Abstract
Although artificial C2-H2 zinc fingers can be designed to recognize specific DNA sequences, it remains unclear to which extent nuclear receptor C4 zinc fingers can be tailored to bind novel DNA elements. Steroid receptors bind as dimers to palindromic response elements differing in the two central base pairs of repeated motifs. Predictions based on one amino acid—one base-pair relationships may not apply to estrogen receptors (ERs), which recognize the two central base pairs of estrogen response elements (EREs) via two charged amino acids, each contacting two bases on opposite DNA strands. Mutagenesis of these residues, E203 and K210 in ERα, indicated that both contribute to ERE binding. Removal of the electric charge and steric constraints associated with K210 was required for full loss of parental DNA-binding specificity and recognition of novel sequences by E203 mutants. Although some of the new binding profiles did not match predictions, the double mutation E203R-K210A generated as predicted a mutant ER that was transcriptionally active on palindromes of PuGCTCA motifs, but not on consensus EREs. This study demonstrates the feasibility of designing C4 zinc finger mutants with novel DNA-binding specificity, but also uncovers limitations of this approach.
INTRODUCTION
Nuclear receptors form a superfamily of ligand-inducible transcription factors that is characterized by two conserved domains, the DNA-binding domain (DBD) composed of two C4 type zinc fingers, and the ligand-binding domain (LBD), which also contains a dimerization interface (1,2). Nuclear receptors can bind DNA as homo- and/or heterodimers, and recognize response elements arranged as direct repeats, palindromes or inverted palindromes of conserved motifs (3–5). Each motif is bound by the DBD of a single monomer, the two zinc fingers of the DBD combining into a single structural fold with a DNA recognition helix and variable dimerization interfaces (6,7).
Consensus estrogen response elements (EREs, Figure 1A), which are palindromes of PuGGTCA motifs with a three base-pair spacer (8–10), are bound by estrogen receptors (ERs) with highest affinity in vitro. Perfect or imperfect EREs present at promoter-proximal locations (11–13) or, as revealed by genome-wide screens, at large distances from the transcriptional start site of estrogen-regulated genes (14,15), are bound by ERα in vivo and mediate regulation of estrogen target genes. Other steroid receptors, including the androgen receptor (AR) and glucocorticoid receptor (GR), also bind palindromes with a three base-pair spacer, but the repeated motifs are PuGNACA sequences (16,17). Non-steroid receptors also recognize PuGGTCA or related motifs, but these motifs are arranged as direct repeats or everted repeats with variable spacing. The affinity and selectivity of nuclear receptors for single PuGGTCA motifs is generally low, but can be increased by receptor-specific recognition of additional 5′ flanking bases (5,13). Thus, nuclear receptors have achieved selectivity in DNA recognition while interacting with only two main types of motifs. The fact that few variations have been observed in the base-contacting amino acids of the 48 human nuclear receptors (18) suggests that this type of protein–DNA recognition has been conserved throughout evolution, possibly because it affords the most favorable combination of affinity and selectivity. Interestingly, however, nuclear receptor homologs identified in Caenorhabditis elegans offer a considerably wider variety of amino acid composition in the DNA recognition helix. Numerous mutations have also been described in the DBD of some nuclear receptors such as VDR and AR, but few changes in DNA-binding patterns have been reported for these mutant receptors (19).
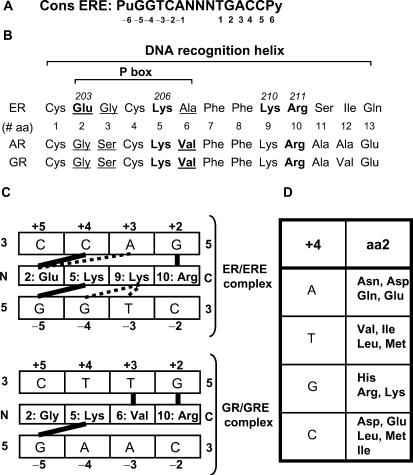
Figure 1.
Model for the selectivity of steroid receptor–DNA interactions. (A) Sequence of the consensus ERE and base numbering used in this study. (B) DNA recognition helix of the estrogen (ER), glucocorticoid (GR) and androgen (AR) receptors. The position of amino acids in the DNA recognition helix is indicated by numbers from 1 to 13. The numbering of base-interacting residues in ERα is also indicated. Base-interacting residues in each receptor are in bold, and P box amino acids are underlined. (C) Models of the amino acid–base interactions underlying specific recognition of the estrogen (top) or glucocorticoid (bottom) response element by their cognate steroid receptors. Interactions considered in the model proposed by Suzuki and Yagi (29) are in bold, while interactions described in the ER-ERE crystal structure (7) but not in the model are in dashed lines. (D) Predicted interactions between amino acid replacement at position 2 of the DNA recognition helix and bases at position +4 [from (29)].
The first clues to the molecular basis of specific DNA recognition by steroid receptors were provided by mutagenesis experiments of the ER and GR, which led to the identification of amino acids responsible for discrimination between the two types of recognition motifs bound by these receptors. Exchanging three amino acids in the ER DBD for the corresponding ones in the GR DBD resulted in a receptor mutant capable of transactivating glucocorticoid target promoters (20). The converse experiment also demonstrated that amino acids at the same three positions (P box, Figure 1B, underlined residues) in the GR were crucial for discriminating between glucocorticoid response elements (GREs) and EREs (21,22). In addition, a loop in the second zinc finger was found to be responsible for specific recognition of two motifs arranged as palindromes with 3 bp spacing (21–23). Crystallographic analyses of complexes between the ER or GR DBDs and their response elements have uncovered the amino acid–base-pair interactions established by these two receptors (7,24). Two residues of the P box, V at the third position in the GR and E at the first position in the ER (E203), contact the central discriminating bases in the ERE and GRE motifs (Figure 1C). In addition, an invariant K residue located further C-terminal in the DNA recognition helix (K210 in ERα) binds the two central bases on the opposite strand with respect to bases contacted by E203 in the ERα–ERE complex, but does not participate in contacts in the GR–GRE complex (Figure 1C). Other bases common to the ERE and GRE are contacted by amino acids conserved in ER and GR (K206 and R211 in ERα). In addition, these interactions are buttressed by a complex network of amino acid–amino acid interactions and amino acid–phosphate interactions.
The specificity of response motif recognition by steroid receptors is thus determined by a small number of specific interactions established by 3–4 amino acids. As a consequence, it may be expected that changing the identity of interacting amino acids in the DNA recognition helix would alter the selectivity of receptor–DNA interaction as can be achieved with artificial C2H2 zinc fingers (25–28). A simple model for amino acid–base interactions within the structural framework of the steroid receptor DBD has been proposed (29). This model relies on chemical rules for possible pairing of amino acid side chains and DNA bases through hydrogen bonding or hydrophobic interactions, and also incorporates stereochemical contraints specific to steroid receptors, based on the position of the DNA recognition helix with respect to the major groove of the DNA. Small, medium or large chains may thus be prefered depending on the position of the interacting amino acids in the DNA recognition helix with respect to the bases (see Figure 1D for possible interactions involving amino acid at position 2). Study of a spontaneous mutation in the first amino acid of the P box of the AR has revealed changes in DNA-binding specificity compatible with the predictions of this model (19). Replacement of G at the first P box position in the AR DBD by R resulted in a mutant that could only bind a subset of the PuGNACA motifs normally bound by this receptor, and this according to the chemical preferences of the R residue with respect to the base at the third position of the motif. However, it remains unclear whether novel types of DNA specificities can be achieved through rational design of nuclear receptor mutants, especially in the case of the ERs, which appear to have more complex determinants of motif recognition than other steroid receptors. The purpose of this study was to examine whether mutating ERα residues that interact with the two central base pairs of EREs either separately or in pairs could generate artificial nuclear receptor DBDs with novel DNA-binding specificities.
