Abstract
Understanding patterns of rRNA evolution is critical for a number of fields, including structure prediction and phylogeny. The standard model of RNA evolution is that compensatory mutations in stems make up the bulk of the changes between homologous sequences, while unpaired regions are relatively homogeneous. We show that considerable heterogeneity exists in the relative rates of evolution of different secondary structure categories (stems, loops, bulges, etc.) within the rRNA, and that in eukaryotes, loops actually evolve much faster than stems. Both rates of evolution and abundance of different structural categories vary with distance from functionally important parts of the ribosome such as the tRNA path and the peptidyl transferase center. For example, fast-evolving residues are mainly found at the surface; stems are enriched at the subunit interface, and junctions near the peptidyl transferase center. However, different secondary structure categories evolve at different rates even when these effects are accounted for. The results demonstrate that relative rates and patterns of evolution are lineage specific, suggesting that phylogenetically and structurally specific models will improve evolutionary and structural predictions.
INTRODUCTION
RNA molecules fold into defined structures that are critical for their biological functions. During RNA evolution, the structure is much more conserved than the sequence (1,2). The sequence variations that contribute to differences between species are those that preserve the structure and function of the RNA molecule.
An important model for studying RNA evolution is the ribosomal RNA (rRNA). The ribosome is a large complex of both RNA and protein, but it is the RNA component that catalyzes one of the most fundamental and most highly conserved biochemical activities: protein synthesis (3). Some universally conserved regions of the rRNA might date back to the RNA world, a hypothetical stage of evolution in which RNA performed all major biochemical reactions (4). In particular, the peptidtyl transferase center, which catalyzes peptide bond synthesis, has been independently recovered by artificial selection from random-sequence pools (5), suggesting that it would have been relatively easy to ‘discover’ after the evolution of RNA (6).
The rRNA is present in all extant species and presumably dates back to the earliest forms of life. It thus reflects the evolutionary history of life itself, and can be used to establish the evolutionary relationships between all species on earth (7). Because reconstruction of phylogeny depends on the evolutionary model that is assumed, it is important to understand how rRNA actually evolves.
The most widely accepted model of rRNA evolution is a ‘rates across sites’ model, in which a multiple sequence alignment is used to assign rates of evolution to each position in the rRNA (8). Secondary structure is expected to influence evolutionary rates primarily through compensatory mutations in stems. Because stems are assumed to be largely structural, any substitution of one base pair for another should typically be acceptable. In contrast, unpaired regions are thought to depend more specifically on their sequence. For example, tetraloops fall into only a few families (9). This view was promoted by the paradoxical finding that most of the highly conserved regions, i.e. regions with no or small variability at the sequence level, in the bacterial small subunit (SSU) rRNA were in unpaired, rather than in paired, regions (10–17). This finding suggested the then-revolutionary view that base pairing is a weak constraint on sequence compared to other influences on the sequence near the active site of the ribosome. This idea is further supported by two additional observations: it is often possible to experimentally swap one base pair for another while preserving function, and paired regions change faster than unpaired regions when the GC content of each region is plotted against total GC content (18).
The assumption that RNA evolution is composed predominantly of compensatory mutations in paired regions suggests that specific rate matrices should be used to describe paired regions for evolutionary studies. RNA violates the assumption of site independence that underlies many evolutionary models, because maintaining base pairing requires the bases at two interacting sites to change in a correlated fashion. Currently, many models of RNA evolution incorporate the nonindependence of sites in paired regions by allowing correlated mutations (12, 19–24), including noncanonical base-pair interactions represented as isostericity matrices (25). The special treatment of paired regions is a more accurate model of RNA evolution than using a single four-state rate matrix. However, these models could potentially be refined further with detailed knowledge about the rates of change in different unpaired regions (hairpin loops, bulges, and multi-helix junctions) and in different taxonomic groups.
Although the standard model of fast-evolving stems is widely accepted (22,26,27), there are three good reasons to believe that the paired–unpaired dichotomy provides a limited view of RNA evolution.
First, although many base pairs in many molecules can be changed experimentally without disrupting function, the same is true for unpaired regions. For example, replacing large or poorly structured loops with tetraloops is commonly performed to improve crystallization of RNAs [see for example (28)]. Accordingly, it is unclear whether, on average, changes in stems can be tolerated more often than changes in unpaired regions.
Second, the early observation that many highly conserved bases in rRNA are unpaired (10) need not imply that most unpaired bases in rRNA are highly conserved. For example, the conservation maps from the comparative RNA web site (29) show that 44 and 35% of the nucleotide positions in bacteria and eukaryotes, respectively (both large subunit (LSU) and SSU) are conserved in more than 98% of the sequences in the alignment. Of these more than 98% conserved positions, only 50–54% are unpaired. Because there are more paired positions than unpaired positions in the rRNA, on average about 50% of the unpaired positions and 30% of the paired positions are highly conserved (more than 98%). The other half of the unpaired positions are thus free to evolve at higher rates. (Note that only positions that are present in at least 95% of the sequences are counted, excluding about 8% of the positions in the bacterial model and about 30% in the eukaryotic model, and that differences in the definition of ‘highly conserved’ can change the figures substantially.)
Third, we recently showed that even random sequences that have never been exposed to selection show different rates of change in the GC contents of paired and unpaired regions as the GC content of the whole molecule changes, suggesting that different bases have different intrinsic propensities for base pairing (30). Consequently, the paradigm introduced by Muto and Osawa for detecting selection as a different response to changes to GC content in different parts of the molecule, which works well for coding regions (31,32), is not valid for rRNA.
The aim of this article is to test the commonly accepted hypotheses that compensatory mutations in paired regions quantitatively dominate RNA evolution, and that the unpaired regions form a single category that can be treated as homogeneous. Specifically, we address the following questions:
Do the different unpaired categories (hairpin loops, bulges, and multi-helix junctions) change at the same rate over a wide range of species? There are several reasons why we might expect these structural elements to evolve at different rates. First, they are subject to different structural and functional requirements. Second, they have different patterns in terms of nucleotide composition (30), which suggests that they are under distinct evolutionary constraints. Third, in a study that distinguishes between these structural elements in six mammalian rRNAs, they are shown to evolve at different rates (33). Incorporating structure-specific rates of change should make current models of RNA evolution more accurate.
Do paired regions always evolve fastest, and is the general pattern of substitution rates shared among all three phylogenetic domains (the archaea, the bacteria and the eukaryotes) and both ribosomal subunits? Rates of change vary considerably among taxa. We might expect to see a change in the relative substitution rates in eukaryotes, because they distinguish themselves from archaea and bacteria in several ways. First, they have longer rRNA sequences (34). Most if not all eukaryotic insertions are on the ribosome surface (35), where substitution rates are known to be higher than in the ribosomal center (36). The eukaryotes also show different trends in the base composition of different structural components of the rRNA (30). Finally, it has been shown in a small sample of eukaryotic sequences that loops and bulges evolve as fast as stems [33].
