Abstract
Meganucleases are sequence-specific endonucleases with large cleavage sites that can be used to induce efficient homologous gene targeting in cultured cells and plants. These enzymes open novel perspectives for genome engineering in a wide range of fields, including gene therapy. A new crystal structure of the I-CreI dimer without DNA has allowed the comparison with the DNA-bound protein. The C-terminal loop displays a different conformation, which suggests its implication in DNA binding. A site-directed mutagenesis study in this region demonstrates that whereas the C-terminal helix is negligible for DNA binding, the final C-terminal loop is essential in DNA binding and cleavage. We have identified two regions that comprise the Ser138–Lys139 and Lys142–Thr143 pairs whose double mutation affect DNA binding in vitro and abolish cleavage in vivo. However, the mutation of only one residue in these sites allows DNA binding in vitro and cleavage in vivo. These findings demonstrate that the C-terminal loop of I-CreI endonuclease plays a fundamental role in its catalytic mechanism and suggest this novel site as a region to take into account for engineering new endonucleases with tailored specificity.
INTRODUCTION
Meganucleases are sequence-specific enzymes which recognize large (12–45 bp) DNA target sites. These enzymes are often encoded by introns or inteins behaving as mobile genetic elements. They recognize sites that usually correspond to intron-free or intein-free genes, where they produce a DNA double-strand break (DSB). Eventually, DSB repair by homologous recombination with an intron- or intein-containing gene results in the insertion of the intron or intein where DSB occurred in specific loci in living cells (1). Meganucleases are used to stimulate homologous recombination in the vicinity of their target sequences in cultured cells and plants (2–6). These results present new perspectives for genome engineering in a wide range of applications, such as the correction of mutations linked with monogenic inherited diseases or the bypass of risks due to the randomly inserted transgenes used in current gene therapy approaches (7).
The use of meganuclease-induced recombination has long been limited by the repertoire of natural meganucleases. In nature, meganucleases are essentially represented by homing endonucleases (HEs), a family of endonucleases encoded by mobile genetic elements, whose function is to initiate DSB-induced recombination events in a process referred to as homing (8). Several hundreds of HEs have been identified in bacteria, eukaryotes and archaea (8); however, the probability of finding a HE cleavage site in a chosen gene is low. Thus, the making of artificial meganucleases with custom-made substrate specificity is an intense area of research (9–12). Lately, zinc-finger DNA-binding domains (13) could be fused with the catalytic domain of the FokI endonuclease, to induce recombination in various cell types, including human lymphoid cells (14–16). However, these chimeric proteins showed high toxicity in cells (14,17), probably due to a low level of specificity. Given their biological function and their exquisite specificity, HEs represent ideal scaffolds to engineer the substrate specificity of proteins that cleave or recombine DNA (18–21).
Sequence homology has been used to classify HEs into four families, the largest one having the conserved LAGLIDADG sequence motif (22). HEs with only one such motif, such as I-CreI (23), function as homodimers. In contrast, larger HEs containing two motifs, such as I-SceI (18) or I-DmoI (19), are single-chain proteins. The 3D structures solved for several LAGLIDADG endonucleases (8, 20, 21, 24–32) indicate that these proteins adopt a similar active conformation as homodimers or as monomers with two separate domains. The LAGLIDADG motifs form structurally conserved α-helices tightly packed at the center of the interdomain or intermonomer interface (8,20, 21, 24–30). On either side of the LAGLIDADG α-helices, a four-stranded β-sheet provides a DNA-binding interface that drives the interaction of the protein with each one of the half sites of the target DNA sequence (8,24). The last acidic residue of the LAGLIDADG motif participates in the DNA cleavage by a metal-dependent mechanism of phosphodiester hydrolysis (22).
The I-CreI structure has been solved without DNA before. In this study, the protein crystallized with only one monomer in the asymmetric unit (33). We have solved the structure of the I-CreI dimer without DNA; its comparison with the DNA-bound crystal structure (8) depicts a different conformation for the C-terminal loop which connects the last two helices α5 and α6. These two structural elements (C-terminal loop and α6 helix) are hereafter referred to as the C-loop and the C-helix. The sequence of the C-terminal loop is highly conserved among dimeric LAGLIDADG HEs and it is involved in contacts with the DNA backbone (33,34). To unravel the function of this region in the endonuclease mechanism of I-CreI, we designed trimmed, double and single mutants in this area. Binding and cleavage experiments illustrate the important role of the residues located at the C-terminal loop in DNA binding and cleavage. Even though different regions that define novel target DNA specificities have been identified in the I-CreI scaffold (35), this work reveals a novel site essential for binding and cleavage which can also be engineered to generate novel specificities.
MATERIALS AND METHODS
Protein expression, purification and crystallization
All the constructions used in this work are based on I-CreI pET24-d(+) plasmid used in (36). Protein expression and purification were performed as in (11). Site-directed mutagenesis was performed using the Quickchange XL site-directed mutagenesis kit from STRATAGENE (www.stratagene.com). All the mutations were checked by DNA sequencing (data not shown). An initial screening for I-CreI crystallization conditions was performed in 96-well plates by vapor-diffusion methods using the Hampton crystal screening using drops containing 1 μl protein solution (7 mg/ml in 20 mM HEPES pH 7.5) and 1 μl precipitant solution equilibrated against 50 μl of reservoir solution at 20°C. Crystals were obtained under several conditions (Crystal Screen 1 conditions 10, 22, 33, 40, 41 and Crystal Screen 2 condition 32). Crystal was made by hanging-drop vapor-diffusion methods using VDX plates; optimization experiments led to the following conditions for crystallization: 1 μl protein at 7 mg/ml in 20 mM HEPES pH 7.5 and 1 μl precipitating buffer containing 20% PEG 4000, 0.1 M HEPES pH 7.5, 10% Iso-propanol, 10% ethylene glycol and 0.01 M magnesium acetate equilibrated against 500 μl precipitating buffer at 20°C. Rod-shaped crystals grown in 4–8 days and were directly collected and frozen in liquid nitrogen.
