 μm could be generated. In contrast to contact microarray printing methods, this approach preserves the fragile pore structure of the membrane required for microsphere-based detection. Patterned nitrocellulose sheets were cut into 3-mm wide strips and then assembled with conjugate release pads in a custom designed plastic housing. An example of the resulting device is shown in Figure 2A. Hybridization-mediated capture of analyte at the cognate capture element of the microarray and non-overlapping hybridization to dyed microsphere conjugated detection oligonucleotide generates a colorimetric signal arising from an increased local concentration of dyed microsphere particles. In the absence of hybridization, microspheres are sufficiently dispersed that additional washing steps are not required to reduce background signal levels. A schematic representation of the hybridization sandwich assay scheme is depicted in Figure 2B.
Figure 2.
(A) A compact plastic housing was designed to a carry conjugate release pad and a LFM membrane. A small port is used to introduce the 10-μl sample volume and a rectangular window allows direct visualization of the microarray capture features. The device is 39 × 5 mm. (B) A schematic representation of the hybridization sandwich assay used for LFM-based nucleic acid detection. Carboxyl-polystyrene dyed microspheres are linked to amine-modified detection oligonucleotide R-57-76-3TN. The microsphere/analyte complex forms by hybridization as sample solution liberates dried microspheres from the conjugate release pad. This complex is captured from solution by hybridization to immobilized capture probes as capillary flow transports the sample/bead solution through the large-pore nitrocellulose matrix. The resulting increase in local microsphere concentration, at capture features complementary to the target analyte, rapidly produces a colorimetric signal visible to the naked eye and easily detected at low concentrations using widely available flatbed scanners. The hybridization based nature of the assay render it well suited for multiplexed detection.
Lateral flow hybridization sandwich assay optimization
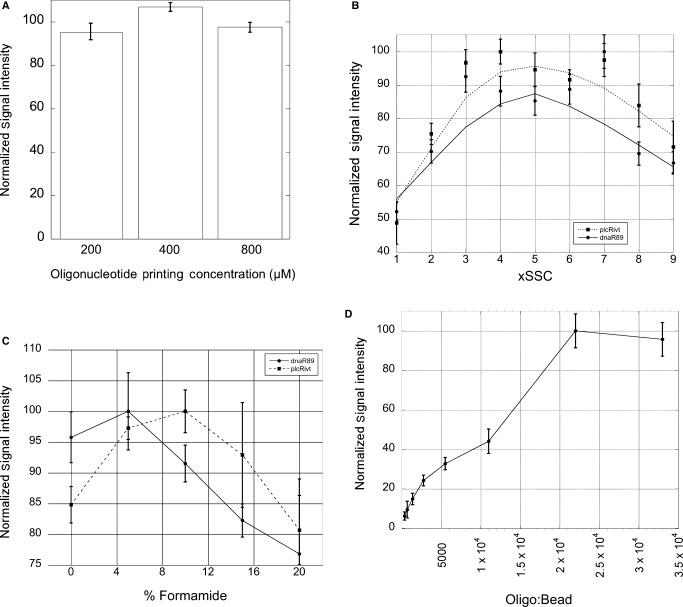
LFMs were fabricated using varying concentrations of capture oligonucleotide to determine optimum printing concentrations. Following lateral flow of 25 fmol dnaR89 in 4× SSC, 5% formamide, 1.4% Triton X-100, 0.1% SDS containing 0.1% R-57-76-3TN coupled microspheres LFMs were scanned on a flatbed scanner and the resulting images quantified. For all capture sequences examined, 400-μM oligonucleotide-printing concentrations provided the most favorable signal intensity (Figure 3A). Standard hybridization conditions employed for these and other characterization studies were determined through an iterative set of optimization experiments that examined the effects of ionic strength, formamide concentration and detection oligonucleotide to bead cross-linking ratios.
Figure 3.
(A) LFM substrates patterned with different concentrations of capture oligonucleotides R-77-96, R-36-55, and R-24-43 were used to detect dnaR89 with R-57-76-3TN microspheres. Signals generated at microarray capture features printed at 200, 400 and 800 μM were quantified following lateral flow of samples containing 5, 10 and 20 fmol dnaR89. Signals were normalized for each capture probe and target concentration. Average signal intensities were calculated and presented in this bar graph. 400 μM printing concentrations consistently provided the strongest signal independent of capture sequence or dnaR89 concentration. Error bars are the 95% confidence interval (two tailed, n = 12). (B) Scatter plot of normalized signal intensity versus SSC concentration. LFM running buffer was optimized for SSC concentration using R-57-76-3TN to detect dnaR89 (circles) or plcRivt (squares). (C) Line plot of normalized signal intensity versus formamide concentration. Formamide concentrations between 0 and 20% in LFM running buffer based on 4× SSC were evaluated for dnaR89 (circles) and plcRivt (squares). Five percent formamide provided near optimal detection of both dnaR89 and plcRivt. (D) Line plot of normalized signal intensity versus the R-57-76-3TN to microsphere ratio. Oligonucleotides/bead of 2.2 × 104 in coupling reactions provided the best performing conjugated microsphere populations as judged by hybridization sandwich assay signal intensity. For parts B–D error bars are the 95% confidence interval (two tailed, n = 4).