How does the distribution of different structural components in the 3D structure of the ribosome affect their evolution? It is known that parts of the rRNA further from the center of the ribosome evolve more quickly (36). Can the generally accepted faster rate of evolution of stems be explained by their spatial distribution in the ribosome, e.g. because stems more frequently occur at the ribosomal surface? Now that we have a 3D structure, understanding the relationship between variability and 3D structure could potentially help both structure prediction and the development of better models of rRNA evolution for phylogeny.
METHODS
Ribosomal sequence and structure data
There are many ribosomal sequences and structural models available, allowing a detailed analysis of evolutionary rates. The first complete rRNA sequences for both the SSU and LSU were determined for E. coli shortly after the Sanger sequencing method became available (37,38). Today, a wealth of aligned sequence data is available. The European rRNA database (RDB) (39) and the comparative RNA web site (CRW) (29) provide alignments containing up to several hundred LSU sequences per phylogenetic domain (about 400 bacterial LSU sequences), and thousands of SSU sequences (about 12 000 bacterial SSU sequences in the RDB).
Soon after the first full-length rRNA sequences were determined, the first covariation-based secondary structure models were developed (10,11,40–44). These models predicted the secondary structure in terms of Watson–Crick and G–U wobble base pairs. As the amount of sequence data has increased, the structural models have repeatedly been refined. Over time, they have matured into complex models that also incorporate non-standard base pairs and tertiary interactions (2,45–49). These models are available on the CRW (29). The RDB (39) provides a similar, independently developed, set of structural models. These models were originally derived by comparing 14 SSU rRNA sequences and surveying existing structural models (50), and have been successively refined (51–58).
In general, the available secondary structure models are of high quality. The bacterial secondary structure model is especially well established and is consistent with chemical experiments (59) and crystal structures of the ribosome (60). The eukaryotic structural model has been accurately determined for the more conserved regions, but the structure of some of the variable regions is still disputed (61). Thus far, there is no crystal structure to resolve these controversial regions.
In this study, we used sequence and structure information from three sources: the European RDB (39), the CRW (29), and the RCSB Protein Data Bank (62,63). Table 1 provides details about the model organisms, alignments, sequence accession numbers, and crystal structures used.
Table 1.
Sources of sequence and structure information
| Source | Model species | Subunit | Seqs | Acession | Ref. | Crystal structure |
|---|---|---|---|---|---|---|
| RDB | E. coli | SSU | 500 | J01695 | (13) | |
| RDB | E. coli | LSU | 71 | J01695 | (13) | |
| CRW | E. coli | SSU | 4214 | J01695 | (29) | |
| CRW | E. coli | LSU | 436 | J01695 | (29) | |
| RDB | T. thermophilus | SSU | 3407 | M26923 | (36) | 1GIX (70,74) |
| RDB | T. thermophilus | LSU | 184 | X12612 | (36) | 1GIY (70,74) |
| RDB | T. thermophilus | 5S | 310 | * | (79) | 1GIY (70,74) |
| RDB | S. cerevisiae | SSU | 500 | J01353 | (80) | |
| RDB | S. cerevisiae | LSU | 77 | U53879 | (17) | |
| CRW | S. cerevisiae | SSU | 1939 | U53879 | (29) | |
| CRW | S. cerevisiae | LSU | 116 | U53879 | (29) |
Source: RDB — European rRNA database, CRW – Comparative RNA web site. Seqs: number of sequences in the alignment from which conservation/variability is calculated. Accession: accession number of the structural model. Ref: reference. Crystal structure: crystal structure we used for 3D calculations. *: structural model derived from 5S rRNA database.
Structural classification
RNA secondary structure is a collection of base pairs, interspersed with unpaired bases. Base pairs can either be nested or non-nested. Two base pairs, one between positions i and j and the other between positions j′ and j′ (where i<j < i′ < j′ and i<i′) are nested if either i<i′ < j′ < j or i<j < i′ < j′. Pseudoknots are non-nested base pairs between a loop of one stem and residues outside that stem (64).
RNA secondary structures can be decomposed into distinct structural classes. A fully nested structure without pseudoknots can be represented as a tree, and thus each position can be classified into either stem, loop, bulge, junction, end or flexible (30). In this study, we did not remove pseudoknots from the structural models, which required us to combine some of the structural classes for simplicity. We distinguished stem, loop, bulge and ‘junction/other’ [essentially the same as in (33)]. The ‘junction/other’ category includes the categories junction, end and flexible in the fully nested structures, and pseudoknotted regions. Most bases in this class are from multi-helix junctions.
In summary, the class ‘stem’ contains all base-paired positions, the class ‘loop’ contains all unpaired positions connecting two halves of a helix, the class ‘bulge’ contains all unpaired bases connecting exactly two helices and all other positions are classified as ‘junction/other’.
Calculating rates of change from large alignments
We used two types of ‘variability maps.’ In these maps, variability is calculated from a large alignment of rRNA sequences (separated by phylogenetic domain and subunit) and superimposed onto a structural model. First, we used the RDB variability as calculated by the substitution rate calibration method (65,66), available from the European rRNA database (39). Second, we used the CRW secondary structure conservation maps, provided on the comparative RNA web site (29), where conservation is calculated based on the nucleotide distribution at a particular alignment position.
The substitution rate calibration method classifies each position as one of six rate categories (seven in more recent publications). Sites that are absent in 75% or more of the sequences in the alignment are considered too variable to be classified and are excluded from the analysis. On the CRW conservation diagrams, only four rate categories are distinguished, ranging from more than 98% conserved to less than 80% conserved. For a base to be classified in one of the four categories, it has to be present in at least 95% of the sequences in the alignment. The CRW conservation method thus uses stricter requirements for classifying residues. The two different measures of conservation agree well (average r2 = 0.824 when using a sliding window); see Supplementary Figures 1 and 2 for the strength of association between the two measures.
Figure 1.
In bacteria, stems dominate in high-rate categories, unpaired regions in low-rate categories. For each structural category, we calculated the percent of positions (y-axis) in each rate category (x-axis). For example, of all the positions in stems, 8% is in rate category 1, 22% is in rate category 2, etc. The graph contains four different series, one for each structural category. Within one series the values add up to 100%. The data is calculated from the RDB variability categories superimposed on the RDB secondary structure model of E. coli.
Figure 2.
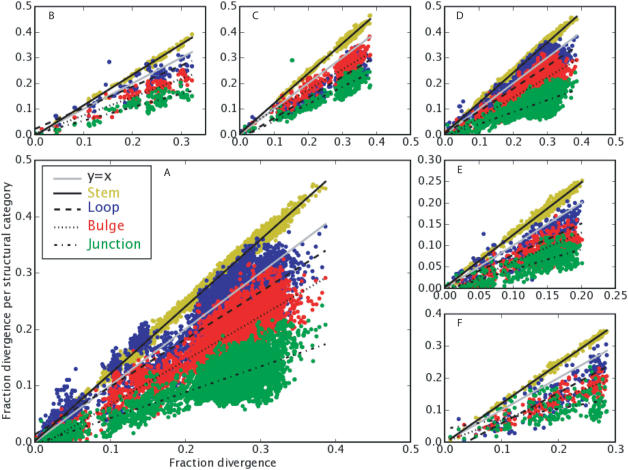
Pairwise sequence comparisons for bacteria and archaea. Stems evolve fastest, both in the complete set of bacteria and in individual lineages. However, the different classes of unpaired regions always evolve at significantly different rates. The scatterplots show the fraction divergence per structural category (y-axis) versus the fraction divergence overall (x-axis); see Methods for definition. (A) SSU bacteria. (B) SSU archaea. (C) LSU bacteria. (D) Bacteria without Actinobacteria. (E) Proteobacteria. (F) Firmicutes.