MATERIALS AND METHODS
Plasmids
The bacterial expression vector pET3.1-HE81 containing the ERα DBD has been described previously (23). The bacterial expression vector pET3.1-ERβ-DBD was constructed by PCR amplification of a cDNA fragment corresponding to amino acids 140–246 of ERβ and subcloning between the KpnI and XhoI sites of a pET3.1, a pET3 (Novagen, San Diego, CA, USA) derivative modified by insertion of a linker containing the KpnI and XhoI sites (23). All ERα DBDs mutants were constructed by insertion between the KpnI and XhoI sites of the pET3.1 vector of a fragment obtained by site-directed mutagenesis of pET3.1-HE81 using PCR amplification.
The wild-type ERα and ERβ expression vectors pSG5-HEG0, pCMV-SPORT-ERβ have been described previously (15,30). Expression vectors for ERαE203A, ERαE203N, ERαE203H, ERαE203R, ERαK210A, ERαE203A-K210A, ERαE203N-K210A, ERαE203H-K210A, ERαE203R-K210A have been constructed by substitution of the fragment between the KpnI and XhoI sites of pSG1-HE80 (23) by a fragment containing the mutation excised from the corresponding pET3.1 vector.
The tk-CAT reporter plasmids containing one copy of the consensus ERE or of palindromes containing base replaments (pAT-tkCAT, pCT-tkCAT) were derived from the pBL-CAT8+ reporter vecteur (8) by insertion of double stranded oligonucleotides containing the response elements flanked with BamHI-BglII sites at the BamHI site upstream of the thymidine kinase promoter. The TATA-CAT reporter vectors containing two copies of the consensus ERE or of the CT palindrome were prepared by substitution of the three EREs in pERE3-TATA-CAT (31) by double-stranded oligonucleotides containing two tandem response elements and flanked by BamHI and BglII sites.
Expression in Escherichia coli of ERα and ERβ DBDs and derivatives thereof
Escherichia coli BL21 DE3 cells were transformed with pET3.1 expression vectors containing the cDNAs for the DBDs of ERα, or of mutants of ERα or ERβ and expression was induced in exponentially growing cultures by IPTG (0.5 mM final) for 2 h. Whole bacterial extracts were prepared by sonication in extraction buffer (Tris-HCl pH 7.4, 25 mM; EDTA pH 8.0, 0.1 mM; NaCl 400 mM; glycerol 10%; DTT 1 mM; PMSF 10 mM and protease inhibitors) followed by centrifugation (at 10 000g for 15 min) of lysates.
To determine the levels of expression of ER DBDs, aliquots (1 ml) were taken from each culture before induction with IPTG. Bacteria were isolated by centrifugation and resuspended in M9 medium containing each amino acid except methionine and cysteine (0.01% weight/volume each). Rifampicin was added (200 μg/ml final) to inhibit bacterial RNA polymerase and expression of the T7 polymerase was induced with IPTG (0.5 mM final) for 30 min. [35S]-methionine was then added and bacteria were further incubated at 37°C for 5 min, collected by centrifugation, resuspended in Laemmli buffer and boiled for 5 min. Labeled proteins were analyzed by electrophoresis on 12% polyacrylamide–SDS gel and visualized by fluorography.
Cell culture and transfection
HeLa cells were maintained in DMEM supplemented with 5% fetal bovine serum (FBS). Cells were switched 3 days before transient transfection to medium without phenol red containing 5% FBS pretreated with activated charcoal to remove traces of hormones. For gel shift assays, cells were transiently transfected with ER expression vectors (5 μg completed to 15 μg with carrier DNA in 10 cm dish) using the calcium phosphate method (32). For CAT assays, HeLa cells were electroporated (107 cells, 0.24 kV, 950 μF in a Bio-Rad Gene Pulser II apparatus) with varying amounts of expression vectors for wt ERα or for different ERα mutants, 4 μg of pCMV-βGal, and 4 μg of tk-CAT reporter vectors containing single copies of different palindromic response elements. DNA mixes were completed to 80 μg with salmon sperm DNA in a final volume of 100 μl of NaCl 210 mM. Electroporated cells were plated in duplicate for parallel immunoblot and CAT assays.
Gel shift assays
Two days post-transfection, HeLa cells were treated for 1 h with 25 nM estradiol and whole cell extracts were prepared by three cycles of freeze-thawing in extraction buffer (Tris HCl pH 7.6, 20 mM; glycerol 20%; KCl 500 mM; DTT 1 mM; EDTA 0.1 mM; PMSF 10 mM and protease inhibitors) followed by centrifugation for 10 min at 13 000 g. Cell extracts were diluted to a final KCl concentration of 125 mM. Samples were preincubated with 2 μg poly(dIdC) for 15 min on ice before addition of [32P]-labeled double-stranded oligonucleotide probes (300 000 c.p.m. per sample). The consensus ERE used for gel retardation assays is derived from the Xenopus vitellogenin A2 gene. Reactions were incubated at 25°C for 15 min then loading buffer (0.1% bromophenol blue, 60% glycerol) was added (1/5 V/V). Complexes were separated by electrophoresis on 5% polyacrylamide gels in 0.25× TBE (45 mM Tris–HCl, 45 mM boric acid and 1 mM EDTA) and visualized by autoradiography. Amounts of bound and free probe were quantified using a Phosphorimaging screen and the Quantity One software from Bio-Rad.
Gels shift assays with whole bacterial extracts containing the ERα or ERβDBD were performed as described above except that extracts were diluted to a final NaCl concentration of 80 mM and 7% polyacrylamide gels in 0.25 × TBE were used to separate the complexes.
CAT assays
Immediately after seeding, cells were treated with estradiol (25 nM) or vehicle (EtOH) for 40 h. Cells were then harvested and extracts were prepared by three cycles of freeze–thawing in CAT extraction buffer (Tris HCl pH8.0, 0.25 M; DTT 1 mM and protease inhibitors). CAT assays were performed and standardized over β-galactosidase activity as described (31).
Immunoblot analyses
Protein concentrations in whole cell extracts prepared for gel shift or CAT assays were estimated using a Bradford assay. Proteins (50 μg) in Laemmli buffer were separated by electrophoresis on 8% polyacrylamide–SDS gels and transferred onto a PVDF membrane. Blots were incubated in blocking solution (PBS 1X, Tween 20 0.05%, BSA 3%) for 1 h and probed with anti-ERα mouse monoclonal antibody B10 (obtained from Prof. P. Chambon) at dilution 1:5000. Membranes were then washed and incubated with a horseradish peroxidase-coupled secondary antibody and visualized with an ECL detection kit.