Data collection, structure solution, model building and refinement
All data were collected at cryogenic temperatures using synchrotron radiation at 100 K. I-CreI crystals were mounted and cryoprotected. The data sets were collected using synchrotron radiation at the ID14-4 beamline at the ESRF (Grenoble), and at the PX beamline at the SLS (Villigen). Diffraction data were recorded on an ADSC-Q4 or Mar225 CCD detectors depending on the beamline. The best data set (Table 1) was collected using a Δϕ = 1 and a wavelength of 0.97 Å. Processing and scaling were accomplished with HKL2000 (37). Statistics for the crystallographic data are summarized in Table 1. The structure was solved using the molecular replacement method as implemented in the program MOLREP (38). The search model was based on a polyalanine backbone derived from the PDB entry 1GZ9. The coordinates from the DNA were deleted in the search model. A refined 2Fo–Fc map showed clear and contiguous electron density for the protein backbone and for many of the side-chains. ARP/wARP and REFMAC5 were applied for automatic model building and refinement to 2.0 Å (Table 1). The coordinates have been deposited in the PDB (2O7M).
Table 1.
Data collection and refinement statistics
| Data collection | |
|---|---|
| Space group | P43 |
| Number of crystals | 1 |
| Temperature (K) | 100 |
| Wavelength (Å) | 0.97 |
| Cell dimensions (Å, °), | a = b = 69.088, c = 93.040 α = β = γ = 90° |
| No. molecules asymmetric unit | 2 |
| Data collection environment, beamline | ADSC-Q4, ID14-4 ESRF |
| Completeness (%) | 93.2 |
| Multiplicity | 5.3 |
| Rsym(%)a | 6 |
| Refinement | |
| Number of reflections | 25 943 |
| Resolution range (Å) | 50–2.00 |
| R-factor/R-free (%) | 18/23 |
| Number of protein atoms (Average B, Å2)b | 2484/33 |
| Number of water molecules (Average B, Å2)b | 262/44 |
| r.m.s. bond length (Å) | 0.029 |
| r.m.s. bond angle (°) | 2.137 |
| Ramachandran plot outliers (number)c | 0 |
aRsym = Ση Σt|Iη,t − < Iη >/Ση Σt|Iη,t|
b Calculated using MOLEMAN.
c Calculated using PROCHECK.
Circular dichroism thermal analysis
Data were acquired with a Jasco 810 model dichrograph, previously calibrated with d-10-camphorsulfonic acid, and equipped with a Jasco Peltier thermoelectric temperature controller CDF-426S. Experiments were performed in phosphate buffer saline (PBS, 137 mM NaCl, 10 mM Na2HPO4·2H2O, 2.7 mM KCl, 2 mM KH2PO4, pH 7.4) at 1°C/min intervals. The protein concentration was 10 μM. The ellipticity at 222 nm was followed from 5 to 95°C in a 2 mm Hellma 110-QS cell.
Analytical ultracentrifugation
Sedimentation equilibrium experiments were performed at 20°C in an Optima XL-A (Beckman-Coulte, Inc.) analytical ultracentrifuge equipped with UV–visible optics, using an An50Ti rotor, with 3 mm double-sector centerpieces of Epon charcoal. Protein concentration was 200 μM in PBS buffer. Short column (23 µl), low-speed sedimentation equilibrium was performed at three successive speeds (11 000, 13 000 and 15 000 r.p.m.), the system was assumed to be at equilibrium when successive scans overlaid and the equilibrium scans were obtained at a wavelength of 280 nm. The base-line signal was measured after high-speed centrifugation (5 h at 42 000 r.p.m.). Whole-cell apparent molecular weight of the protein was obtained using the program EQASSOC (39). The partial specific volume of I-CreI was 0.7436 ml/g at 20°C, calculated from the amino acid composition with the program SEDNTERP (downloaded from the RASMB server) (40).
The sedimentation velocity experiment was carried out in an XL-A analytical ultracentrifuge (Beckman-Coulte, Inc.) at 42 000 r.p.m. and 20°C, using an An50Ti rotor and 1.2 mm double-sector centerpieces. Absorbance scans were taken at 280 nm. The protein concentration was 50 μM in PBS. The sedimentation coefficients were calculated by continuous distribution c(s) Lamm equation model (41) as implemented in the SEDFIT program. These experimental sedimentation values were corrected to standard conditions to get the corresponding s20,w values using the SEDNTERP program (40). Further hydrodynamic analysis (i.e. calculation of frictional coefficient ratio) was performed with the SEDFIT program to obtain de c(M) distribution (41).