Both CRW conservation and RDB variability data are available for SSU/LSU E. coli as bacterial model and S. cerevisiae as eukaryotic model. The structural models from both sources are not exactly the same, but share on average 80.5% of their base pairs (see Supplementary Data). We used the variability maps to assign each position to a rate category.
Calculating rates of change from pairwise comparisons
Calculating the rate of change from pairwise sequence comparisons circumvents two important problems that arise from calculating variability values from large alignments. The first problem is that many positions have ambiguous structural classifications. This problem arises because the variability values calculated from the large alignments are superimposed on a single structural model that is representative of a whole phylogenetic domain. Because not all sequences in the alignment fold into exactly the same structure, the structural category at many positions will only be valid for a subset of the alignment. For example, a position that is in a bulge in the model species, might be in a multi-helix junction in another species in the alignment because of a stem-loop insertion. In the pairwise comparison method, we avoid this problem because we do not assume a single structural model for every sequence in the alignment. Instead, each sequence has its own model (available on the CRW), which is used in the comparison. Positions with ambiguous structural classifications can be averaged over the different possibilities, or excluded from the analysis. In addition, limiting the comparison to more closely related species also reduces the number of ambiguous positions. The second problem is that the strict presence/absence requirements in the CRW calculations result in many unclassified positions. This problem mainly affects alignments of eukaryotic sequences, because these alignments contain many insertions that are only present in a few species. Pairwise comparisons avoid this problem because variable regions are not excluded.
In the pairwise comparison method, we counted the base changes between the sequences in each structural element. The neutral assumption would be that all the changes in the molecule are distributed equally over the different structural elements, and thus that each structural element absorbs the same percentage of change. We applied two different counting methods. First, counting point mutations only, in which we only counted positions with a non-degenerate base in both sequences and the same structural classification. Second, counting insertions and deletions (indels) in addition to point mutations, in which we counted positions with a non-degenerate base in both sequences or a non-degenerate base in one sequence and a gap in the other. Especially in comparisons over larger evolutionary distances, incorporating indels is very important because they make up a large part of the sequence divergence.
From the raw counts, we calculated the fraction divergence overall and per structural category as the fraction of positions with a different base in both sequences divided by the total number of positions that we were counting. As mentioned before, not all positions were counted. Positions where both sequences had a gap were eliminated before the counting process started, and positions that did not meet the criteria for being included in the counting process were ignored. For example, these could be positions that contained degenerate bases or that were not sequenced. We also calculated the fraction of comparisons in which the hairpin loops were changing faster than the stems. Optionally, we could split the counts when the structural classification was ambiguous (otherwise ignored), and we could limit the allowed fraction of divergence or ignored positions. The tables containing results of these calculations will specify the counting method and chosen options and limits.
For the pairwise comparisons, we used the sequences, structural models and a high-quality alignment from the CRW web site (all downloaded in June 2006). Initially, we did a large-scale comparison within each phylogenetic domain and subunit, where we compared all sequences for which there was a structural model and an entry in the CRW alignment. When looking over the whole range of diversity, many positions were ignored because of conflicting structural information. Focusing on an individual lineage reduced the structural differences, because the species were more closely related and thus less structural changes had occurred since the time of divergence. Not all sequences for which a structural model was available had an exact match in the alignment. For these groups, we aligned the sequences with MUSCLE (67) and inserted the gaps into the corresponding structural classifications. These alignments were of high quality, because the species were closely related (see Supplementary Data). Since there were at most very small differences between the data calculated from the CRW alignment or from the MUSCLE alignment, we reported the results for the largest data set.
3D structural calculations
We performed structural calculations using the PDB files 1GIX and 1GIY, corresponding to the crystal structures of the ribosomal subunits from Thermus thermophilus solved at 5.5 Å. These files provide the 3D coordinates for the phosphorus atom in each residue. It has previously been shown that the average variability of residues increases with distance from the center of the ribosome (36), but it is unclear that the geometric center is the correct reference point. We calculated distances between the P atom of each residue and the following locations: the distance from the peptidyl transferase center (‘PTC’), defined as the P atom of residue A2451; the distance from the tRNA path (‘path’), defined as the distance from the closest P atom of any of the three tRNAs or any of the two mRNA codons included in the crystal structure; the distance from the closest protein (‘protein’), defined as the distance from the closest C-α atom of any protein attached to the same subunit; the distance from the subunit interface (‘interface’), defined as the distance from the closest P atom in the other subunit, as well as the distance from the center (‘center’), defined as the distance from average of the coordinates of all P atoms in the ribosome (including SSU, LSU and 5S rRNA). We then correlated each of these distances for each residue with evolutionary rate and structural category, as calculated above.
RESULTS
We find that structural categories in the ribosomal RNA evolve at different rates, and that these rates vary across phylogenetic domains. Although it is true that highly conserved regions tend to be unpaired, the converse, that unpaired regions are more conserved, is not always true (although it is widely assumed).
Stems indeed dominate rRNA evolution in bacteria and archaea
In bacteria, stems contain many fast-evolving positions and few slow-evolving positions compared to the three different unpaired categories. Figure 1 shows this relationship using evolutionary rates derived from a large alignment of SSU bacterial sequences, superimposed on the E. coli secondary structure. The data shown are for the RDB evolutionary rates and structural model, but any combination of CRW and RDB rates and structures gives essentially identical results (see Supplementary Data for further comparisons). This observation is consistent with previous reports of fast-evolving stems in bacterial SSU rRNA (10–17). For each structural category, the distribution of rates appears to be bimodal with residues evolving at intermediate rates being rare. This pattern is also observed in the eukaryotes, perhaps suggesting that residues in the ribosome are either under strong selection or under no selection.
Directly counting the changes between sequences through pairwise comparisons revealed a similar pattern (Table 2). In both bacterial and archaeal SSU sequences, we observed that stems evolve fastest, the three unpaired regions are slower, with hairpin loops being faster than bulges and the junctions being slowest (Figure 2A and B). In both groups, counting only point mutations or including indels did not alter the observation. Including indels slightly increased the fraction of divergence in hairpin loops at larger overall evolutionary distances (data not shown). The fastest evolving structural elements change 2.7-fold faster than the slowest evolving elements (measured at half of the maximum divergence).
Table 2.