Given the profound impact ionic strength has on the stringency of DNA hybridization, microsphere dispersion and lateral flow characteristics, SSC concentration was varied from 1 × to 9 × and assay performance evaluated by densitometry of LFMs following hybridization sandwich assays conducted using 25 fmol of the synthetic target dnaR89 or ∼200 fmol of plcRivt (35,36). Figure 3B summarizes the results of SSC concentration optimization experiments. Near optimal signal intensity was obtained for both dnaR89 and plcRivt at SSC concentrations between 2× and 7×. For use in standard LFM running buffer 4× SSC was selected as it provided sensitive hybridization-based detection of plcR derived sequences, good capillary lateral flow characteristics, and favorable microsphere dispersion and release-pad liberation.
Formamide is known to reduce the melting temperature of DNA and RNA duplexes and may facilitate capture and detection probe accessibility to binding sites within the target through a destabilization of analyte secondary structure (37). To determine the optimum concentration of formamide in LFM running buffer, a series of LFM experiments were conducted at varying formamide concentrations using both dnaR89 and plcRivt. 10 μl of 4× SSC, 1.4% Triton X-100 and 0.1% SDS containing 25 fmol dnaR89 or ∼200 fmol plcRivt and varying concentrations of formamide, as indicated in Figure 3C, were subjected to LFM analysis and the resulting hybridization signals quantified by densitometry. These experiments revealed a slight but reproducible increase in signal intensity at 5% formamide. All subsequent studies presented here were performed using 4× SSC, 1.4% Triton X-100, 0.1% SDS and 5% formamide.
Given that higher stock concentrations of synthetic oligonucleotide dnaR89 could be obtained, which allowed high confidence quantification of this synthetic target relative to what could be achieved with comparatively dilute solutions of the in vitro transcription product plcRivt, subsequent LFM characterization studies made use of dnaR89. The similarity of buffer optima displayed by dnaR89 and by plcRivt synthetic targets supported the assertion that dnaR89 could be used as an accurate proxy for the performance of LFM assays for NASBA product detection. Others have reported similar findings concluding that appropriately designed DNA oligonucleotides can be used as synthetic targets for the development of assays ultimately used for NASBA product detection (38). Therefore, subsequent LFM assay optimization and characterization was conducted using dnaR89.
To determine the optimum ratios for cross-linking detection oligonucleotides to dyed polystyrene microspheres, we examined populations of beads coupled to oligonucleotide at varying ratios. The 3′-amine modified detection oligonucleotide R-57-76-3TN was covalently linked to polystyrene dyed microspheres using EDAC. The resulting bead/oligonucleotide complexes were evaluated for their ability to mediate detection of dnaR89 in a hybridization sandwich assay. Coupling reactions using a 2.2 × 104:1 oligonucleotide to bead ratio were found to provide optimum signal as determined by densitometry (Figure 3D).
Characterization of LFM-based nucleic acid detection sensitivity
The detection oligonucleotide R-57-76-3TN carried a 3′-spacer region consisting of 18 T residues to increase the accessibility of bead bound oligonucleotides for hybridization. R-57-76-3N, which carried the same analyte complementary sequence as R-57-76-3TN but without the T18 spacer, was found to exhibit significantly reduced hybridization to dnaR89 consistent with prior reports that a poly(dT) spacer sequence increases hybridization efficiency to solid-phase coupled oligonucleotides (data not shown) (39,40). T18 spacers were not incorporated into LFM immobilized capture oligonucleotides as they were found to be dispensable for hybridization.