NMR data acquisition
NMR spectra were recorded at 25°C in a Bruker AVANCE 600 spectrometer equipped with a cryoprobe. Protein samples were 500 μM in PBS buffer plus 5% 2H2O. 2,2-Dimethyl-2-silapentane-5-sulfonate sodium salt (DSS) was used as internal proton chemical shift reference.
Band-shift assay conditions and complex formation titration
Band-shift assays were performed in 10 mM Tris-HCl pH 8, 50 mM NaCl, 10 mM CaCl2 or MgCl2, 1 mM DTT incubated 1 h at room temperature using 5 μM (0.0793 μg/μl) 6-FAM duplex and 20 μM (0.463 μg/μl) protein. After incubation, the samples were subjected to electrophoresis using a 15% acrylamide-TBE gel.
Titration curves were performed with 500 nM 6-FAM DNA duplex and 0–10 000 nM protein (monomer concentration). Assuming that one molecule of duplex DNA binds to one molecule of I-CreI protein dimer, the dissociation constants (KD) (Table 2) were determined by data fitting (Origin, Microcal) to the equation: [DNA–P] = [DNA–P]max X ((KD + [DNA] + [P] – sqrt((KD + [DNA] + [P])2 – 4 × [DNA] × [P])))/(2X[DNA]), where [DNA–P] is the concentration of the DNA–protein complex, [DNA–P]max is the maximum possible concentration of complex, [DNA] is the total concentration of 6-FAM DNA duplex (500 nM), [P] is the total concentration of the protein (dimer protein concentration). The value of [DNA–P] was calculated from the reduction in the intensity of the lower band in the gel (free DNA) as the intensity of the shifted band (bound DNA) was smaller than the loss of intensity in the lower band, due to fluorescence quenching or dampening by the bound protein (gels are provided in Supplementary Data). The adjustable parameters during the fitting were [DNA–P]max and KD.
Table 2.
Dissociation constants for the binding of 6-FAM duplex (see Figure 3) to I-CreI C-helix mutants in the presence of Ca2+
| Protein | KD (nM) |
|---|---|
| I-CreI | 0.2 |
| I-CreI T143G | 5 |
| I-CreI K142G | 40 |
| I-CreI S138A | 250 |
| I-CreI K139M | 110 |
| I-CreI Δ2 | 170 |
| I-CreI GG | 50 |
| I-CreI AM | 1240 |
| I-CreI Δ1 | No binding detected |
In vitro cleavage assay conditions
Cleavage assays were performed at 37°C in 10 mM Tris-HCl (pH 8), 50 mM NaCl, 10 mM MgCl2 and 1 mM DTT. The amount of enzyme and target were: 100 ng for the XmnI-linearized DNA substrate (pGEM-T Easy 10AAA_5GTC_P) (11,36) and 0.25–120 ng dilutions for I-CreI and helix mutant proteins, in 25 μl final volume reaction. The linearized target plasmid has 3 kb and after cleavage yields two smaller bands of 2 and 1 kb. Reactions were stopped after 1 h by the addition of 5 μl of 45% glycerol, 95 mM EDTA (pH 8), 1.5% (w/v) SDS, 1.5 mg/ml proteinase K and 0.048% (w/v) bromophenol blue (6× buffer stop), incubated at 37°C for 30 min and electrophoresed in a 1% agarose gel. The gels were stained using SYBR Safe DNA gel staining (INVITROGEN) and the intensity of the bands observed upon UV light illumination were quantified with the ImageJ software (http://rsb.info.nih.gov/ij/). The percentage of cleavage was calculated with the following equation: percentage cleavage = 100 X (I2kb + I1kb)/(I3kb + I2kb + I1kb), where I1kb, I2kb and I3kb are the intensities of the 1-, 2- or 3-kb bands. The cleavage rate was calculated using the enzyme concentration needed to cut 50% of the target DNA (C50) (Table 3).
Table 3.
C50 protein concentration needed to cleave 50% of the target DNA in the in vitro assay (Figure 4)
| Protein | C50 (nM) |
|---|---|
| I-CreI | 2.8 |
| I-CreI T143G | 3.8 |
| I-CreI K142G | 4.5 |
| I-CreI S138A | 3.4 |
| I-CreI K139M | 1.8 |
| I-CreI Δ2 | 41.8 |
| I-CreI GG | 476 |
| I-CreI AM | 64 |
| I-CreI Δ1 | No cleavage detected |
Construction of target clones
The 10AAA_5GTC_P 24-bp target sequence (TCAAAACGTCGTACGACGTTTTGA) is a palindrome of a half-site of the natural I-CreI target (TCAAAACGTCGTGAGACAGTTTGG). 10AAA_5GTC_P is cleaved as efficiently as the I-CreI natural target in vitro and ex vivo in both yeast and mammalian cells. The palindromic targets, derived from 10AAA_5GTC_P, were cloned as previously described (11) into the reporter vectors: the yeast pFL39-ADH-LACURAZ (using the Gateway protocol, Invitrogen), containing a I-SceI target site as a control. Yeast reporter vectors were transformed into Saccharomyces cerevisiae strain FYBL2-7B (MAT a, ura3Δ851, trp1Δ63, leu2Δ1, lys2Δ202).