Stems evolve fastest in bacteria and archaea
| Lineage | SU | Aln | Mode | FD | FI | Split | Cmp | MD | L>S | Plot |
|---|---|---|---|---|---|---|---|---|---|---|
| Archaea | SSU | CRW | P | 1.0 | 1.0 | F | 171 | 30.1 | 5.26 | |
| Archaea | SSU | CRW | I | 1.0 | 1.0 | F | 171 | 32.2 | 10.5 | ⋆ |
| Bacteria | SSU | CRW | P | 1.0 | 0.3 | F | 10011 | 34.1 | 4.58 | |
| Bacteria | SSU | CRW | I | 1.0 | 0.3 | F | 10011 | 38.7 | 6.19 | ⋆ |
| Bacteria - ** | SSU | CRW | P | 1.0 | 0.3 | F | 9108 | 34.1 | 0.53 | |
| Bacteria - ** | SSU | CRW | I | 1.0 | 0.3 | F | 9108 | 38.7 | 2.22 | ⋆ |
| Bacteria | LSU | CRW | P | 1.0 | 0.2 | F | 787 | 34.7 | 1.27 | |
| Bacteria | LSU | CRW | I | 1.0 | 0.10 | F | 729 | 38.2 | 1.37 | ⋆ |
| Bacteria | LSU | CRW | P | 0.06 | 0.1 | F | 12 | 4.35 | 75.0 | |
| Bacteria | LSU | CRW | I | 0.06 | 0.1 | F | 12 | 4.45 | 75.0 | |
| α-Proteobacteria | SSU | MUS | I | 1.0 | 1.0 | F | 55 | 16.5 | 7.27 | ⋆ ⋆ |
| β-Proteobacteria | SSU | MUS | I | 1.0 | 0.1 | F | 21 | 15.2 | 4.76 | ⋆ ⋆ |
| γ-Proteobacteria | SSU | MUS | I | 1.0 | 1.0 | F | 496 | 20.1 | 2.42 | ⋆ ⋆ |
| γ-Proteobacteria | LSU | MUS | I | 1.0 | 0.1 | F | 136 | 34.1 | 2.94 | |
| Firmicutes | SSU | MUS | P | 1.0 | 1.0 | F | 190 | 25.9 | 1.58 | |
| Firmicutes | SSU | MUS | I | 1.0 | 1.0 | F | 190 | 28.5 | 2.63 | ⋆ |
| Firmicutes | SSU | MUS | I | 1.0 | 0.15 | F | 153 | 28.5 | 3.27 |
In archaea and bacteria, the percentage of comparisons in which the hairpin loops dominate (column L>S) is very low. The table contains the following columns: Lineage (‘Bacteria - **’ refers to the comparisons among the bacteria where all comparisons between two Actinobacteria are excluded), SU (subunit), Aln (type of alignment, CRW = alignment from comparative RNA web site, MUS = MUSCLE alignment), Mode (P = point mutations only, I = Indels added), FD (limit on fraction divergence), FI (limit on fraction ignored), Split (whether the counts at positions with ambiguous structural classification are split between the two structural categories; if false, the positions are ignored), Cmp (number of pairwise comparisons), MD (maximum divergence observed), L>S (percent of comparisons in which loops change faster than stems), and plot (data plotted in Figure 2, ⋆⋆ entries are plotted in one graph).
The rates of substitution observed in the whole bacterial domain reappeared when focusing on clusters of more closely related species. In proteobacteria (Figure 2E) and firmicutes (Figure 2F), stems typically evolved faster than any unpaired categories. There did not seem to be a difference between the bacilli and the mollicutes when inspected individually, despite the extreme changes in GC content in the mycoplasmas (68). Among spirochetes, the pattern also seemed typical (data not shown).
In both the archaea and bacteria, some sequences appeared to escape the general pattern. Among the archaea, about 5% of the comparisons when counting only point mutations (10% with indels) contradicted the general observation that stems change faster than loops. In all of these comparisons one of the sequences came from either the Aeropyrum or Pyrococcus genus. In the comparisons within bacterial SSU sequences, about 6% had higher rates of change for loops than for stems. Most of these outliers were comparisons among the Actinobacteria. When all pairwise comparisons between two species within this group were excluded, the fraction of comparisons that contradicted the general pattern was reduced to 0.5% (Table 2 and Figure 2D) (see Discussion for further interpretation of these patterns).
As for SSU sequences, in bacterial LSU sequences all unpaired categories changed slower than the stems (Figure 2C). However, unlike in SSU sequences, LSU bulges changed faster than loops, when considering all bacteria or when focusing just on the LSU γ-proteobacteria (data not shown).
Hairpin loops evolve faster than other structural categories in eukaryotes
In eukaryotic SSU rRNA, loops are the fastest evolving structural element, and evolve 1.37-fold faster than stems. Eukaryotes thus do not follow the typical pattern of evolutionary rates observed in archaea and bacteria. This deviation from the bacterial pattern is consistent across analyses.
Superimposing the variability values derived from large alignments onto a single structural model is only partially useful for eukaryotes. The first reason is that there is controversy over the structure of a particular region (positions 634–861 in the eukaryotic RDB model for accession J01353) (61), in the CRW model all these positions are unpaired, in the RDB model these positions are in a complex pseudoknotted region. The other problem is that eukaryotic sequences have large insertions or deletions with respect to each other. In the CRW conservation diagrams, any position that is present in less than 95% of the sequences of the alignment is considered variable and not characterized further. This applies to about 30% of the positions in both the SSU and LSU eukaryotic alignment. With these caveats in mind, we examined the fraction of positions in each rate class for all structural elements.
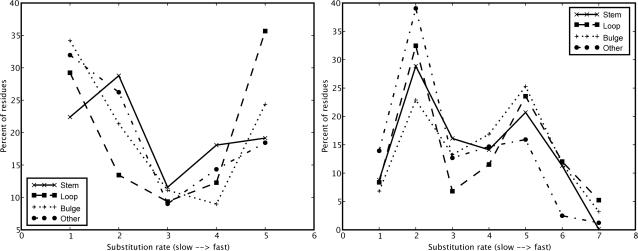
When we used the CRW conservation values and CRW structural model and ignored all unclassified positions, loops, bulges and stems had the same fraction of positions in rate class 3 (out of 4) and stems have the highest fraction of positions in the fastest evolving class. This observation could no longer be made when we included the unclassified positions. In that case, stems dominated in rate class 3 and 4 (but also in class 2, which contains moderately conserved positions), but the unpaired regions dominated in the class with the most variable positions. More than 45% of the junctions fell in this class due to the long unstructured region. In fact, when we ignored this region, hairpin loops dominated that class (Figure 3 left). The combination of RDB variability scores and the RDB structural model showed the change more clearly. When we ignored unclassified positions, loops and bulges dominated in the two fastest rate classes. The positions in the unclassified regions were all unpaired, and hairpin loops dominated this class independent of excluding the structurally controversial region (Figure 3 right). In summary, the results from the large alignment are not clear-cut, but they strongly suggest that hairpin loops contain the highest fraction of fast evolving positions.
Figure 3.