Modeling
Modeling was performed interactively, using the InsightII/Discover package (Version 2000, Accelrys Inc., San Diego, CA, USA). The X-ray structure of the ER DBD bound to DNA (7) was used as a starting conformation. Each model was submitted to unrestrained energy minimization using the AMBER forcefield (33) until an energy minimum was reached. The presence or absence of particular pair-wise amino acid–base interactions in the final structure was treated as a possibility or impossibility to form a particular interaction in a given structural context. Distance measurements between atoms were performed with InsightII tools using a Silicon Graphics O2 computer.
RESULTS
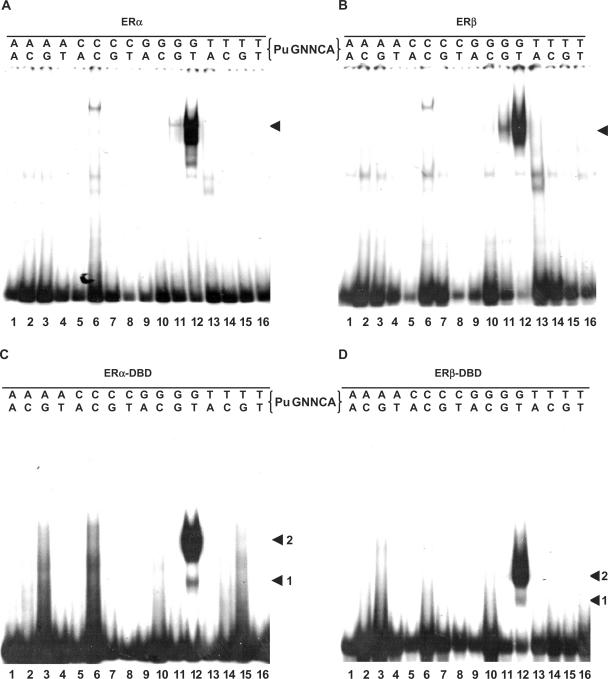
Patterns of response element binding by isolated ERα and ERβ DBDs are similar to each other and to those of the full-length receptors
Estrogen receptors bind with high affinity the consensus palindromic EREs consisting of two PuGGTCA motifs separated by a three base-pair spacer (Figure 1A). Although natural response elements are often imperfect palindromes (12,13), base-pair replacements usually result in a loss of affinity (12,14). To verify that binding patterns are mainly derived from the DBDs of ERs, we have compared the effect of symmetric substitutions at each position of the consensus ERE on binding by full-length ERs transiently expressed in HeLa cells (ERα and ERβ, Figure 2A and B, respectively) or by the ERα or ERβ DBD (ERα DBD and ERβDBD, Figure 2C and D, respectively). The wild-type ERE was bound by either the full-length ERα receptor or the corresponding DBD with the highest affinity (note the higher degree of free probe depletion with wt ERE, Figure 2A and C, lanes 19). Consistent with our previous observations that the DBDs of steroid receptors are sufficient to discriminate between EREs and GREs (23), ER DBDs selectively bound the array of probes in a pattern similar to that of the full-length receptors, although all probes were less efficiently bound with isolated DBDs. This is consistent with a loss of affinity, but not specificity, resulting from the absence of the strong dimerization interface in the LBD (34). In addition, all replacements introduced in both arms of the ERE reduced binding to the same extent for ERα and ERβ (compare panels A and B, and C and D), which share a high degree of conservation in their DBDs [90% in region C as defined in (35)]. Although the expression levels of transiently expressed full-length ERα and ERβ could not be compared, note that the DBDs were expressed to similar levels as assessed by [35S]Met incorporation (data not shown).
Figure 2.
Effect of replacements in both arms of the ERE palindrome on complex formation with full-length ERs and isolated DBDs. (A and B) Gel shift assays performed with whole cell extracts of HeLa cells transiently transfected with expression vectors for ERα (B) or ERβ (C), and a panel of probes corresponding to all base possibilities introduced at each position of the response elements in both arms of the palindrome. (C and D) Gel shift assays performed with bacterial extracts containing the DBDs of ERα (C) or ERβ (D), using the same panel of probes as in A and B.
Point mutations in the consensus ERE destabilize interaction with ERs according to chemical compatibility and steric constraints with interacting amino acids
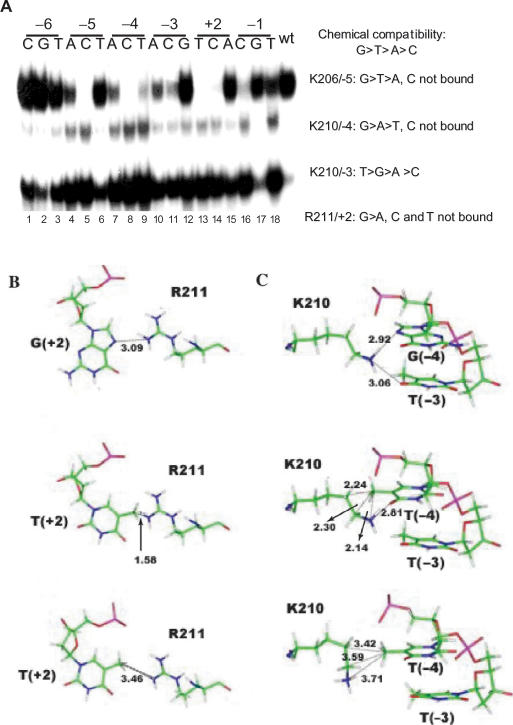
The DNA recognition helix formed by the C-terminal part of the first zinc finger of ERα contains several basic amino acids involved in contacts with bases in the target motifs (Figure 1B, residues in bold; underlined residues are part of the P box). Nucleotides G (−5) and G (+2) interact with residues at position 5 (K206) and 10 (R211) of the DNA recognition helix, respectively (Figure 1C). In addition, K210 (position 9 in the DNA recognition helix) interacts with both G (−4) and T (−3) through direct and water-mediated contacts (Figure 1C). Thus, basic residues are involved in recognition of all positions in the ERE except −/+6 and −/+1, which are not directly contacted in the crystal structure, although both display a preference for purines (Figure 2 A–D, lanes 1–3 and 16–18). These direct interactions involving basic amino acids conform to general chemical rules, with G and T, which present only negatively charged groups in the major groove, being preferred over A, which presents both a positively and a negatively charged group, and C, which contains only a positively charged group, leading to unfavorable electrostatic interactions. Accordingly, replacement of G by C in a single motif at position −4 or −5 was sufficient to abolish binding (Figure 3A, lanes 5 and 8). Of note however, the order of the preferred bases is not identical for each contacting basic amino acid. Most noticeably, replacement in a single motif of G at position +2 by T was sufficient to abolish binding (Figure 3A, lanes 13). Molecular modeling suggests that lack of binding to T at position +2 results from steric hindrance due to the methyl group of T + 2, which prevents productive interaction between the amines of R211 and O4 of T + 2 (Figure 3B).
Figure 3.