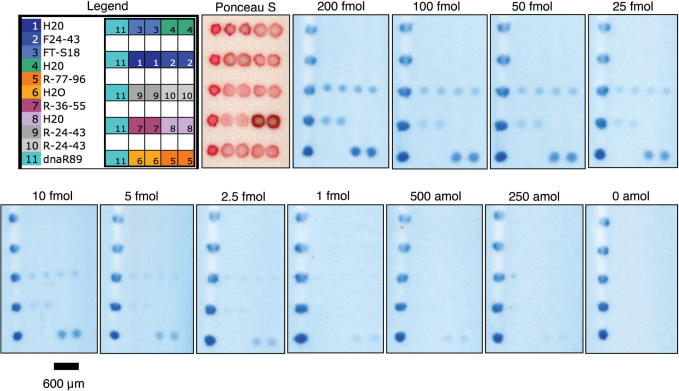
To determine the relative performance of hybridization sandwich assays making use of capture oligonucleotides with complementarity to different locations of the target sequence, three capture oligonucleotides were synthesized and compared using sandwich assays employing detection oligonucleotide R-57-76-3TN coupled dyed microspheres. R-77-96 was designed to participate in base stacking with R-57-76-3TN when hybridized to the target. Base stacking has been reported to stabilize hybridization and allow efficient capture of dilute nucleic acids by hybridization (41–45). The binding sites for the three capture oligonucleotides examined (R-77-96, R-36-55 and R-24-43) are illustrated in Figure 1B. Varying quantities of synthetic target dnaR89, between 0 and 200 fmol, were used for these studies. Figure 4 depicts LFM membranes following detection of the indicated amounts of target oligonucleotide dnaR89. LFMs carried dnaR89, which hybridizes directly to the microsphere conjugated detection probe, as a positive hybridization control. Positive control features were printed as the left most element of each LFM row to assist in feature identification. Negative hybridization controls, F24-43 and FT-S18, were based on the reverse complement of R-77-96 and an unrelated F. tularensis derived sequence, respectively. Additionally, to confirm that no carryover contamination occurred during printing, H2O containing Ponceau S was printed on LFM substrates between positive control and capture oligonucleotide deposition. No signal was detectable in either hybridization negative controls or H2O negative control microarray elements.
Figure 4.
Representative LFMs are shown following detection of the indicated amounts of dnaR89. The microarray physical layout is provided in the color legend. The panel labeled ‘Ponceau S’ is an LFM prior to sample addition. Ponceau S allows visualization of successful oligonucleotide deposition but migrates away from the capture zone during sample transport across the substrate. Contrast was adjusted using the Auto Contrast function in Photoshop CS2 to increase reproduction contrast. Auto Contrast adjustment was not used for images subjected to quantification. The bar is 600 µm for all LFM panels.
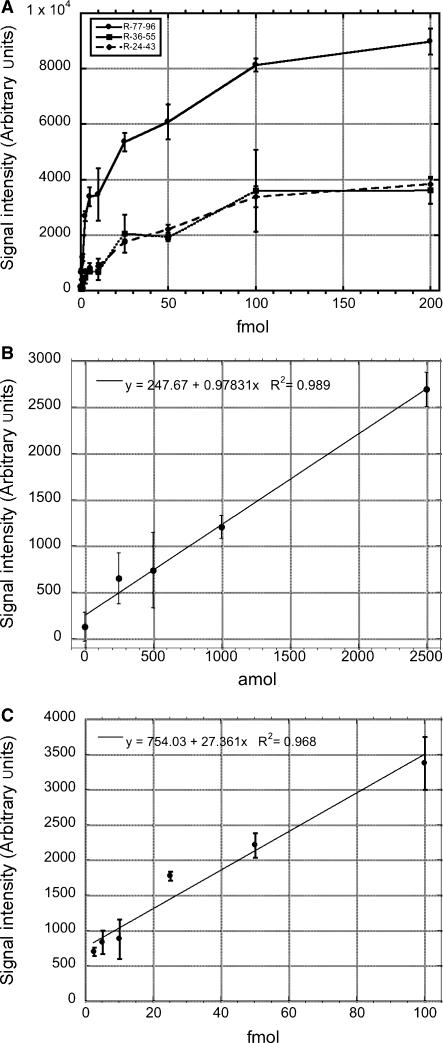
Background corrected signal intensity was determined from LFM images using GenePix Pro 6.0 microarray data extraction software. The results, presented in Figure 5A, reveal R77-96 produces significantly higher hybridization signals than R-36-55 or R-24-43 for all examined quantities of dnaR89, suggesting a significant contribution of base stacking effects to LFM hybridization sandwich assay sensitivity.
Figure 5.
(A) The relative performance of three different capture oligonucleotides (R-77-96, circle/solid line; R-36-55, square/solid line; R-24-43, diamond/dashed line) was determined using varying amounts of dnaR89 from 0 to 200 fmol. The capture probe R-77-96 provides significantly more sensitive detection than the other capture sequences evaluated using R-57-76-3TN coupled microspheres. (B) R-77-96 signal intensity versus amol dnaR89 from 0 to 2500 amol is plotted with a linear regression line (R2 = 0.989). (C) R-24-43 signal intensity versus fmol dnaR89 from 2.5 to 100 fmol plotted with a linear regression line (R2 = 0.968). For all parts error bars are the 95% confidence interval (one tailed, n = 6).