Comparison between rates inferred using CRW and RDB rate categories showing that loops dominate high-rate classes in eukaryotes. The graphs show the fraction of positions (y-axis) per rate class (x-axis) for each structural element. Both graphs show data for S. cerevisiae. Left: CRW conservation values, CRW structural model, excluding the controversial region, including unclassified positions as highest rate class. Right: RDB variability scores, RDB structural model, including the controversial region and unclassified positions. These two figures present essentially the same data. The difference is caused by the different rate categories used by the two data sources. Rate categories 1 and 5 in CRW correspond roughly to 1/2 and 5/6/7 in RDB, respectively.
The results from the pairwise sequence comparisons leave no doubt that hairpin loops dominate rRNA evolution in eukaryotes (Figure 4A and Table 3). When we counted insertions and deletions in addition to point mutations, hairpin loops changed at a faster rate than stems in more than 90% of the comparisons. The fact that indels are important for eukaryotic evolution was indicated by the high fraction of ignored positions when only counting point mutations (and hence the limitation on the maximum fraction of ignored positions). This was emphasized by the increase in the percentage of pairwise comparisons in which hairpin loops change faster than stems when we added indels to the counts (from about 80–93%).
Figure 4.
Pairwise sequence comparisons for eukaryotes. Hairpins (blue) evolve fastest, both overall and in each lineage individually. The scatterplots show the fraction divergence per structural category (y-axis) versus the fraction divergence overall (x-axis); see Methods for definition. (A) SSU eukaryotes. (B) Viridiplantae and Metazoa. (C) Alveolata. (D) Fungi. (E) Stramenopiles. (F) LSU eukaryotes.
Table 3.
Hairpin loops dominate evolution in eukaryotes
| Lineage | SU | Aln | Mode | FD | FI | Split | Cmp | MD | L > S | Plot |
|---|---|---|---|---|---|---|---|---|---|---|
| Eukaryotes | SSU | CRW | P | 1.0 | 0.3 | F | 2061 | 27.7 | 74.8 | |
| Eukaryotes | SSU | CRW | P | 0.1 | 0.2 | F | 163 | 9.97 | 89.6 | |
| Eukaryotes | SSU | CRW | I | 0.35 | 0.15 | F | 620 | 35.0 | 94.4 | ⋆ |
| Eukaryotes | SSU | CRW | I | 0.1 | 0.15 | F | 115 | 9.90 | 93.0 | |
| Eukaryotes | SSU | CRW | I | 0.2 | 0.15 | T | 259 | 19.9 | 93.8 | |
| Viridiplantae | SSU | MUS | P | 1.0 | 1.0 | F | 136 | 15.8 | 100 | ⋆⋆ |
| Viridiplantae | SSU | MUS | I | 1.0 | 1.0 | F | 136 | 17.5 | 99.3 | |
| Metazoa | SSU | MUS | P | 1.0 | 1.0 | F | 36 | 20.6 | 100 | ⋆⋆ |
| Metazoa | SSU | MUS | I | 1.0 | 1.0 | F | 36 | 30.6 | 97.2 | |
| Alveolata | SSU | MUS | P | 1.0 | 0.15 | F | 190 | 17.8 | 86.3 | |
| Alveolata | SSU | MUS | I | 1.0 | 0.1 | F | 209 | 33.9 | 98.1 | ⋆ |
| Alveolata | SSU | MUS | I | 0.1 | 0.02 | F | 78 | 6.34 | 96.2 | |
| Fungi | SSU | MUS | P | 1.0 | 0.1 | F | 105 | 17.5 | 46.7 | |
| Fungi | SSU | MUS | I | 1.0 | 0.06 | F | 119 | 22.5 | 87.4 | ⋆ |
| Stramenopiles | SSU | MUS | P | 1.0 | 1.0 | F | 703 | 17.1 | 65.4 | |
| Stramenopiles | SSU | MUS | I | 1.0 | 1.0 | F | 703 | 19.9 | 91.2 | ⋆ |
| Stramenopiles | SSU | MUS | I | 1.0 | 1.0 | T | 703 | 20.8 | 93.9 | |
| Eukaryotes | LSU | CRW | P | 0.15 | 0.15 | F | 19 | 15.0 | 47.4 | |
| Eukaryotes | LSU | CRW | I | 0.15 | 0.15 | F | 18 | 13.5 | 61.1 | ⋆ |
The table contains the following columns: Lineage, SU (subunit), Aln (type of alignment, CRW = alignment from comparative RNA web site, MUS = MUSCLE alignment), Mode (P = point mutations only, I = indels added), FD (limit on fraction divergence), FI (limit on fraction ignored), split (whether the counts at positions with ambiguous structural classification are split between the two structural categories; if false, the positions are ignored), Cmp (number of pairwise comparisons), MD (maximum divergence observed), L>S (percent of comparisons in which loops change faster than stems) and plot (data plotted in Figure 4, ⋆⋆ entries are plotted in one graph).
The results from comparisons within several eukaryotic lineages corroborated the observation over all eukaryotes that hairpin loops evolve fastest (Figure 4B-E). In plants and animals, the pattern held for every single comparison (when only counting point mutations). In the fungi, stramenopiles and alveolata, insertions and deletions played an important role, and resulted in a 30–40% increase in the fraction of comparisons in which loops evolve faster than stems.
Although too few eukaryotic LSU sequences were available for the same analysis as above, we employed this data by comparing the three sequences for which we had a structural model and an aligned sequence (Arabidopsis thaliana, Saccharomyces cerevisiae and Oryza sativa) to all other sequences in the eukaryotic LSU alignment. In these comparisons, we classified positions using the secondary structure of the first sequence. Examining sequences up to 15% divergence from these reference sequences, hairpin loops evolved faster than stems in 46% (just point) and 56% (indels) of the comparisons (Figure 4F). These results suggest that the pattern holds between SSU and LSU, although the small data set makes this conclusion tentative.
To control for differences in GC content, which can affect the distribution of different structural categories (30), we limited the range of GC content to be consistent between bacteria and eukaryotes (45–55%). This produced essentially identical results to those shown, indicating that they are not an artifact of GC content (data not shown).
The distribution of structural elements in 3D structure only partially explains differences in rate
Crystal structures of the complete ribosome have recently become available (69–75), allowing us to relate evolution of the rRNA to specific structural features. Since the complete ribosomal structure is only available for bacterial species, we limit this analysis to that phylogenetic domain. We correlated the distance from each of several structural features (see Methods) in the T. thermophilus sequence with rate category and structural category in the bacterial alignment. This comparison was revealing; the distribution of structural categories within the ribosome is seen to be highly non-random.
Distance from features within the ribosome strongly affects conservation in all structural categories
We tested whether the conservation of residues varied systematically with distance from several different features within the ribosome, and whether these variations were consistent across structural categories. Figure 5 shows the distribution of rate categories as a function of distance from the PTC in the bacterial LSU (the PTC is located within the LSU). Moving away from the PTC, the slower rate categories rapidly decrease in abundance, whereas the faster rate categories rapidly increase, showing a clear relationship between distance from the PTC and evolutionary rate (the proportion of bases changing at intermediate rates seems to be relatively constant in each distance bin). For example, 85.7% of the residues are in rate class 1 at 0–10 Å and none are in rate class 7; at >110 Å 9.68% of the residues are in rate class 1 and 41.9% are in rate class 7.