Mating of meganuclease expressing clones and screening in yeast
For screening homodimer mutants, we used the protocol described previously (11). Briefly, mating was performed using a colony gridder (QpixII, Genetix). Mutants were gridded on nylon filters covering YPD plates, using a high gridding density (∼20 spots/cm2). A second gridding process was performed on the same filters to spot a second layer consisting of 64 or 75 different reporter-harboring yeast strains for each variant. Membranes were placed on solid agar YPD-rich medium, and incubated at 30°C for one night, to allow mating. Next, filters were transferred to synthetic medium, lacking leucine and tryptophan, with galactose (1%) as a carbon source, and incubated for 5 days at 37°C (30°C for I-CreI), to select for diploids carrying the expression and target vectors. After 5 days, filters were placed on solid agarose medium with 0.02% X-Gal in 0.5 M sodium phosphate buffer, pH 7.0, 0.1% SDS, 6% dimethyl formamide (DMF), 7 mM β-mercaptoethanol, 1% agarose, and incubated at 37°C to monitor β-galactosidase activity. Results were analyzed by scanning and quantification was performed using proprietary software.
RESULTS AND DISCUSSION
Structural differences between the bound and unbound I-CreI DNA structures
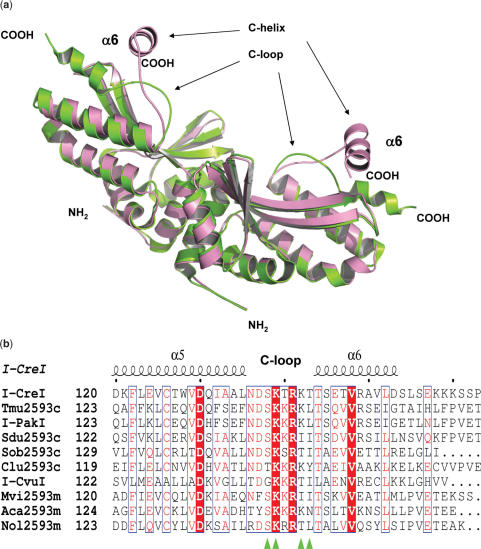
The structure of the I-CreI was solved by molecular replacement and refined to 2.0 Å resolution. The dimer without DNA allowed us to observe the protein conformational changes upon DNA binding after comparison with the protein–DNA complex (PDB code 1G9Z) (Figure 1a). The most striking differences could be observed in the conformation of the C-terminal region. Whereas in the DNA-bound structure the C-helix and the C-loop are aligned with the DNA, in the unbound structure both elements are located on top of the cavity where the DNA binds, suggesting that the loop and the C-helix could work as a lock opening and closing the DNA-binding groove. This region was not observed in a previous structure of I-CreI with only one monomer in the asymmetric unit (33). Besides, the C-terminal domain of I-CreI is well conserved among other members of its family (42) indicating its important role in this meganuclease group working mechanism (Figure 1b). A detailed view of the protein–DNA interactions in the C-terminal area showed that Ser138, Lys139, Lys142 and Thr143 at the SKTRKT motif are involved in hydrogen bonds with the DNA backbone (34) (Figure 1c). The position of these residues is completely different in the unbound DNA state (Figure 1d), indicating that a conformational change is needed to bind the nucleic acid. Although these interactions were described before (34) and the amino acids are conserved, there is little information about their role during meganuclease action (12,43).
Figure 1.
Superposition of the bound and unbound DNA I-CreI structure. (a) Cα ribbon representation of I-CreI (pink) and the I-CreI-DNA (green) structures. DNA has been omitted for clarity. (b) Sequence alignment of the C-terminal region from members of the I-CreI family (42). The position of the mutated residues in the SKTRKT motif is indicated with a green triangle (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). (c) Detailed view of S138, K139, K142 and T143 contacts with the DNA backbone. (d) Comparison of the positions of S138, K139, K142 and T143 between the bound (green) and unbound (pink) DNA structures.
Design of I-CreI mutants
To unravel the role of the C-terminal region of I-CreI, a series of trimmed, double and single mutants were designed based on the structural differences between the bound and unbound DNA structures. The two truncated mutants were designed to clarify the role of the C-loop and the C-helix. I-CreI Δ1 (amino acids 1–137) lacked both the C-loop and the C-helix whereas I-CreI Δ2 (amino acids 1–144) contained the C-loop. Based on the contacts with the DNA backbone in the SKTRKT motif, the double mutants I-CreI AM (S138A and K139M) and I-CreI GG (K142G and T143G) were produced, as well as their single variants I-CreI S138A, I-CreI K139M, I-CreI K142G and I-CreI T143G. The S138A and K139M mutations were designed to change the polar character of the corresponding side chains with minimal alteration of their sizes. On the other hand, The K142G and T143G mutants were designed to maintain the polarity and the flexibility of the loop, whereas the size of the side chain was reduced.
I-CreI mutants are dimers as the wild type
To demonstrate that the effect in meganuclease activity was due to the mutations and not to structural changes, we analyzed their structure stability and oligomerization state. Thermal denaturation followed by circular dichroism of all the mutants showed cooperative, sigmoidal transitions similar to that of the wild type (Supplementary Data Figure 1a). Even though the denaturation is irreversible and causes protein precipitation, this result is consistent with the presence of a folded tertiary structure, although with small changes in stability as indicated by the narrow range of apparent mid point temperature values. This tertiary structure is the same for all the mutants and the WT as shown by the same set of dispersed signals observed in (Supplementary Data Figure 1b) their 1D 1H NMR spectra. It is well known that the I-CreI family of meganucleases binds DNA as homodimers, therefore to analyze the oligomerization state of the mutants they were subjected to analytical ultracentrifugation. The experiment showed that all the mutants behaved as dimers independently of the mutation (Supplementary Data Figure 1c). Altogether, these experiments indicate that the mutants are folded and conserve the I-CreI scaffold involved in meganuclease activity.