To define the detection limit of the LFM assay, a one-tailed t-test was used to determine quantities of dnaR89 that generated signal intensities significantly above 0 amol negative controls. Signals generated at R-77-96 capture features with 250 amol and greater quantities of dnaR89 were significantly >0 amol dnaR89 controls (P < 0.05, n = 6). By the same criterion, 1 fmol dnaR89 detection limits were obtained for both R-24-43 and R-36-55 (P < 0.05, n = 6). Figure 5B depicts the performance of LFM detection over 0 to 2500 amol dnaR89 range using the R-77-96/R-57-76-3TN capture/detection probes. LFM detection exhibited excellent linearity, R2 = 0.989, over this 10-fold range of target molecules. While capture probe R-24-43 exhibited less sensitivity than R-77-96, this capture probe displayed excellent signal linearity between 2.5 and 100 fmol dnaR89, R2 = 0.968 (Figure 5C). These findings demonstrate that the LFM capacity to display multiple capture sequences can be used to simultaneously provide sensitive detection and extend assay linearity through the use of capture probes with differing hybridization characteristics.
LFM detection time course studies
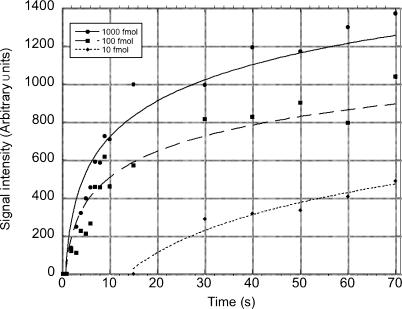
The small sample volumes used for LFM detection and the reduced surface area traversed during capillary lateral flow significantly reduces detection times for the LFM relative to traditional lateral flow devices. To quantitatively present the speed of LFM nucleic acid hybridization-based detection, we used digital video to follow hybridization sandwich assay-mediated detection of synthetic target molecule dnaR89. Individual frames were isolated from video datasets and quantified for relative signal intensity over the course of capillary lateral flow across the LFM substrate. The resulting signal data was plotted versus time in seconds as shown in Figure 6. For time measurements, t0 was defined as the time when the sample front reached the first row of LFM features. Signal was detectable for 1000-fmol target in 2 s following sample transport across R-77-96 capture elements. Within 4 s 100 fmol dnaR89 was detectable while 10 fmol was clearly detectable by 30 s as defined by the earliest time point at which 90% of the pixels composing the R-77-96 microarray features were greater than one standard deviation above background. Lateral flow transport of the 10-μl sample was complete by 120 s.
Figure 6.
Time course of LFM detection: 10-μl samples containing either 1000 fmol (circle), 100 fmol (square) or 10 fmol (diamond) dnaR89 were run on appropriately patterned LFMs. Video data were collected and colorimetric signal intensity measured from video frames at R-77-96 capture features. Capillary transport of the 10-μl sample was complete by 120 s. Lines represent logarithmic curve fits to the data.
Allele-specific Isothermal Amplification and LFM-based Product Detection
Prior reports have described a single nucleotide polymorphism (SNP) present in B. anthracis but not close phylogenetic near neighbors including B. cereus and B. thuringiensis (29,30). This SNP has been used as the basis for a sensitive and highly discriminatory real-time PCR assay for B. anthracis (30). To determine the utility of LFM technology for detecting minority nucleic acids in complex samples, NASBA primers were designed to amplify the plcR allele of B. anthracis. P1 and P2 primer sequences, plc-P1 and plc-P2, used for NASBA amplification are provided in Table 1 and their binding positions illustrated in Figure 1.
Varying amounts of total cellular RNA isolated from B. anthracis or 2 ng of B. thuringiensis HD 621 RNA as a negative control were introduced to 1 μg of total human cellular RNA isolated from HeLa S3 cells. The resulting mixtures were subjected to NASBA amplification using plc-P1 and plc-P2 primers. Human RNA was included in NASBA amplification reactions to approximate the nucleic acid complexity expected in human diagnostic specimens. An aliquot of 2 μl of NASBA reaction mixture was removed after a 60-min incubation at 41°C, mixed with 8 μl of LFM running buffer and assayed for plcR amplicon by LFM. Dyed microspheres cross-linked to R-57-76-3TN were used for detection of NASBA amplicons captured on LFMs carrying R-77-96. Data from these studies are presented in Figure 7. Following 60 min of NASBA amplification, as little as 0.5 pg for total cellular B. anthracis RNA could be detected in a background matrix of 1 μg of human total RNA. These studies closely approximate the conditions expected for complex human diagnostic samples and reveal the capacity of the LFM platform to specifically detect NASBA reaction products generated from mixed samples where the target sequence is a minority species. While the number of plcR mRNA copies in a B. anthracis cell has not been determined, an estimate of LFM assay sensitivity, in terms of B. anthracis cells, can be calculated based on total RNA yields. Total RNA yields from vegetative B. anthracis were in the range of ∼167–250 fg RNA/cell. Using this value, an estimate of LFM sensitivity corresponds to the detection of approximately to 2–3 B. anthracis cells.