Figure 5.
Distribution of rate categories as a function of distance from the PTC. Each bar graph shows the fraction of atoms in each rate category (y-axis) versus the distance from the PTC [x-axis; last bin contains all atoms >100 Å (or 140 Å on the SSU graph) away from the PTC]. The fractions within a distance bin (vertical column) add up to 1.0. (A) LSU all structural elements. (B) SSU all structural elements. (C) LSU stem. (D) LSU loop. (E) LSU bulge. (F) LSU junction/other.
The rate categories show clear patterns as a function of distance from the core structural elements. Figure 6A shows two shells of residues in the LSU, the inner shell being within 20 Å of the PTC and the outer shell being at least 80 Å from it. The inner shell is primarily composed of slow-evolving residues (cool colors), reflecting the fact that this is one of the most conserved regions within the ribosome; in contrast, the outer shell consists primarily of fast-evolving sites (warm colors).
Figure 6.
Distribution of structural elements and rate categories in the ribosome. Each panel shows the T. thermophilus ribosome, with residues highlighted according to proximity to specific structural features and colored either by rate or by structural category. Panel (A) shows the large subunit with residues within 20 Å of the PTC (near the tRNA ends) and residues >80 Å away from the PTC (outer shell) highlighted. The residues are colored by rate category (fast evolving sites in orange/red, slow evolving sites in cyan/blue, sites changing at an intermediate rate in gray). The small subunit, 5S rRNA, and proteins are hidden. The PyMol script that generates these figures is available as Supplementary Data to allow interactive exploration of these features. Panel (B) shows all residues in rate category 1 colored blue, all residues in rate category 7 and 8 colored in red colors. Panel (C) shows all residues within 15 Å from PTC (right), within 15 Å from the tRNA path in the SSU (left) and within 10 Å on both sides of the subunit interface (middle), colored by structural element. Stem in yellow, loop in blue, bulge in red and junction in green.
We see a similar trend for each of the other distances, including distance to the nearest protein and distance to the path of the tRNA within the ribosome. We also find that these patterns hold for each ribosomal feature and for both subunits (Figure 5B; note that because both the center and the PTC are located within the LSU, the closest approach to these features in the SSU is at least 49.5 Å). The results also hold for the structural categories individually, to the limits of sampling error (Figure 5 C–F show the LSU PTC distances broken down by structural category). These results were statistically highly significant (P-value ranged from 3.03× 10−82 to 0.002 in a G test for independence between distance bin and structural category). The effect was strongest for the PTC (85.7% of bases within 10 Å in rate category 1) and weakest for proteins and the subunit interface (about 25% of bases within 10 Å in rate category 1 and intermediate for the tRNA path). Thus, as expected, proximity to the PTC and, to a lesser extent, interactions with tRNAs and proteins exhibit strong selective influences on residues within the rRNA. These influences are reproducible for each individual structural category; it is not true that the stems, but not the loops, are influenced by the distance to the nearest protein. Interestingly, the effects do not saturate after a few angstroms but continue out to the surface of the molecule. For example, the residues 60 Å from the PTC in the LSU evolve slower than the residues 80 Å from the PTC, suggesting that the influence of specific functional sites may extend over long distances.
Fast-evolving sites (in all structural categories) thus tend to be common near the surface of the ribosome and rare in the interior, as previously observed by many investigators [see for example (36)]. Figure 6B shows the slowest rate category from the RDB data (category 1) in blue, and the fastest categories (7 and 8) in red. The fast categories clearly cluster near the surface. Figure 6B thus directly illustrates the trend that fast-evolving sites are more abundant on the outside of the ribosome in both subunits and across all structural elements.
Structural categories are unequally distributed throughout the rRNA
We then tested whether proximity to important functional elements of the ribosome was associated with specific structural categories. Three important structural features are highlighted in Figure 6C: the PTC, the tRNA path and the SSU/LSU subunit interface. The cluster of residues within 15 Å of the PTC (right-hand side of the figure, clustered around the end of the tRNA) is almost entirely composed of junctions (shown in green), consistent with previous findings (76). The cluster of residues within 15 Å of the tRNA path in the small subunit (top-left of the diagram, near the anticodon loop of the tRNA) is also primarily composed of junctions, with some stems (yellow) and a few bulges (red). The cluster of residues within 10 Å of the subunit interface (center) consists primarily of stems.
Stems are most equally distributed in both the SSU and the LSU, making up about 60% of the atoms in each distance bin. In the SSU, the area close to the tRNA path is mainly made up of stems (30–60%) and junctions (30–45%), and almost no bulges (5–8%) and loops (5–15%) (Figure 7A). In the LSU, this area is mostly stems (50–60%), loops (9–17%) and bulges (16–20%), with junctions comprising just 10–15% of nearby residues. Interestingly, in the LSU, the region around the PTC is composed almost entirely of junctions (71% of the residues within 10 Å fall into this category, and no bulges are present); the proportion of residues that are junctions falls steeply with increasing distance from the PTC, dropping to less than 25% above 20 Å (Figure 7B). Figure 7C and D shows that the residues participating in inter-subunit RNA–RNA contacts at the subunit interface are mostly in stems, loops and bulges (with only 9.6% and 6.5% junctions within 10 Å of the subunit interface in SSU and LSU, respectively). All the differences in structural category representation are highly significant (P ranges from 1.37× 10−20 to 0.01 in G tests for independence).
Figure 7.
Distribution of structural elements in the ribosome as a function of distance from specific structural features. Each bar graph shows the fraction of atoms in each structural element (y-axis) versus the distance from a particular feature (x-axis; last bin contains all atoms more than that distance away). The fractions within a distance bin (vertical column) add up to 1.0. (A) SSU, tRNA path. (B) LSU, PTC. (C) SSU, subunit interface. (D) LSU, subunit interface.
The distribution of structural elements as a function of other features can be found in the Supplementary Data. Visualization of this distribution in 3D is very enlightening. For example, one can color the residues at both sides of the subunit interface by structural element or rate category to find that these are mostly stems, loops and bulges, and in general highly conserved, or changing at intermediate rates. We encourage readers to explore the structure using the PyMol script we supply.
Variation in rates in different structural categories is not fully explained by proximity to functional elements
Because both the rates and the structural categories are strongly influenced by the overall structure of the ribosome, the differences in overall evolutionary rate we observe in different structural categories might be solely due to unequal distances from, for example, the PTC. Can we predict the distribution of rate categories in the stems purely from the distance data? The null hypothesis we are testing here is that the structural category has no influence on evolutionary rate, and that distance from functionally important regions is the sole factor that affects evolutionary rate. If this null hypothesis were true, we would be able to calculate the rate of evolution of each structural category as the weighted average of the products of the rate at each distance and the fraction of bases in the structural category that is found at that distance.