Chemical and steric constraints determine the specificity of complex formation with wt ERα. (A) Gel shift assays performed with the isolated ERα DBD and a panel of probes containing all possible base replacements introduced at each position of a single motif of the consensus ERE. Predicted and observed patterns of interactions are described on the right-hand side of the figure. (B) Interaction between R211 and G + 2 as seen in the crystal structure of the ERDBD/ERE (7) (top panel). Note the H bond formed between the amino group of R211 and the N7 atom of G + 2. Replacement of G + 2 (top panel) by T + 2 (middle panel) results in steric conflict between the amino groups of R211 and the methyl group of T + 2; energy minimization shows that steric hindrance can be relieved, but R211 cannot interact in the major groove with O4 of T + 2. (C) Interaction between K210 and G-4 and T-3 as seen in the crystal structure of the ERDBD–ERE complex (24) (top panel); note the H bonds formed between the amino group of K210 and the N7 atom of G-4 and the O4 atom of T-3; replacement of G-4 (top panel) by T-4 (middle panel) results in steric conflicts between the side chain of K210 and the methyl group of T-4; energy minimization shows that displacement of the K210 side chain to accomodate this methyl group prevents K210 from interacting with charged groups of T-4 and T-3.
The pattern of recognition of bound response elements carrying replacements at positions −4 (G > A > T > C) and −3 (T > G > A > C) also differed from that predicted from charge preference due to interaction with basic amino acid K210 (G > T > A > C). Because E203 interacts with bases at positions +3 and +4, i.e. on the other DNA strand, in the crystal structure (7), we compared the roles of both K210 and E203 in modulating patterns of base recognition. While E203 has an opposite and complementary type of chemical selectivity for bases (C > A > T > G) compared to K210, the complex mode of recognition of the two central base pairs by two amino acids may be expected to result in increased steric constraints. At position −4, replacement in a single motif by T drastically reduced binding (Figure 3A, lane 9) and replacement in both motifs of the ERE completely eliminated complex formation (Figure 2A–D, lanes 9) and transactivation [data not shown, see also (36)]. This contrasts with the capacity of other nuclear receptors such as RAR, RXR and VDR to bind to PuGTTCA motifs [(36,37); see also (5) for a review]. Energy minimization indicates that movement of K210 to avoid the methyl group of T-4 prevents interaction with DNA (Figure 3C). In addition, E203 is not capable of interacting with the amino group of A at position +4 (not shown). At position −3, G was less favorable than T (compare lanes 12 and 19 in Figure 3A and Figure 2A–D), contrary to what is observed at the other positions recognized by basic residues (Figure 3A). Modeling indicates that C + 3 would create packing problems with E203, preventing interaction of this amino acid with C + 4, and K210 is too distant to interact with G−3 (not shown). Thus, our experimental and modeling data suggest that both E203 and K210 contribute to the selectivity of response element recognition with respect to the two central bases of the ERE.
Roles of E203 and K210 in the specificity and affinity of ER interaction with palindromic response elements
To better analyze the respective roles of E203 and K210 in determining the specificity and affinity of ER interaction with response elements, we further characterized the ER-binding specificity with respect to the two central base pairs of the palindrome motifs using a panel of 16 probes representing all combinations of these central bases introduced in both arms of the palindrome. Full-length ERs and isolated DBDs bound with high affinity only to the GT (response elements are designated by bases found on the minus strand at position −4/−3) combination found in the wt ERE and, with lower affinity, to the GG element (Figure 4A–D). Note that complexes formed on element CC with full-length receptors (lane 6) were also observed with extracts from cells transfected with the parental expression vector, and that the smears observed in some lanes with bacterially expressed ER DBDs also appeared with extracts from bacteria transformed with the parental expression vector (not shown).
Figure 4.
ERs bind with high selectivity to PuGNNCA palindromes. (A) and (B) Extracts from transiently transfected HeLa cells expressing full-length ERα (A) or ERβ (B) were incubated with 32P-labeled oligonucleotide probes containing PuGNNCA palindromes and complexes were separated on 5% polyacrylamide gels. The position of the dimeric complex is indicated by an arrow. (C) and (D) Bacterial extracts containing the ERα (C) or ERβ (D) DBDs were incubated with the same probes and complexes were separated by electrophoresis on 7% polyacrylamide gels. The position of specific complexes containing monomers (1) or dimers (2) of ER DBDs are indicated in the figure.
The very high selectivity of ERs for the two central base pairs in their response elements differs from that of the GR or AR, whose DBDs could both recognize all four PuGNACA-containing palindromes [(20) and data not shown]. Crystallographic analysis of the GR DBD indicates that contacts are established only with the base at position +3 through a V residue (position 6 in the DNA-binding helix). E203 is replaced by a G residue in GR and AR, and the K residue corresponding to K210 in ERα does not contribute to GRE recognition. The higher selectivity of ER interaction with response elements could therefore result either from the fact that E203 interacts with both adjacent +4/+3 bases, and/or from the contribution of K210 in recognizing the −4/−3 bases on the opposite strand of DNA.
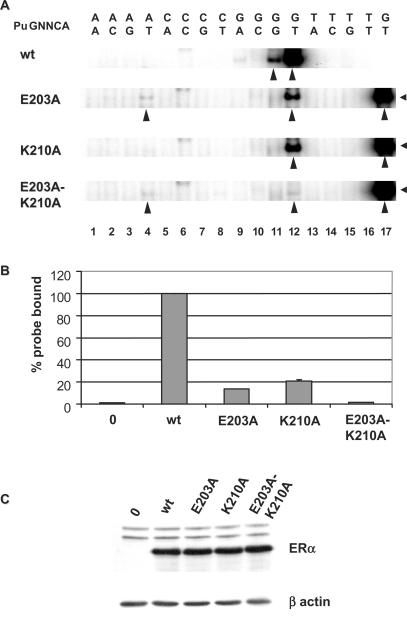
To investigate the respective roles of these two amino acids to the affinity and/or selectivity of binding, we replaced either E203 or K210 by A residues in ERα and examined complex formation with the panel of 16 probes corresponding to all possible variants at the two central positions of the repeated motifs. Both mutants still bound with highest efficiency to the consensus ERE (Figure 5A, lanes 12), although the intensity of the retarded band was ∼5-to 10-fold lower than with the wt ERα (compare Figure 5A, lanes 12 and 17; see also Figure 5B). However, the K210A mutation was active in transactivation assays using a reporter gene containing the consensus ERE (Figure 8C). Intriguingly, peak transcriptional activity was ∼60% higher for K210A than for the wt receptor, suggesting that K210 plays a negative role in transcriptional activation.
Figure 5.
Role of K210 and E203 in the affinity and specificity of receptor–DNA complex formation. (A) Gel shift assays performed with wt ERα or the K210A, E203A and E203A-K210A mutants expressed in HeLa cells and a panel of probes containing all possible base combinations at the two central positions of each PuGGTCA motifs in the consensus ERE. Arrows indicate the main specific complexes. Binding of the wt ERα to the consensus ERE (GT) is shown for comparison in lane 17. (B) Quantitation of the levels of binding of the transfected receptors to the consensus ERE probe, expressed as bound over bound plus free probe. Results are an average of three experiments, and error bars indicate SD. (C) Levels of expression of the different transfected receptors as assessed by western analysis using the monoclonal B10 antibody.