Figure 7.
(A) Indicated amounts of total cellular RNA from B. anthracis Sterne strain 7702 or, as a negative control, 2 ng B. thuringiensis strain HD 621 RNA (0 fg panel) were introduced to 1 μg of total human cellular RNA isolated from HeLa S3 cells. RNA mixtures were subjected to NASBA amplification for 60 min after which 2 μl aliquots of the NASBA reactions were mixed with 8 μl of LFM running buffer and introduced to LFMs. Enlarged LFM sub-regions are shown following cropping, grayscale conversion and Auto Contrast adjustment in Photoshop. The legend indicates microarray element identities: (+) dnaR89 as a positive hybridization control, (–) R-57-76-3N as negative hybridization control, (24–43) capture probe R-24-43, (36–55) capture probe R-36–55, (77–96) capture probe R-77-96. (B) Graph of quantified signals from B. anthracis and B. thuringiensis challenged LFMs with linear regression line (R2 = 0.970). 0 fg B. anthracis total cellular RNA data point contains 2 ng B. thuringiensis total cellular RNA in addition to 1 μg human total cellular RNA. Error bars depict measurement SD (three determinations).
DISCUSSION
Lateral flow detection of DNA or RNA amplification reaction products provides one means of simplifying nucleic acid detection. Indeed, a lateral flow platform may offer many significant advantages for employing nucleic acid assays under conditions where a fully equipped molecular biology laboratory infrastructure is not available or desirable. Such situations would include resource poor settings, point-of-care, battlefield deployments and scenarios where first responders must quickly determine the threat presented by an unknown substance. To date, however, lateral flow devices have predominantly been fabricated using one or a few capture lines thus limiting the information capacity of the device to one or a few analytes (14,27). As a step toward higher information-content lateral flow nucleic acid detection, we have developed nitrocellulose-patterning methods that enable microarray feature density to be attained on lateral flow compatible substrates. Making use of a non-contact peizo actuated picoliter deposition system, we have patterned lateral flow compatible nitrocellulose membranes with features similar in size and spacing to those typically found on spotted glass microarrays.
The sensitivity of lateral flow nucleic acid detection methods previously reported in the literature has been in the order of 1 fmol (25). We find that the LFM platform provides rapid detection of as little as 250 amol of target using a low-cost and widely available flatbed scanner, a standard personal computer system and a commercially available microarray data extraction suit or free image analysis software. This detection limit is similar to the sensitivity reported for fluorescence and chemiluminescence microarray detection strategies (9,46). While the LFM implementation reported here exhibits excellent linearity (R2 = 0.989), the linear dynamic range is less than that commonly associated with fluorescence-based detection. Alternative LFM detection schemes and the use of capture probes of differing hybridization characteristics should enable greater dynamic range while retaining the simplicity of the LFM approach. Indeed, examining the signals generated by R-77-96 and by R-24-43, the effective linear range of the LFM assay extends over a 400-fold range of target from 250 to 100 fmol (Figure 5B and C). The information density of the LFM offers the capacity for additional capture probes of varying hybridization potential to be included that should allow this dynamic range to be extended further. The uniformity of sample flow exhibited by the LFM suggests that larger capture probe sets can be accommodated without complications arising from physical factors. For example, concentrations of analyte 40-fold above the linear range of R-77-96 did not adversely impact the linearity of R-24-43 signal at LFM elements situated directly downstream (with respect to sample flow) of R-77-96 capture features (Figure 5B and C). Only at artificially high microsphere capture densities, such as those produced by the positive control hybridizations in Figure 4, are signal gradients observed as a function of physical location on the LFM, presumably due to physical occlusion of membrane pores by high local accumulations of microspheres.