Ser138-Lys139 and Lys142-Thr143 constitute two new sites essential for DNA binding
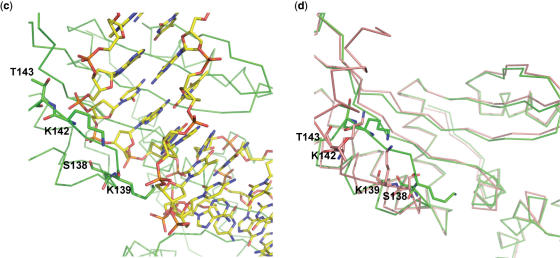
Electrophoretic mobility shift assays (EMSAS) in the presence of Mg2+ and Ca2+ were used to analyze qualitatively the behavior of the C-terminal mutants in DNA binding (Figure 2). Whereas the presence of Ca2+ allows DNA binding, Mg2+ is indispensable to bind and cleave DNA (12). Even though the binding capability of I-CreI was abolished in the Δ1 mutant, the Δ2 one was able to bind the labeled DNA probe demonstrating that the C-loop is essential in DNA binding while the C-helix is not. In addition, binding was detected in the presence of both cations as in the wild-type I-CreI.
Figure 2.
Electrophoretic mobility shift assays of the C-terminal truncated, double and single mutants in the presence of Mg2+ and Ca2+ (Gels with the experimental data for the KD measurements are available as Supplementary Data).
On the other hand, both I-CreI AM and I-CreI GG double mutants were affected in their DNA-binding properties independently of the cation present, indicating that Ser138, Lys139, Lys142 and Thr143 contacts with the DNA backbone are crucial to bind the nucleic acid. Therefore, these residues in the SKTRKT motif constitute two new sites essential for I-CreI DNA binding.
To define the distinct properties of each site in the C-loop, the single mutants were assayed by EMSA in the same conditions. The binding of the labeled probe to the single mutants is less affected than to the double ones; however, they displayed differences depending on the cation present in the assay. Whereas a strong dependence of Mg2+ could be observed in the Ser138–Lys139 site, the single mutants in the Lys142–Thr143 site could bind DNA notwithstanding the cation present in the mobility assay.
These differences could be observed quantitatively after the measurement of the dissociation constants for all the different mutants in the presence of Ca2+ (Table 2). The affinity of the wild-type is similar to that measured by others (44), however all the mutants presented affinities that are reduced by a factor of 24 to 6000, the effect being larger for the double mutants as compared to the corresponding single ones, indicating a synergy between the two residues in each region for DNA binding.
Ser138–Lys139 and Lys142–Thr143 regions are needed for DNA cleavage activity
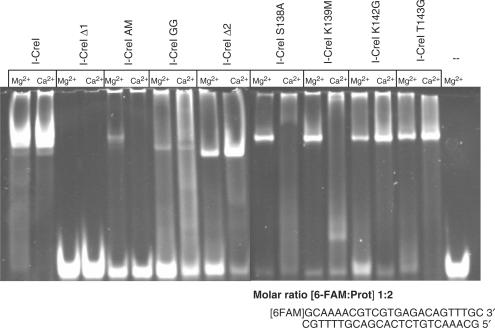
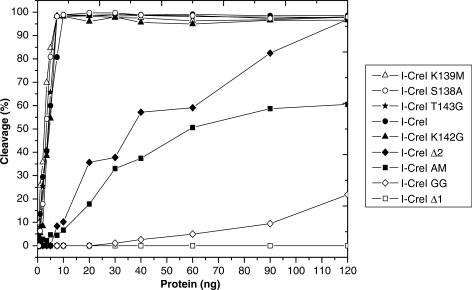
The analysis of the different mutants in the DNA-binding assays has clear implications for DNA cleavage activity, consequently an examination of their cleavage properties on a wild-type DNA sequence was carried out. Figure 3 displays a graph representing the percentage of cleavage against the amount of HE (the original gels are available as Supplementary Data). The mutants can be divided into two groups based on the comparison of their cleavage properties to the wild-type HE; the first is composed of the truncated mutants I-CreI Δ1 and I-CreI Δ2 and the double mutants I-CreI AM and I-CreI GG, whereas the single mutants I-CreI S138A, I-CreI K139M, I-CreI K142G and I-CreI T143G form the second. Members of the first group displayed a reduced cleavage activity when compared to the wild-type I-CreI (Figure 3 and Table 3). Although I-CreI Δ1 and I-CreI GG cleavage properties are abolished or severely affected, I-CreI Δ2 and I-CreI AM showed a reduced activity (Table 3) that is increased when higher HE amounts are used (Figure 3). However, the cleavage properties of the single mutants that composed the second group are similar to the wild type (Figure 3 and Table 3).
Figure 3.
Summary of the gel in vitro cleavage assay of the C-terminal truncated, double and single mutants. The gels with the experimental data for all the cleavage experiments are available as Supplementary Data.