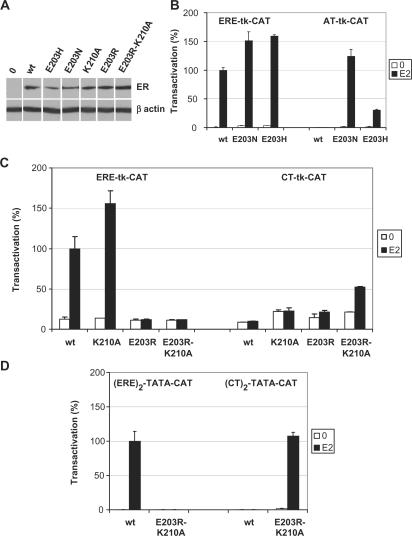
Figure 8.
Altered patterns of reporter vector transactivation by ER mutants. HeLa cells were transiently transfected with variable concentrations of expression vectors for the wt ERα or mutants (10 ng–3 μg per 10 cm plate), tk-CAT reporter vectors containing consensus ERE, AT or CT elements (2 μg per plate), and an internal control CMV-βGal reporter vector (2 μg per plate). Cells were treated with estrogen or vehicle for 40 h and duplicate extracts were assayed ER protein expression levels in western analysis and for β-galactosidase/CAT activity. (A) Standardized expression levels of wt ER and different mutants. Note that some mutants (E203N and E203R-K210A) were transfected at a 3-fold higher concentration (3 μg) than other expression vectors (1 μg) to obtain similar expression levels. (B) CAT activity of wt, E203N and E203H, shown at peak transactivation levels for all plasmids (corresponding to 300 ng for wt and E203H and 1 μg for E203N). (C) Same as in B. except that expression vectors for wt or the K210A, E203R or E203R-K210A mutants were used together with tk-CAT reporter vectors containing the consensus ERE or the CT element. Peak transactivation was obtained at 300 ng (wt or K210A) or 1 μg (E203R-K210A). Other plasmids (K210A, E203R), used here at 300 ng, were inactive even at the highest tested concentration (3 μg; not shown). (D) Same as in C except that expression vectors for the wt ER or the E203R-K210A mutant were used together with TATA-CAT reporter vectors containing two copies of the consensus ERE or the CT element. Maximal transactivation levels, obtained at 1 μg (wt) or 3 μg (E203R-K210A), are shown.
Mutant E203A bound with reduced efficacy to the consensus ERE compared to the wt ERα (Figure 5A and B), and formed a weak complex with the PuGATCA probe (AT, Figure 5A, lane 4), which was not detected with the K210A mutation. Finally, the double mutant K210A–E203A bound very weakly to the consensus ERE probe, and also formed a detectable complex with the AT probe (Figure 5A, lanes 4, 8, 12 and 16). Binding of E203A to the AT probe suggests that the main role in restricting binding to this element in the wt receptor is played by E203 rather than K210. The absence of high affinity complexes observed with the K210A mutant suggests that the role of K210 in the selectivity of response element recognition is redundant with the role played by E203.
The effects of mutations of E203 on selective DNA binding are not predictable from simple chemical and steric compatibility rules
A simple DNA recognition model for steroid receptors has been proposed previously (8), based on both the general rules of chemical compatibility between amino acids and base pairs, and stereochemical constraints due to the position of the DNA-binding helix across the major groove as derived from the crystal structures of the ER and GR DBDs (Figure 1D for predicted interactions at amino acid position 2). Note that this model is based on a one amino acid–one base interaction relationship, and in particular does not take into account recognition of the two central bases by both K210 and E203 (Figure 1C, dotted lines), considering only the role of E203 with C + 4 (Figure 1C, bold line). Our finding that E203 and K210 cooperate for selective binding to GT palindromes incited us to test the predictions of this model for replacement of E203 by other amino acids. Replacement of E by N is predicted to switch recognition from C at position +4 to A (TN elements), and replacement by H or R should lead to specific recognition of a G at this position (CN elements, Figure 1D). These mutations were introduced in ERα full length and in the isolated DBD. Similar expression levels were obtained for all full-length mutants and the wild-type receptor as determined by western analysis (data not shown). Whole cell extracts containing the mutant receptors E203N and E203H formed complexes mainly with the wt ERE (GT probe, Figure 6A and B, lanes 13 and Figure 6D and E, lanes 12). While binding to the wt ERE was not abolished, the patterns of probe recognition were altered with both mutants, which bound to the AT element (Figure 6A and B, lanes 5 and Figure 6D and E, lanes 4). These interactions are transcriptionally productive, as demonstrated by transient cotransfection of CAT reporter vectors containing the corresponding response elements cloned upstream of the thymidine kinase promoter with expression vectors for the different mutants (Figure 8B, ERE-tk-CAT and AT-tk-CAT reporter vector). Titration curves were performed using varying concentrations of transfected plamids, and protein levels were measured by western analysis (Figure 8A). Surprisingly, saturation was reached at identical protein concentrations for all receptors on either response element, indicating that the difference in efficiency of complex formation in vitro is not observable in our reporter assay (data not shown). This is possibly due to cooperative effects with other transcription factors bound to the promoter, or to chromatin organization. Peak transactivation levels were similar for the wt and mutant receptors on the consensus ERE, reflecting intact transcription activation properties for the mutants (Figure 8B). On the other hand, mutant E203H, but not E203N, had ∼6-fold lower peak levels of transactivation on the AT element than on the consensus ERE (Figure 8B; peak levels on the two response elements were obtained at the same protein concentration). The differential levels of maximal transcriptional activation by the E203H mutant on the two response elements may be related to the previously reported allosteric effect of the DNA sequence on ERα transcriptional activation properties (38,39).
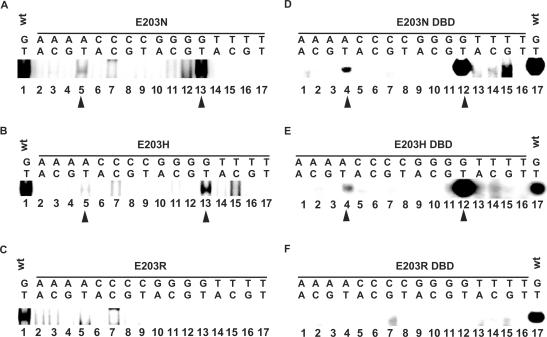
Figure 6.
Mutations of E203 do not generate expected switches in DNA-binding specificity. (A, B and C) HeLa cells were transiently transfected with expression vectors for ERα mutant E203N (A) E203H (B) or E203R (C). Whole cell extracts were used in a gel shift assay with the sixteen PuGNNCA probes and complexes were separated by electrophoresis on 5% polyacrylamide gels. The complexes formed are indicated by an arrowhead. For comparison, the complex formed between wt ERα and the consensus ERE (GT) is loaded on each gel (lane 1). (D, E and F) Similar assays were performed using bacterial extracts containing mutant ER DBDs carrying the E203N, E203H or E203R mutations.