LFMs offer several advantages arising directly from the miniaturization of the system without sacrificing detection sensitivity. While traditional lateral flow assays make use of sample volumes in the order of hundreds of microliters to milliliters, the miniaturization approach we have developed reduces sample volume to 10 μl. This reduced sample volume significantly decreases the consumption of reagents required for amplification. Here we have made use of 2 μl of a NASBA reaction diluted to 10 µl in running buffer. By reducing standard NASBA reaction volumes from 20 to 2 μl, a one order of magnitude reduction in enzyme consumption is realized. It should also be noted that other amplification schemes, such as those that make use of microfluidic systems or lab-on-a-chip technologies, could be integrated with a miniaturized lateral flow-based detection system to provide a rapid and cost-effective means of detecting analytes.
A further benefit of miniaturization is the time required to detect analyte following introduction of amplified material to the LFM. While the procedures used here employed NASBA amplification and traditional RNA isolation protocols requiring ∼90 min to complete, more recent advances in nucleic acid preparation and amplification have reported significant reduction in sample processing times (47). As amplification protocols become more rapid, the speed with which amplicons can be detected, without complex optical systems and fluorescent detection, becomes critical to realizing the potential of these technologies. The LFM methods described here detect nucleic acid analytes in less than 2 min. Given that 250 amol is equivalent to 1.5 × 108 molecules, efficient amplification methods that offer 109-fold amplification, widely cited amplification levels for PCR- and NASBA-based techniques (22,48), would theoretically enable the detection of single-copy targets by LFM following amplification. Future systems that couple advanced amplification technologies and compatible streamlined nucleic acid preparation modalities with rapid LFM detection will allow significant decreases in sample-to-answer times without costly or complex instrumentation.
ACKNOWLEDGEMENTS
We thank Paige Pardington for technical assistance and Lonna R. Atkeson for helpful discussions. This article was reviewed prior to release from Los Alamos National Laboratory and approved for unlimited distribution (LA-UR-07-0843). The work was funded by the U.S. Department of Homeland Security and by the U.S. Department of Agriculture. Funding to pay the Open Access publication charges for this article was provided by U.S. Department of Agriculture.
Conflict of interest statement. None declared.
REFERENCES
- 1.Huckle D. Point-of-care diagnostics: will the hurdles be overcome this time? Expert Rev. Med. Devices. 2006;3:421–426. doi: 10.1586/17434440.3.4.421. [DOI] [PubMed] [Google Scholar]
- 2.Yang S, Rothman RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004;4:337–348. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin CD, Linder V, Sia SK. Lab-on-a-chip devices for global health: past studies and future opportunities. Lab. Chip. 2007;7:41–57. doi: 10.1039/b611455e. [DOI] [PubMed] [Google Scholar]
- 4.Koch WH. Technology platforms for pharmacogenomic diagnostic assays. Nat Rev. Drug Discov. 2004;3:749–761. doi: 10.1038/nrd1496. [DOI] [PubMed] [Google Scholar]
- 5.Mackay IM. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 2004;10:190–212. doi: 10.1111/j.1198-743x.2004.00722.x. [DOI] [PubMed] [Google Scholar]
- 6.Cirino NM, Musser KA, Egan C. Multiplex diagnostic platforms for detection of biothreat agents. Expert Rev. Mol. Diagn. 2004;4:841–857. doi: 10.1586/14737159.4.6.841. [DOI] [PubMed] [Google Scholar]
- 7.Petrik J. Diagnostic applications of microarrays. Transfus. Med. 2006;16:233–247. doi: 10.1111/j.1365-3148.2006.00673.x. [DOI] [PubMed] [Google Scholar]
- 8.Heller MJ. DNA microarray technology: devices, systems, and applications. Annu. Rev. Biomed. Eng. 2002;4:129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 9.Peytavi R, Raymond FR, Gagne D, Picard FJ, Jia G, Zoval J, Madou M, Boissinot K, Boissinot M, et al. Microfluidic device for rapid (<15 min) automated microarray hybridization. Clin. Chem. 2005;51:1836–1844. doi: 10.1373/clinchem.2005.052845. [DOI] [PubMed] [Google Scholar]