These results indicate that the trimmed and double mutants whose DNA binding is abolished or affected do not cleave DNA or they need higher amounts of HE to cleave the plasmid. It is noteworthy that the I-CreI Δ2 mutant which conserves the wild-type amino acids in the C-loop but lacks the C-helix, even though its cleavage activity is reduced (Figure 3 and Table 3), is able to cleave the target, while the Δ1 is not, indicating that the C-loop is essential for cleavage but not the C-helix. On the other hand, the single mutants depict an activity similar to the wild type.
In vivo assays demonstrate that the two regions in the C-loop are essential for DNA cleavage and influence target specificity
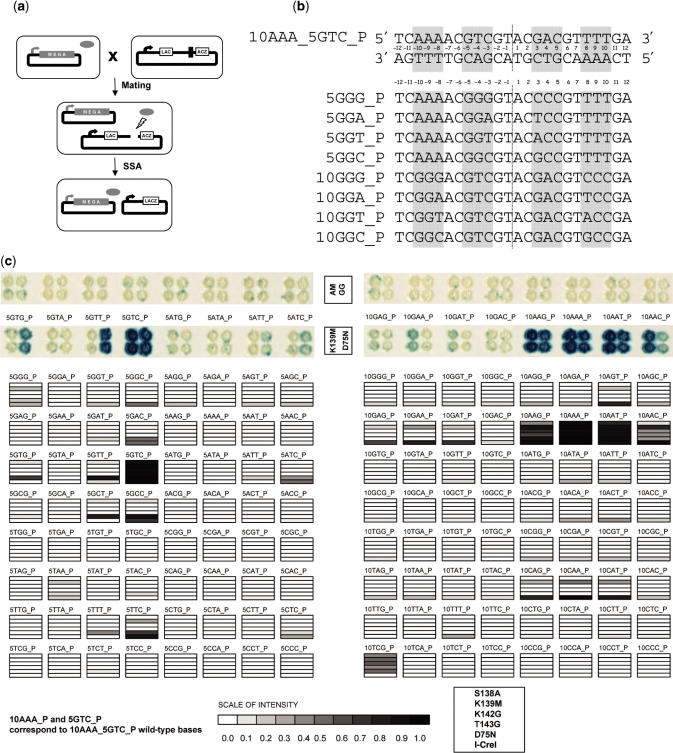
To confirm our results in vivo, we performed cleavage assays with all the mutants as in (45), including the I-CreI and I-CreI D75N proteins, as well with the 128 possible 5NNN_P and 10NNN_P targets (Figure 4c). The use of the D75N mutation decreases the toxicity of I-CreI in overexpression experiments, and the wild type and its D75N mutant display similar in vitro activities and levels of specificity (45). Cleavage patterns were similar to those previously found with I-CreI and I-CreI D75N (45). None of the trimmed or double mutants, whose binding and cleavage activities were affected by the mutations in vitro, displayed activity on any of these targets. As an example, the I-CreI AM and I-CreI GG are shown (Figure 4c, upper panel). Interestingly, the four single mutants cleave in vivo the 5GTC_P target (Figure 4). Whereas I-CreI K139M is able to cleave 5GGG_P, 5TAA_P and 5TTC_P targets, the I-CreI S138A cleaves 5GTG_P, 5GTT_P, 5GCT_P and 5GCC_P targets displaying a very similar profile to that of I-CreI D75N (Figure 4c). In contrast, all the single mutants showed activity on 10AAG_P and 10AAT_P target, in addition to the wild-type 10AAA_P. Lower levels of cleavage could also be observed with these four mutants with 10TCG_P and 10AAC_P. In addition, the I-CreI K139M mutant was also able to cleave seven additional targets, as it can be observed in Figure 4c. The profile of the I-CreI K139M mutant is very similar to that of I-CreI (without its toxicity) while the three other single mutants are closer to I-CreI D75N (45). It is noteworthy that I-CreI S138A, the only position of the four mutated residues that makes contacts with the 5NNN_P region of the DNA backbone (34), shows a 5NNN_P cleavage profile similar to I-CreI D75N.
Figure 4.
Profiling of single mutants. (a) Yeast screening assay principle. A strain harboring the expression vector encoding a single mutant is mated with a strain harboring a reporter plasmid. In the reporter plasmid, a LacZ reporter gene is interrupted with an insert containing one of the target sites of interest, flanked by two direct repeats. Upon mating, the meganuclease (gray oval) generates a double-strand break at the site of interest, allowing restoration of a functional LacZ gene by single-strand annealing (SSA) between the two flanking direct repeats. (b) DNA targets. The 10AAA_5GTC_P target (top) is a palindrome cleaved by I-CreI. The different name (The 10AAA_P or 5GTC_P) indicates whether it was used in the 5NNN_P or 10NNN_P experiment. All targets used in this study are palindromes derived from 10AAA_5GTC_P by substitution of six nucleotides in ±8, ±9 and ±10 or in ±3, ±4 and ±5. A few examples are shown (bottom). The 10GGG_P target differs from the 10AAA_P target by the GGG triplet in –10, –9, –8 and CCC in 8, 9, 10, etc. The 5GGG_P target differs from 10AAA_5GTC_P by the GGG triplet in −5, −4, −3 and CCC in 3, 4, 5, etc. (c) Mutant target profiles. Each mutant was profiled in yeast on a series of 128 palindromic targets (5NNN_P or 10NNN_P). An example of cleavage activity of 15 targets in yeast is presented for a single mutant (K139M) and I-CreI D75N. As described previously (11), blue staining indicates cleavage. Additionally a representation of the cleavage profile of all single mutants compared to I-CreI D75N and I-CreI. Gray levels reflect the intensity of the signal. I-CreI is toxic in yeast (11) and profiles have been established at 30°C instead of 37°C. All other mutants were studied at 37°C.