For example, suppose that the distance from the PTC were the only factor that influenced the rate of evolution. If all bases near the PTC evolved slowly, but few of these bases are stems, stems would appear to evolve rapidly simply because they are, on average, far from the PTC. We can account for this effect by binning the residues into distance classes (e.g. every 10 Å) from the feature of interest. For each distance, we multiply the fraction of all bases that are in each rate category by the fraction of all stems that appear at that distance. For example, if 4% of all stems were within 10 Å of the PTC, and residues within 10 Å of the PTC were 75% in rate category 1, 20% in category 2 and 5% in category 3, the contribution of residues within 10 Å of the PTC to the overall rate of evolution in stems would be 75% × 4% = 3% for category 1, 20% × 4% = 0.8% for category 2 and 5% × 4% = 0.2% for category 3. Repeating this calculation for the other distances and summing the results gives the predicted fraction of bases in stems that fall into each rate category. We can then test whether this prediction matches the overall distribution of stems among rate categories. See the Supplementary Data for a description on calculating the correlations between predicted and actual rate distributions.
We predicted the rate distribution from each structural feature (center, tRNA path, etc.). Out of all correlations between predicted and actual rate distributions, only the prediction based on the distance to the tRNA path has a statistically significant correlation with the actual distribution (see Supplementary Data for graphs and additional discussion). For example, using the distance from the tRNA path in the LSU, r2 = 0.33 and P = 0.001 for the relationship between observed and expected deviations. The effect is thus highly significant, but relatively small, explaining only a third of the variance. The distance from the center of the ribosome is not predictive (r2 = 0.07 − 0.11, P>0.05).
Thus, most of the variation in the rate of evolution in the different structural categories is not explained by the differential distribution of these structural categories throughout the ribosome. For example, the fast evolution of stems in bacteria cannot be simply explained by a high abundance of stems on the ribosome surface. We therefore reject the null hypothesis that the distance from functionally important regions is the sole factor that affects evolutionary rate, and instead conclude that structural category itself influences the evolutionary rate.
DISCUSSION
We have demonstrated that different structural elements change at different rates in different lineages. In bacteria and archaea, we observe the generally accepted pattern of fast-evolving stems. However, this pattern differs markedly in eukaryotes, where hairpin loops actually evolve considerably faster than stems do. This result is not primarily due to insertions and deletions in non-conserved surface loops in the eukaryotes, because it persists when these regions are excluded from the analysis. The different types of unpaired regions always behave differently from one another, underscoring the importance of moving beyond the paired–unpaired dichotomy in studies of evolutionary rates in rRNA.
To minimize the effects of errors in the structural models, the alignments, and the rate inference procedure, we used several complementary methods that agreed well with one another. The general trends we identified are supported by existing conservation maps and secondary structure models calculated by two different research groups (RDB and CRW), and by direct inference of the amount of change in each structural category from pairs of sequences. We verified that the choice of whether to include or exclude gaps in calculations of evolutionary distance, and use of either automated MUSCLE alignments or hand-curated alignments from CRW, produced similar results. No matter which metric is used to measure the substitution rates, hairpin loops evolve substantially faster than stems in the eukaryotic lineage, and these results hold both over short and long evolutionary distances.
There is a small but significant effect of the distribution of structural elements throughout the ribosome: for example, the region around the PTC is largely made up of junctions, whereas the subunit interface and the regions near proteins (subject to the limits of the 5.5 Å resolution of the crystal structure) are largely made up of stems. We believe that differences in evolutionary rate between structural categories are not due to these differences in distribution because we can calculate the distribution of rates in each structural category that would be expected if distance from functionally important regions were the only factor, and these distributions of rates do not match. Thus, the differences in rates are likely to be meaningful and are not simply an artifact of the composition of the most conserved regions.
The distribution of structural categories in the ribosome was influenced more strongly by proximity to defined structural features, such as the PTC and the tRNA path, than by proximity to the geometric center. These results suggest that the factors driving the distribution of structural elements within the ribosome are primarily adaptive rather than consequences of, say, the physics of helix packing. However, the results contrast strikingly with proteins, in which hydrophobic residues preferentially assort themselves into the core of the molecule. Thus, secondary structure (and, presumably, nucleotide composition) is likely to be a poor guide to predicting whether a particular region of the rRNA is buried or surface exposed.
Relative rates of evolution of different structural categories, especially the ratio of changes in stems to loops, differ drastically in different lineages. These results suggest that the influence of each structural category on the rate of evolution is not universally consistent, diminishing the plausibility of using differences in rates in different regions to infer properties of the secondary structure. However, the results do suggest that models of rRNA evolution that are specific to particular lineages will be important for making the best alignments and phylogenies. For example, the knowledge that loops evolve rapidly in eukaryotes would lead us to give changes in these regions of the sequence less weight for phylogenetic inference. With the vast number of sequences now flooding the databases (∼300 000 SSU sequences deposited in the Ribosomal Database Project as of this writing, and pyrosequencing able to produce 100 000–300 000 sequence fragments in a single 4-h run), detailed models of specific groups of organisms will become increasingly feasible.
Outliers from the general pattern of rRNA evolution may suggest interesting biology. For example, the Actinobacteria appear to resemble the eukaryotic pattern more than the general bacterial pattern. It is possible that ecological factors such as multicellularity, or molecular features such as linear rather than circular chromosomes, in this lineage (77) cause them to resemble eukaryotes more than other bacteria in factors influencing rRNA evolution. This group has relatively high GC content, contrasting with the low GC content in eukaryotes overall, suggesting that differences in base composition are not the main factor. Similarly, in the archeal SSU, about 5% of the comparisons (when counting only point mutations) or 10% (when adding indels), do not support the general conclusion that stems evolve faster on average outside the eukaryotes. Loops evolve faster than stems in comparisons between Aeropyrum pernix and Sulfolobus, Thermoproteus, Methanothermobacter or Methanobacterium, and between Pyrococcus and Sulfolobus. Aeropyrum and Pyrococcus are very similar in this respect. Aeropyrum is thought to be among the deepest diverging aerobic archaea, which may suggest some convergence with the eukaryotic pattern.
CONCLUSIONS
This work has several implications for future analyses. For example, when constructing phylogenetic trees, different models of RNA evolution should be adopted (provided that sufficient sequences are available to infer the parameters robustly). These models should be both specific for structural categories, including treating the different types of unpaired regions separately; they should also be specific for particular phylogenetic groups. For example, the general substitution model for bacteria does not fit the Actinobacteria well. Similarly, methods for comparing microbial communities, such as Fst (78), are based on diversity in an rRNA alignment. These methods may be improved by adding masks that weight more or less variable regions differently. Weighting by structural category may be an important first step for relatively unconserved regions.
The differences in the distributions of different structural categories appear to be driven primarily by proximity to functional features in the ribosome, rather than assorting by geometric configuration such as the ribosome center. This observation, combined with the lineage specificity of the rates of evolution of the different structural categories, suggest that the findings outlined here are likely to vary by lineage rather than reflecting universal characteristics of RNA evolution. Interestingly, the model that most change in functional RNAs comes from compensatory mutations in stems is not universally true. In this context, we eagerly await the availability of the structure of a eukaryotic ribosome for comparison with the results presented here for the bacterial ribosome.