- 10.Wei CW, Cheng JY, Huang CT, Yen MH, Young TH. Using a microfluidic device for 1 microl DNA microarray hybridization in 500 s. Nucleic Acids Res. 2005;33:e78. doi: 10.1093/nar/gni078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim DV, Simpson JM, Kearns EA, Kramer MF. Current and developing technologies for monitoring agents of bioterrorism and biowarfare. Clin. Microbiol. Rev. 2005;18:583–607. doi: 10.1128/CMR.18.4.583-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glynou K, Ioannou PC, Christopoulos TK, Syriopoulou V. Oligonucleotide-functionalized gold nanoparticles as probes in a dry-reagent strip biosensor for DNA analysis by hybridization. Anal. Chem. 2003;75:4155–4160. doi: 10.1021/ac034256+. [DOI] [PubMed] [Google Scholar]
- 13.Rule GS, Montagna RA, Durst RA. Rapid method for visual identification of specific DNA sequences based on DNA-tagged liposomes. Clin. Chem. 1996;42:1206–1209. [PubMed] [Google Scholar]
- 14.Dineva MA, Candotti D, Fletcher-Brown F, Allain JP, Lee H. Simultaneous visual detection of multiple viral amplicons by dipstick assay. J. Clin. Microbiol. 2005;43:4015–4021. doi: 10.1128/JCM.43.8.4015-4021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozwich D, Johansen KA, Landau K, Roehl CA, Woronoff S, Roehl PA. Development of a novel, rapid integrated Cryptosporidium parvum detection assay. Appl. Environ. Microbiol. 2000;66:2711–2717. doi: 10.1128/aem.66.7.2711-2717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuiderwijk M, Tanke HJ, Sam Niedbala R, Corstjens PL. An amplification-free hybridization-based DNA assay to detect Streptococcus pneumoniae utilizing the up-converting phosphor technology. Clin. Biochem. 2003;36:401–403. doi: 10.1016/s0009-9120(03)00057-2. [DOI] [PubMed] [Google Scholar]
- 17.Zijlmans HJ, Bonnet J, Burton J, Kardos K, Vail T, Niedbala RS, Tanke HJ. Detection of cell and tissue surface antigens using up-converting phosphors: a new reporter technology. Anal. Biochem. 1999;267:30–36. doi: 10.1006/abio.1998.2965. [DOI] [PubMed] [Google Scholar]
- 18.Duck P, Alvarado-Urbina G, Burdick B, Collier B. Probe amplifier system based on chimeric cycling oligonucleotides. Biotechniques. 1990;9:142–148. [PubMed] [Google Scholar]
- 19.Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 21.Kievits T, van Gemen B, van Strijp D, Schukkink R, Dircks M, Adriaanse H, Malek L, Sooknanan R, Lens P. NASBA isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J. Virol. Methods. 1991;35:273–286. doi: 10.1016/0166-0934(91)90069-c. [DOI] [PubMed] [Google Scholar]
- 22.Malek L, Sooknanan R, Compton J. Nucleic acid sequence-based amplification (NASBA) Methods Mol. Biol. 1994;28:253–260. doi: 10.1385/0-89603-254-x:253. [DOI] [PubMed] [Google Scholar]
- 23.Fong WK, Modrusan Z, McNevin JP, Marostenmaki J, Zin B, Bekkaoui F. Rapid solid-phase immunoassay for detection of methicillin-resistant Staphylococcus aureus using cycling probe technology. J. Clin. Microbiol. 2000;38:2525–2529. doi: 10.1128/jcm.38.7.2525-2529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baeumner AJ, Schlesinger NA, Slutzki NS, Romano J, Lee EM, Montagna RA. Biosensor for dengue virus detection: sensitive, rapid, and serotype specific. Anal. Chem. 2002;74:1442–1448. doi: 10.1021/ac015675e. [DOI] [PubMed] [Google Scholar]
- 25.Baeumner AJ, Pretz J, Fang S. A universal nucleic acid sequence biosensor with nanomolar detection limits. Anal. Chem. 2004;76:888–894. doi: 10.1021/ac034945l. [DOI] [PubMed] [Google Scholar]
- 26.Hartley HA, Baeumner AJ. Biosensor for the specific detection of a single viable B. anthracis spore. Anal. Bioanal. Chem. 2003;376:319–327. doi: 10.1007/s00216-003-1939-5. [DOI] [PubMed] [Google Scholar]
- 27.Zaytseva NV, Montagna RA, Lee EM, Baeumner AJ. Multi-analyte single-membrane biosensor for the serotype-specific detection of Dengue virus. Anal. Bioanal. Chem. 2004;380:46–53. doi: 10.1007/s00216-004-2724-9. [DOI] [PubMed] [Google Scholar]
- 28.Edwards KA, Baeumner AJ. Optimization of DNA-tagged dye-encapsulating liposomes for lateral-flow assays based on sandwich hybridization. Anal. Bioanal. Chem. 2006;386:1335–1343. doi: 10.1007/s00216-006-0705-x. [DOI] [PubMed] [Google Scholar]