CONCLUSIONS
Modifying the substrate specificity of DNA-binding proteins by mutagenesis and screening/selection is a difficult task. This is even harder in the case of HEs whose main characteristic is their large DNA recognition sites. Several laboratories have relied on a semi-rational approach to limit the mutant libraries to be handled, choosing a small set of relevant residues based on structural data. This set is generally composed of residues that in the I-CreI DNA complex structure make direct contacts with the homing site. However, the comparison of the C-terminal region conformations in the I-CreI and the I-CreI DNA complex suggested that this region could be involved in a lock mechanism with clear implications in DNA binding and cleavage (Figure 1).
In this article, we have dissected the role of the C-terminal region of I-CreI in the HE mechanism. After comparison between the I-CreI structures in different DNA-bound states, we identified two regions in the SKTRKT motif involved in DNA binding.
To address the role of these amino acids in the endonuclease mechanism of I-CreI, we designed trimmed mutants in the C-terminal area and double and single mutants in the SKTRKT motif. The finding of these two regions demonstrates that the C-loop plays an essential role in DNA binding (Figure 2) and this function is performed in a concerted manner between the two amino acids in the site because the double mutants were preferentially affected in cleavage, both in vitro and in vivo.
Remarkably, the mutations K142G and T143G bind the DNA probe with higher affinity than the S138A and K139M ones (Table 3). This difference between the two sites is probably due to their distinct proximities to the residues involved in DNA cleavage and metal binding. The well-conserved D137 (Figure 1b) has been proposed to contribute to the organization of the water shell around the scissile phosphate in the metal-binding site (44). Thereby, mutations in the surrounding area such as S138A and K139M, where residues that can promote hydrogen bonds are suppressed, could disturb the water shell in the metal-binding site, turning it more selective towards the physiological cation. This is not the case for the K142G and T143G mutations located ∼23 Å away from the metal-binding site, too far to promote changes in the solvatation in that area.
Even though the I-CreI AM and the I-CreI Δ2 showed reduced cleavage activity in vitro (Figure 3 and Table 3), they did not show any activity in vivo suggesting that this residual activity was due to the large amount of protein used during the assay. The yeast cleavage assay confirmed that double and trimmed mutations abolished cleavage activity whereas the single mutants displayed a clear activity (Figure 4c).
In addition, our results indicate that protein regions lacking base-specific homing site contacts could play an important role not only in binding and cleavage, but also perhaps in target specificity (Figure 4c). New specificities could be achieved by the creation of new base-specific contacts or the release of negative interactions. If wild-type I-CreI makes energetically non-favorable contacts with a particular 10NNN_5NNN_P site, then a mutant endonuclease could interact better with that site simply by eliminating the unfavorable interactions; this seems to be the explanation for the results in Figure 4c. Therefore, the similar cleavage profile exhibited by the I-CreI S138A mutant and the D75N could arise from the release of energetically non-favorable contacts present in wild-type I-CreI.
The use of meganuclease-induced recombination has long been limited by the repertoire of natural meganucleases. For genome-engineering applications, the major advantage of HEs is their exquisite specificity, a feature that becomes essential when engaging into therapeutic applications. Thus, the production of artificial endonucleases with custom specificities is an intense area of research. The finding that specificity of the target DNA could be controlled by amino acids lacking base-specific homing site contacts with the DNA complicates the production of intelligent molecular scalpels that recognize and substitute certain DNA sequences; however, this knowledge can be used to improve even more the specificity of the engineered enzymes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online
ACKNOWLEDGEMENTS
We would like to thank the ESRF (ID14-4) and SLS (PX) biocrystallography beamlines personnel for their help during data collection. We thank Jerome Mikolajczak for the graphical representation of yeast results and Elena Ramos for her helpful technical assistance. Funding to pay the Open Access publication charges for this article was provided by Cellectis CBE1806B.
Conflict of interest statement. None declared.