The observed DNA-binding specificity of these mutants do not correspond to predictions based on the proposed model, as no stable binding was observed with TN motifs for E203N (Figure 6A, lanes 14–17), or CN motifs with E203H (Figure 6B, lanes 6–9). Accordingly, the TT and CT elements did not confer estrogen responsiveness to the tk promoter in the presence of these mutants (data not shown). Note that no specific complexes were observed with the E203R mutant on any of the PuGNNCA probes (Figure 6C and F). Lack of binding to the consensus ERE and lack of transactivation on an ERE-tk-CAT reporter (Figure 8C) suggest that R at this position has a destabilizing effect that is stronger than the absence of side chain (A mutation, see Figure 5). Replacement of E203 by R in the structure of the ER–ERE complex reveals that the R side chain is too bulky to fit in the major groove of DNA, and that the amino groups exert repulsive effects with the positively charged groups of C + 4 and C + 5 and steric conflict with C + 4 (Figure 9C and data not shown). In addition, neither binding to CN elements nor transactivation from reporter vectors containing the CT palindrome (Figure 8C, CT-tk-CAT reporter vector) was obtained, indicating again that the predicted switch in specificity has not occured. These results indicate that a model based on a one amino acid—one base-pair relationship is not an accurate description of the interaction between the ER and its target response element, and suggest that K210 plays a role in modulating the DNA-binding specificity of receptors carrying mutations at position 203.
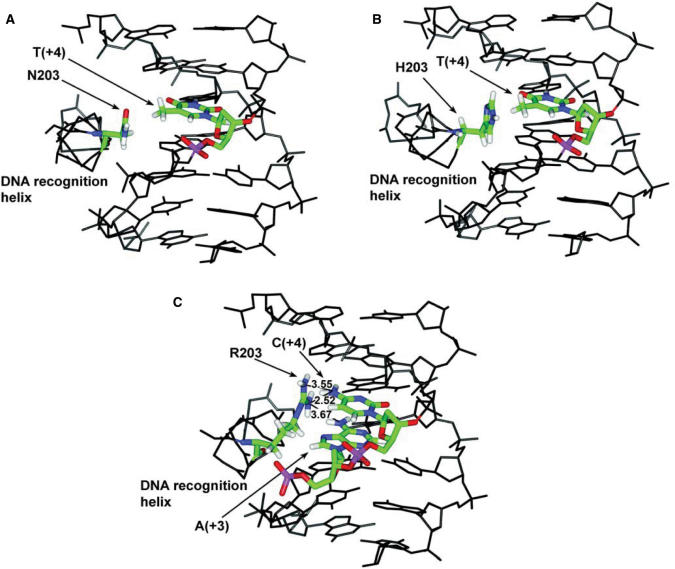
Figure 9.
Modeling of the effect of mutations at position 203 on interaction with palindromic response elements. (A and B) Complexes formed between the ER mutant E203N (A) or E203H (B) and the AT element after replacement of E203 by N or H in the ERαDBD/ERE crystal structure and energy minimization. The position of the N and H side chains are restrained by the presence of S193. (C) Steric conflicts and charge repulsion with the amino groups of C + 4 in the consensus response element resulting from replacement of E203 by R.
K210 restricts changes in DNA-binding specificity of mutants at position 203
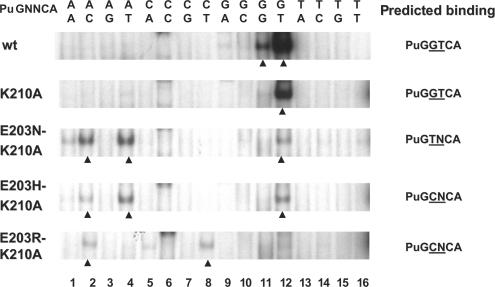
Contrary to the total absence of binding observed with the E203R mutant of ERα, mutation of the corresponding amino acid in the AR (G577) to R led to a different pattern of response element binding than that of the wt receptor. While wt AR bound all four PuGNACA elements, the G577R mutant bound PuGGACA elements very weakly, but interacted with the three other PuGNACA elements, with a preference for the element containing a G at position +4 (PuGCACA). In the context of the GR DBD, and presumably also the AR DBD, the K residue corresponding to K210 (position 9 in the DNA recognition helix) does not bind DNA, but is involved in an interaction with E at position 13 in the DNA recognition helix (Figure 1B). Thus, we investigated whether mutation K210A may facilitate association of the E203 mutants to novel binding sites and/or reduce binding to the consensus ERE. The resulting E203N/K210A and E203H/K210A double mutants had a much reduced binding to this probe (Figure 7), confirming the role of K210 in binding to the consensus element in the absence of a residue at position 203 recognizing C + 4. However, these mutants still did not form stable complexes with probes containing bases predicted to interact with the N or H residue at position 203 (TN, CN, respectively, see Figure 7).
Figure 7.
Mutation K210A introduced in E203 mutants reduces binding to the consensus ERE and reveals the switched specificity of mutant E203R. (A) Gel shift assays performed with double mutants of ERα expressed in HeLa cells and the panel of 16 probes containing all possible base combinations at the two central positions of each PuGGTCA motifs in the consensus ERE. Arrows indicate the main specific complexes formed. Note that similar levels of expression were obtained in western analysis (not shown).
In addition to complexes with AT palindromes already observed with the single mutants, the strongest complexes detected with E203N/K210A and E203H/K210A double mutants were with the AC element (Figure 7). On the other hand, the double mutant E203R-K210A gained binding to the CT element (Figure 7), as could be predicted from chemical compatibility between R at position 203 and G + 4 (Figure 1D). This mutant also bound to the AC element (Figure 7). Thus, the specificity of base recognition expected from replacing the E203 residue by R was revealed in the presence of the K210A mutation, although an additional unpredicted complex was also formed with comparable efficiency. As the level of complex formation on the CT element was much lower than that of the wt receptor on the consensus ERE, we examined whether the interaction between the E203R-K210A mutant and the CT probe is transcriptionally productive. Whereas no estradiol-induced transcription could be detected with wt, K210A or E203R on a CT-tk-CAT reporter vector, the double mutant E203R-K210A gave rise to a detectable increase in estradiol-stimulated transcription (Figure 8C). The double mutant, like the single mutant E203R, was not active on ERE-tk-CAT, confirming the switch in specificity. The complete switch in DNA-binding specificity of the double mutant was confirmed using reporter vectors containing tandem response elements. No transactivation of the reporter vector containing EREs was observed in the presence of the double mutant after estrogen treatment, while peak stimulation of the reporter vector containing CT elements was comparable to that of wt ERα on the reporter containing consensus EREs (Figure 8D).
DISCUSSION
Although C2H2 zinc fingers can be tailored to bind virtually any DNA sequence, nuclear receptors have not demonstrated similar flexibility. A possible reason for success in the rational design of artificial C2H2 zinc finger proteins is the modular structure of this type of DBDs, each finger recognizing three base pairs and multiple zinc fingers extending the length of bound DNA. On the other hand, steroid receptors bind DNA as dimers recognizing palindromic response elements. As dimerization contributes to the affinity of DNA binding, binding to non-palindromic sequences would likely be of low affinity. Nevertheless, the question remains whether steroid receptors can bind other DNA elements than their two natural cognate response elements. A previously proposed model for prediction of specific amino acid–base interactions by the steroid receptor DNA-binding helix (8) incorporates both chemical rules governing amino acid–base interactions and stereochemical constraints resulting from the position of the DNA recognition helix in the major groove of response element as determined by crystallographic studies. However, this model relies on one amino acid—one base-pair relationships and ignores some of the contacts described in the ER–ERE cocrystal structure. In particular, E203 recognizes not only the base at position +4 (Figure 1C, bold line), but also that at position +3 (dotted line), and bases on the opposite strand make contacts with K210. On the other hand, the lysine residue corresponding to K210 in the context of the GR does not bind DNA, and its role in binding of ERs to EREs has not been experimentally confirmed.