- 29.Easterday WR, Van Ert MN, Simonson TS, Wagner DM, Kenefic LJ, Allender CJ, Keim P. Use of single nucleotide polymorphisms in the plcR gene for specific identification of Bacillus anthracis. J. Clin. Microbiol. 2005;43:1995–1997. doi: 10.1128/JCM.43.4.1995-1997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Easterday WR, Van Ert MN, Zanecki S, Keim P. Specific detection of bacillus anthracis using a TaqMan mismatch amplification mutation assay. Biotechniques. 2005;38:731–735. doi: 10.2144/05385ST03. [DOI] [PubMed] [Google Scholar]
- 31.Hill KK, Ticknor LO, Okinaka RT, Asay M, Blair H, Bliss KA, Laker M, Pardington PE, Richardson AP, et al. Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 2004;70:1068–1080. doi: 10.1128/AEM.70.2.1068-1080.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pannucci J, Cai H, Pardington PE, Williams E, Okinaka RT, Kuske CR, Cary RB. Virulence signatures: microarray-based approaches to discovery and analysis. Biosens. Bioelectron. 2004;20:706–718. doi: 10.1016/j.bios.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Deiman B, van Aarle P, Sillekens P. Characteristics and applications of nucleic acid sequence-based amplification (NASBA) Mol. Biotechnol. 2002;20:163–179. doi: 10.1385/MB:20:2:163. [DOI] [PubMed] [Google Scholar]
- 34.Spiro A, Lowe M, Brown D. A bead-based method for multiplexed identification and quantitation of DNA sequences using flow cytometry. Appl. Environ. Microbiol. 2000;66:4258–4265. doi: 10.1128/aem.66.10.4258-4265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albretsen C, Haukanes BI, Aasland R, Kleppe K. Optimal conditions for hybridization with oligonucleotides: a study with myc-oncogene DNA probes. Anal. Biochem. 1988;170:193–202. doi: 10.1016/0003-2697(88)90108-x. [DOI] [PubMed] [Google Scholar]
- 36.Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3:195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- 37.Blake RD, Delcourt SG. Thermodynamic effects of formamide on DNA stability. Nucleic Acids Res. 1996;24:2095–2103. doi: 10.1093/nar/24.11.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baeumner AJ, Leonard B, McElwee J, Montagna RA. A rapid biosensor for viable B. anthracis spores. Anal. Bioanal. Chem. 2004;380:15–23. doi: 10.1007/s00216-004-2726-7. [DOI] [PubMed] [Google Scholar]
- 39.Guo Z, Guilfoyle RA, Thiel AJ, Wang R, Smith LM. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 1994;22:5456–5465. doi: 10.1093/nar/22.24.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Day PJR, Flora PS, Fox JE, Walker MR. Immobilization of polynucleotides on magnetic particles: Factors Influencing hybridization efficiency. Biochem. J. 1991;278:735–740. doi: 10.1042/bj2780735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Meara D, Nilsson P, Nygren PA, Uhlen M, Lundeberg J. Capture of single-stranded DNA assisted by oligonucleotide modules. Anal. Biochem. 1998;255:195–203. doi: 10.1006/abio.1997.2472. [DOI] [PubMed] [Google Scholar]
- 42.Lane MJ, Paner T, Kashin I, Faldasz BD, Li B, Gallo FJ, Benight AS. The thermodynamic advantage of DNA oligonucleotide ‘stacking hybridization’ reactions: energetics of a DNA nick. Nucleic Acids Res. 1997;25:611–617. doi: 10.1093/nar/25.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Meara D, Yun Z, Sonnerborg A, Lundeberg J. Cooperative oligonucleotides mediating direct capture of hepatitis C virus RNA from serum. J. Clin. Microbiol. 1998;36:2454–2459. doi: 10.1128/jcm.36.9.2454-2459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kandimalla ER, Manning A, Lathan C, Byrn RA, Agrawal S. Design, biochemical, biophysical and biological properties of cooperative antisense oligonucleotides. Nucleic Acids Res. 1995;23:3578–3584. doi: 10.1093/nar/23.17.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kieleczawa J, Dunn JJ, Studier FW. DNA sequencing by primer walking with strings of contiguous hexamers. Science. 1992;258:1787–1791. doi: 10.1126/science.1465615. [DOI] [PubMed] [Google Scholar]
- 46.Cheek BJ, Steel AB, Torres MP, Yu YY, Yang H. Chemiluminescence detection for hybridization assays on the flow-thru chip, a three-dimensional microchannel biochip. Anal. Chem. 2001;73:5777–5783. doi: 10.1021/ac0108616. [DOI] [PubMed] [Google Scholar]
- 47.Roper MG, Easley CJ, Landers JP. Advances in polymerase chain reaction on microfluidic chips. Anal. Chem. 2005;77:3887–3893. doi: 10.1021/ac050756m. [DOI] [PubMed] [Google Scholar]
- 48.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]