REFERENCES
- 1.Thierry A, Dujon B. Nested chromosomal fragmentation in yeast using the meganuclease I-Sce I: a new method for physical mapping of eukaryotic genomes. Nucleic Acids Res. 1992;20:5625–5631. doi: 10.1093/nar/20.21.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl Acad. Sci. USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol. Cell. Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiurazzi M, Ray A, Viret JF, Perera R, Wang XH, Lloyd AM, Signer ER. Enhancement of somatic intrachromosomal homologous recombination in Arabidopsis by the HO endonuclease. Plant Cell. 1996;8:2057–2066. doi: 10.1105/tpc.8.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 8.Chevalier BS, Monnat R.J., Jr, Stoddard BL. The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nat. Struct. Biol. 2001;8:312–316. doi: 10.1038/86181. [DOI] [PubMed] [Google Scholar]
- 9.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat R.J., Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gimble FS, Moure CM, Posey KL. Assessing the plasticity of DNA target site recognition of the PI-SceI homing endonuclease using a bacterial two-hybrid selection system. J. Mol. Biol. 2003;334:993–1008. doi: 10.1016/j.jmb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 12.Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 2002;30:3870–3879. doi: 10.1093/nar/gkf495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 14.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 15.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 16.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 17.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 19.Dalgaard JZ, Garrett RA, Belfort M. A site-specific endonuclease encoded by a typical archaeal intron. Proc. Natl Acad. Sci. USA. 1993;90:5414–5417. doi: 10.1073/pnas.90.12.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flick KE, McHugh D, Heath JD, Stephens KM, Monnat R.J., Jr, Stoddard BL. Crystallization and preliminary X-ray studies of I-PpoI: a nuclear, intron-encoded homing endonuclease from Physarum polycephalum. Protein Sci. 1997;6:2677–2680. doi: 10.1002/pro.5560061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva GH, Dalgaard JZ, Belfort M, Van Roey P. Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI. J. Mol. Biol. 1999;286:1123–1136. doi: 10.1006/jmbi.1998.2519. [DOI] [PubMed] [Google Scholar]
- 22.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Kim HH, Yuan X, Herrin DL. Purification, biochemical characterization and protein-DNA interactions of the I-CreI endonuclease produced in Escherichia coli. Nucleic Acids Res. 1997;25:3767–3776. doi: 10.1093/nar/25.19.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurica MS, Monnat R.J., Jr, Stoddard BL. DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol. Cell. 1998;2:469–476. doi: 10.1016/s1097-2765(00)80146-x. [DOI] [PubMed] [Google Scholar]
- 25.Alaverdi N, Shih CC. CD5. Other names: T1, Leu-1, Tp67 (human), and Ly-1 (mouse) J. Biol. Regul. Homeost. Agents. 2000;14:230–233. [PubMed] [Google Scholar]
- 26.Poland BW, Xu MQ, Quiocho FA. Structural insights into the protein splicing mechanism of PI-SceI. J. Biol. Chem. 2000;275:16408–16413. doi: 10.1074/jbc.275.22.16408. [DOI] [PubMed] [Google Scholar]
- 27.Duan X, Gimble FS, Quiocho FA. Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity. Cell. 1997;89:555–564. doi: 10.1016/s0092-8674(00)80237-8. [DOI] [PubMed] [Google Scholar]
- 28.Werner E, Wende W, Pingoud A, Heinemann U. High resolution crystal structure of domain I of the Saccharomyces cerevisiae homing endonuclease PI-SceI. Nucleic Acids Res. 2002;30:3962–3971. doi: 10.1093/nar/gkf523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichiyanagi K, Ishino Y, Ariyoshi M, Komori K, Morikawa K. Crystal structure of an archaeal intein-encoded homing endonuclease PI-PfuI. J. Mol. Biol. 2000;300:889–901. doi: 10.1006/jmbi.2000.3873. [DOI] [PubMed] [Google Scholar]
- 30.Moure CM, Gimble FS, Quiocho FA. The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity. J. Mol. Biol. 2003;334:685–695. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 31.Spiegel PC, Chevalier B, Sussman D, Turmel M, Lemieux C, Stoddard BL. The structure of I-CeuI homing endonuclease: evolving asymmetric DNA recognition from a symmetric protein scaffold. Structure. 2006;14:869–880. doi: 10.1016/j.str.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama H, Shimamura T, Imagawa T, Shirai N, Itoh T, Sako Y, Miyano M, Sakuraba H, Ohshima T, et al. Structure of a hyperthermophilic archaeal homing endonuclease, I-Tsp061I: contribution of cross-domain polar networks to thermostability. J. Mol. Biol. 2006;365:362–78. doi: 10.1016/j.jmb.2006.09.066. [DOI] [PubMed] [Google Scholar]
- 33.Heath PJ, Stephens KM, Monnat R.J., Jr, Stoddard BL. The structure of I-Crel, a group I intron-encoded homing endonuclease. Nat. Struct. Biol. 1997;4:468–476. doi: 10.1038/nsb0697-468. [DOI] [PubMed] [Google Scholar]
- 34.Chevalier B, Turmel M, Lemieux C, Monnat R.J., Jr, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 35.Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, et al. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. Academic Press, New York, 276. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 38.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D. Biol. Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 39.Minton AP. Modern Analytical Ultracentrifugation. Cambridge, MA: Birkhauser Boston, Inc.; 1994. [Google Scholar]
- 40.Laue T.MS, Shah BD, Ridgeway TM, Pelletier SL. Computer-Aided Interpretation of Analytical Sedimentation Data for Proteins. Cambridge, UK: Royal Society of Chemistry; 1992. [Google Scholar]
- 41.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucas P, Otis C, Mercier JP, Turmel M, Lemieux C. Rapid evolution of the DNA-binding site in LAGLIDADG homing endonucleases. Nucleic Acids Res. 2001;29:960–969. doi: 10.1093/nar/29.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seligman LM, Stephens KM, Savage JH, Monnat R.J., Jr Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli. Genetics. 1997;147:1653–1664. doi: 10.1093/genetics/147.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevalier B, Sussman D, Otis C, Noel AJ, Turmel M, Lemieux C, Stephens K, Monnat R.J., Jr, Stoddard BL. Metal-dependent DNA cleavage mechanism of the I–CreI LAGLIDADG homing endonuclease. Biochemistry. 2004;43:14015–14026. doi: 10.1021/bi048970c. [DOI] [PubMed] [Google Scholar]
- 45.Smith J, Grizot S, Sylvain A, Aymeric D, Jean-Charles E, Jesús P, Pilar R, Francisco B, Jerónimo B, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.