Contrary to the GR and AR, which recognize motifs with variable base composition at position −4 conforming to the PuGNACA consensus (20), ERs display a high degree of specificity for bases at position −4/−3. GT was the only element bound with high affinity, while GG was tolerated when introduced in only one motif, such as in the ERE found in the promoter of the rabbit uteroglobin gene (40), but reduced binding efficacy dramatically when introduced in both motifs. This binding pattern is consistent with chemical preferences of E203 for C or A at position +4 and +3, and of K210 for G or T at position −4 and −3, with restrictions imposed by steric constraints with either amino acids. Mutagenesis of each amino acid indicated that both contribute to recognition of the two central base pairs in terms of binding efficiency. E203 also plays a specific role in restricting binding to AT elements, since mutation E203A, but not K210A, allowed formation of complexes. Molecular modeling indicates that this is due to steric conflicts between the carbonyl group of E203 and the methyl group of thymine +4.
The contribution of K210 to ERE binding likely explains the fact that E203N and E203H mutants still interacted with high affinity with the consensus response elements (GT). Interactions with the AT element, which were not predicted by the proposed model, were observed in gel shift and transactivation assays with both E203N and E203H mutants, and likely result from removal of negative constraints exerted by E203 on T + 4, as observed also in the E203A mutant. Our modeling indicates that the methyl group of thymine can be accomodated by side chain rearrangement when E203 is replaced by N or H (Figure 9A and B). On the other hand, lack of binding of the E203R mutant to all tested elements and of transcriptional activity on the consensus ERE and CT element can be explained by conflicts in charge preference between R203 and K210 for recognition of the same base pairs.
The role of K210 in preventing a specificity switch by mutations at position 203 is demonstrated by the observation that the double mutant E203R-K210A could bind the CT element, as predicted by base compatibility between R and G + 4, whereas this interaction was not observed with the single mutation E203R. As noted above, the K residue in AR at the corresponding position does not contact DNA, explaining the capacity of the AR mutant with an R at the position equivalent to 203 to interact with G + 4. Another difference between the two receptors is the contribution of V at position 6 in the DNA recognition helix of AR (Figure 1B) in complex stabilization. This may explain the relatively low affinity of the E203R-K210A mutant for the CT element. However, this interaction was transcriptionally productive. Surprisingly, similar levels of transactivation were obtained with the mutant receptor on the CT element as with the wt receptor on the consensus element at comparable protein concentrations along a range of amounts of transfected plamids, failing to reveal a different efficiency of reporter DNA saturation (data not shown). Titration curves of mutant E203N and E203H on the consensus ERE and AT element also failed to reveal a shift in protein concentrations necessary to reach peak transcriptional activity. This indicates that our in vivo assay may not discriminate between binding sites in the range of affinities that is observable in gel shift assay (within 30-fold of wt ERα-consensus ERE affinity) and may be due to synergism with other transcription factors for binding to our reporter genes in vivo. A higher stringency of in vitro versus in vivo assays is supported by the observation that half-palindromes, which are not bound in our gel shift assay, are enriched, albeit to a much lower degree than high-affinity EREs, in chromatin fragments bound by ERα in a genome-wide analysis (15). On the other hand, our transactivation data confirms the total loss of interaction between the E203R-K210A and the consensus ERE and thus the switch in specificity toward the CT element.
Binding of the E203R-K210A to some unpredicted sites, i.e the AC element, reflects the limitations of simple models in fully accounting for the complex interactions between these residues and the two central base pairs. In addition, the expected binding of the E203H-K210A mutant to the CN palindromes and of the E203N-K210A mutant to the TN elements were not observed. Replacement of E203 by N or H reveals that both residues are too short to interact with charged groups at position 6 or 7 of A (+4) or G (+4), respectively (Figure 9A and B). Finally, mutants E203H-K210A or E203N-K210A still bound the consensus ERE (GT element), contrary to what was observed with mutation E203R-K210A. The total abrogation of binding to the consensus ERE appears due to charge and steric conflict of R with C + 4 (Figure 9C).
Together, our results indicate that simple chemical and stereochemical rules cannot predict accurately the changes in the selectivity of ER–DNA interactions induced by specific mutations in the two central base pairs. A clear limitation is the need to incorporate the contribution of several amino acids to recognition of the same base pairs, and the role of one amino acid in recognizing two adjacent bases. The combined effects of E203 and K210 in interacting with the same bases is apparent both at the level of charge and steric constraints, resulting in the tighter DNA-binding specificity for the two central base pairs observed for ER versus other steroid receptors. Further, steric constraints play an important role in preventing potential interactions. Additional experiments will also be necessary to determine whether the effect of amino acid replacement at other positions in the ERα DNA-binding helix, which are involved in simpler one residue–one base interactions, is more easily predictable. It remains possible that other receptors may be more amenable to the rational design of mutant receptors with altered DNA-binding specificity, due to differences in composition of the DNA-binding helix and/or in the mode of DBD dimerization. In this respect, it will be of interest to investigate the DNA-binding specificity of non-classical C. elegans receptors, which contain widely diverging combinations of amino acids in their DNA recognition helix. Finally, combinatorial approaches as performed for C2H2 zinc fingers (41,42) may help clarify how amino acids of the DNA-binding helix cooperate toward the establishment of novel DNA-binding specificities.
ACKNOWLEDGEMENTS
This work was supported by funding from the Natural Sciences and Engineering Research Council of Canada (NSERC) grant to S.M. J.D. has been supported by an FRSQ fellowship, and V.N.D. by a studentship from the Montreal Centre for Experimental Therapeutics in Cancer—Canadian Institutes for Health Research teaching program. S.V.S. is a CIHR scientist, and S.M. is an FRSQ Chercheur-Boursier Senior and holds the CIBC Breast Cancer Research Chair at Université de Montréal. Funding to pay the Open Access publication charges for this article was provided by NSERC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Green S, Chambon P. Nuclear receptors enhance our understanding of transcription regulation. Trends Genet. 1988;4:309–314. doi: 10.1016/0168-9525(88)90108-4. [DOI] [PubMed] [Google Scholar]
- 2.Beato M, Klug J. Steroid hormone receptors: an update. Hum. Reprod. Update. 2000;6:225–236. doi: 10.1093/humupd/6.3.225. [DOI] [PubMed] [Google Scholar]
- 3.Glass CK. Differential recognition of target genes by nuclear receptor monomers, dimers and heterodimers. Endocr. Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- 4.Rastinejad F, Perlman T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 5.Khorasanizadeh S, Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem. Sci. 2001;26:384–390. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- 6.Baumann H, Paulsen K, Kovacs H, Berglund H, Wright AP, Gustafsson JA, Hard T. Refined solution structure of the glucocorticoid receptor DNA-binding domain. Biochemistry. 1993;32:13463–13471. doi: 10.1021/bi00212a011. [DOI] [PubMed] [Google Scholar]
- 7.Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 8.Klein-Hitpass L, Ryffel GU, Heitlinger E, Cato AC. A 13 bp palindrome is a functional estrogen responsive element and interacts specifically with estrogen receptor. Nucleic Acids Res. 1988;16:647–663. doi: 10.1093/nar/16.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Driscoll MD, Sathya G, Muyan M, Klinge CM, Hilf R, Bambara RA. Sequence requirements for estrogen receptor binding to estrogen response elements. J. Biol. Chem. 1998;273:29321–29330. doi: 10.1074/jbc.273.45.29321. [DOI] [PubMed] [Google Scholar]
- 10.Kulakosky PC, McCarty MA, Jernigan SC, Risinger KE, Klinge CM. Response element sequence modulates estrogen receptor alpha and beta affinity and activity. J. Mol. Endocrinol. 2002;29:137–152. doi: 10.1677/jme.0.0290137. [DOI] [PubMed] [Google Scholar]
- 11.Walker P, Germond J-E, Brown-Luedi M, Givel F, Wahli W. Sequence homologies in the region preceding the transcription initiation site of the liver estrogen-responsive vitellogenin and apo-VLDLII gene. Nucleic Acids Res. 1984;12:8611–8626. doi: 10.1093/nar/12.22.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez R, Nguyen D, Rocha W, White JH, Mader S. Diversity in the mechanisms of gene regulation by estrogen receptors. Bioessays. 2002;24:244–254. doi: 10.1002/bies.10066. [DOI] [PubMed] [Google Scholar]
- 14.Bourdeau V, Deschenes J, Metivier R, Nagai Y, Nguyen D, Bretschneider N, Gannon F, White JH, Mader S. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol. Endocrinol. 2004;18:1411–1427. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- 15.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006 doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 16.Klock G, Strähle U, Schütz G. Oestrogen and glucocorticoid responsive elements are closely related but distinct. Nature. 1987;329:734–736. doi: 10.1038/329734a0. [DOI] [PubMed] [Google Scholar]
- 17.Nordeen SK, Suh BJ, Kühnel B, Hutchinson CA. Structural determinants of a glucocorticoid receptor recognition element. Mol. Endocrinol. 1990;4:1866–1873. doi: 10.1210/mend-4-12-1866. [DOI] [PubMed] [Google Scholar]
- 18.Robinson-Rechavi M, Escriva Garcia H, Laudet V. The nuclear receptor superfamily. J. Cell Sci. 2003;116:585–586. doi: 10.1242/jcs.00247. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen D, Steinberg SV, Rouault E, Chagnon S, Gottlieb B, Pinsky L, Trifiro M, Mader S. A G577R mutation in the human AR P box results in selective decreases in DNA binding and in partial androgen insensitivity syndrome. Mol. Endocrinol. 2001;15:1790–1802. doi: 10.1210/mend.15.10.0709. [DOI] [PubMed] [Google Scholar]
- 20.Mader S, Kumar V, de Verneuil H, Chambon P. Three amino acids of the oestrogen receptor are essential to its ability to distinguish an oestrogen from a glucocorticoid-responsive element. Nature. 1989;338:271–274. doi: 10.1038/338271a0. [DOI] [PubMed] [Google Scholar]
- 21.Umesono K, Evans RM. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 22.Danielsen M, Hinck L, Ringold GM. Two amino acids within the knuckle of the first zinc finger specify DNA response element activation by the glucocorticoid receptor. Cell. 1989;57:1131–1138. doi: 10.1016/0092-8674(89)90050-0. [DOI] [PubMed] [Google Scholar]
- 23.Mader S, Chambon P, White JH. Defining a minimal estrogen receptor DNA binding domain. Nucleic Acids Res. 1993;21:1125–1132. doi: 10.1093/nar/21.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 25.Sera T, Uranga C. Rational design of artificial zinc-finger proteins using a nondegenerate recognition code table. Biochemistry. 2002;41:7074–7081. doi: 10.1021/bi020095c. [DOI] [PubMed] [Google Scholar]
- 26.Blancafort P, Segal DJ, Barbas CF., III Designing transcription factor architectures for drug discovery. Mol. Pharmacol. 2004;66:1361–1371. doi: 10.1124/mol.104.002758. [DOI] [PubMed] [Google Scholar]
- 27.Mandell JG, Barbas CF., III Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papworth M, Kolasinska P, Minczuk M. Designer zinc-finger proteins and their applications. Gene. 2006;366:27–38. doi: 10.1016/j.gene.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki M, Yagi N. DNA recognition code of transcription factors in the helix-turn-helix, probe helix, hormone receptor, and zinc finger families. Proc. Natl Acad. Sci. USA. 1994;91:12356–12361. doi: 10.1073/pnas.91.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tora L, Mullick A, Metzger D, Ponglikitmongkol M, Park I, Chambon P. The cloned human oestrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 1989;8:1981–1986. doi: 10.1002/j.1460-2075.1989.tb03604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barsalou A, Gao W, Anghel S, Carriere J, Mader S. Estrogen response elements can mediate agonist activity of antiestrogens in human endometrial Ishikawa cells. J. Biol. Chem. 1998;273:17138–17146. doi: 10.1074/jbc.273.27.17138. [DOI] [PubMed] [Google Scholar]
- 32.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 33.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TEI, Ferguson DM, Seibel GL, Singh UC, Weiner PK, et al. AMBER 4.1. San Francisco, CA, USA: University of California; 1995. [Google Scholar]
- 34.Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 35.Krust A, Green S, Argos P, Kumar V, Walter P, Bornert JM, Chambon P. The chicken oestrogen receptor sequence: homology with v-erbA and the human oestrogen and glucocorticoid receptors. EMBO J. 1986;5:891–897. doi: 10.1002/j.1460-2075.1986.tb04300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mader S, Leroy P, Chen JY, Chambon P. Multiple parameters control the selectivity of nuclear receptors for their response elements. Selectivity and promiscuity in response element recognition by retinoic acid receptors and retinoid X receptors. J. Biol. Chem. 1993;268:591–600. [PubMed] [Google Scholar]
- 37.Mader S, Chen JY, Chen Z, White J, Chambon P, Gronemeyer H. The patterns of binding of RAR, RXR and TR homo- and heterodimers to direct repeats are dictated by the binding specificites of the DNA binding domains. EMBO J. 1993;12:5029–5041. doi: 10.1002/j.1460-2075.1993.tb06196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood JR, Greene GR, Nardulli AM. Estrogen response elements function as allosteric modulators of estrogen receptor conformation. Mol. Cell. Biol. 1988;18:1927–1934. doi: 10.1128/mcb.18.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol. Endocrinol. 2002;16:469–486. doi: 10.1210/mend.16.3.0814. [DOI] [PubMed] [Google Scholar]
- 40.Slater EP, Redeuilh G, Theis K, Suske G, Beato M. The uteroglobin promoter contains a noncanonical estrogen responive element. Mol. Endocrinol. 1990;4:604–610. doi: 10.1210/mend-4-4-604. [DOI] [PubMed] [Google Scholar]
- 41.Choo Y, Klug A. Toward a code for the interactions of zinc fingers with DNA: selection of randomized fingers displayed on phage. Proc. Natl Acad Sci. USA. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magnenat L, Blancafort P, Barbas CF., III In vivo selection of combinatorial libraries and designed affinity maturation of polydactyl zinc finger transcription factors for ICAM-1 provides new insights into gene regulation. J. Mol. Biol. 2004;341:635–649. doi: 10.1016/j.jmb.2004.06.030. [DOI] [PubMed] [Google Scholar]