We conclude that rates of evolution in different lineages and structural features of the rRNA show an unexpectedly rich and complex pattern, and that better understanding of this pattern will refine the results of a wide range of studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Michael Yarus, Quentin Vicens, Erik Schultes, John Quinn, Catherine Lozupone, and members of the Knight lab for helpful discussions of this work. A Keck RNA Bioinformatics award supported this work in part and was used to pay the Open Access publication charges for this article.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fox GE, Woese CR. The architecture of 5S rRNA and its relation to function. J. Mol. Evol. 1975;6:61–76. doi: 10.1007/BF01732674. [DOI] [PubMed] [Google Scholar]
- 2.Gutell RR, Larsen N, Woese CR. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noller HF, Hoffarth V, Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert W. Origin of life: the RNA world. Nature. 1986;319:618. [Google Scholar]
- 5.Welch M, Majerfeld I, Yarus M. 23S rRNA similarity from selection for peptidyl transferase mimicry. Biochemistry. 1997;36:6614–6623. doi: 10.1021/bi963135j. [DOI] [PubMed] [Google Scholar]
- 6.Yarus M, Welch M. Peptidyl transferase: ancient and exiguous. Chem. Biol. 2000;7:R187–R190. doi: 10.1016/s1074-5521(00)00027-2. [DOI] [PubMed] [Google Scholar]
- 7.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 8.Uzzell T, Corbin KW. Fitting discrete probability distributions to evolutionary events. Science. 1971;172:1089–1096. doi: 10.1126/science.172.3988.1089. [DOI] [PubMed] [Google Scholar]
- 9.Woese CR, Winker S, Gutell RR. Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc. Natl. Acad. Sci. USA. 1990;87:8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, et al. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noller HF, Kop J, Wheaton V, Brosius J, Gutell RR, Kopylov AM, Dohme F, Herr W, Stahl DA, et al. Secondary structure model for 23S ribosomal RNA. Nucleic. Acids. Res. 1981;9:6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rzhetsky A. Estimating substitution rates in ribosomal RNA genes. Genetics. 1995;141:771–783. doi: 10.1093/genetics/141.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.VandePeer Peer, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381–3391. doi: 10.1093/nar/24.17.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultes E, Hraber PT, LaBean TH. Global similarities in nucleotide base composition among disparate functional classes of single-stranded RNA imply adaptive evolutionary convergence. RNA. 1997;3:792–806. [PMC free article] [PubMed] [Google Scholar]
- 15.Abouheif E, Zardoya R, Meyer A. Limitations of metazoan 18S rRNA sequence data: implications for reconstructing a phylogeny of the animal kingdom and inferring the reality of the Cambrian explosion. J. Mol. Evol. 1998;47:394–405. doi: 10.1007/pl00006397. [DOI] [PubMed] [Google Scholar]
- 16.Otsuka J, Terai G, Nakano T. Phylogeny of organisms investigated by the base-pair changes in the stem regions of small and large ribosomal subunit RNAs. J. Mol. Evol. 1999;48:218–235. doi: 10.1007/pl00006461. [DOI] [PubMed] [Google Scholar]
- 17.BenAli A, Wuyts J, DeWachter R, Meyer A, Van de Peer Y. Construction of a variability map for eukaryotic large subunit ribosomal RNA. Nucleic Acids Res. 1999;27:2825–2831. doi: 10.1093/nar/27.14.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang HC, Hickey DA. Evidence for strong selective constraint acting on the nucleotide composition of 16S ribosomal RNA genes. Nucleic Acids Res. 2002;30:2501–2507. doi: 10.1093/nar/30.11.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schöniger M, Haeseler von A. A stochastic model for the evolution of autocorrelated DNA sequences. Mol. Phylogenet. Evol. 1994;3:240–247. doi: 10.1006/mpev.1994.1026. [DOI] [PubMed] [Google Scholar]
- 20.Muse SV. Evolutionary analyses of DNA sequences subject to constraints of secondary structure. Genetics. 1995;139:1429–1439. doi: 10.1093/genetics/139.3.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tillier ERM, Collins RA. Neighbor-joining and maximum likelihood with RNA Sequences: addressing the inter-dependence of sites. Mol. Biol. Evol. 1995;12:7–15. [Google Scholar]
- 22.Tillier ERM, Collins RA. High apparent rate of simultaneous compensatory base-pair substitutions in ribosomal RNA. Genetics. 1998;148:1993–2002. doi: 10.1093/genetics/148.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tillier ERM. Maximum likelihood with multiparameter models of substitution. J. Mol. Evol. 1994;39:409–417. [Google Scholar]
- 24.Higgs PG. RNA secondary structure: physical and computational aspects. Q. Rev. Biophys. 2000;33:199–253. doi: 10.1017/s0033583500003620. [DOI] [PubMed] [Google Scholar]
- 25.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler WC, Honeycutt RL. Paired sequence difference in ribosomal RNAs: evolutionary and phylogenetic implications. Mol. Biol. Evol. 1988;5:90–96. doi: 10.1093/oxfordjournals.molbev.a040480. [DOI] [PubMed] [Google Scholar]
- 27.Higgs PG. Compensatory neutral mutations and the evolution of RNA. Genetica. 1998;102–103:101. [PubMed] [Google Scholar]
- 28.Golden BL, Podell ER, Gooding AR, Cech TR. Crystals by design: a strategy for crystallization of a ribozyme derived from the Tetrahymena group I intron. J. Mol. Biol. 1997;270:711–723. doi: 10.1006/jmbi.1997.1155. [DOI] [PubMed] [Google Scholar]
- 29.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smit S, Yarus M, Knight R. Natural selection is not required to explain universal compositional patterns in rRNA secondary structure categories. RNA. 2006;12:1–14. doi: 10.1261/rna.2183806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muto A, Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl. Acad. Sci. USA. 1987;84:166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight RD, Freeland SJ, Landweber LF. A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-4-research0010. RESEARCH0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vawter L, Brown WM. Rates and patterns of base change in the small subunit ribosomal RNA gene. Genetics. 1993;134:97–608. doi: 10.1093/genetics/134.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 35.Doudna JA, Rath VL. Structure and function of the eukaryotic ribosome: the next frontier. Cell. 2002;109:153–156. doi: 10.1016/s0092-8674(02)00725-0. [DOI] [PubMed] [Google Scholar]
- 36.Wuyts J, Van de Peer Y, De Wachter R. Distribution of substitution rates and location of insertion sites in the tertiary structure of ribosomal RNA. Nucleic Acids Res. 2001;29:5017–5028. doi: 10.1093/nar/29.24.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brosius J, Dull TJ, Noller HF. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1980;77:201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brosius J, Palmer ML, Kennedy PJ, Noller HF. Complete nucleotide sequence of a 16S ribosomal RNA gene from. Escherichia coli. Proc. Natl. Acad. Sci. USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wuyts J, Perriere G, Van De Peer Y. The European ribosomal RNA 0database. Nucleic Acids Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stiegler P, Carbon P, Zuker M, Ebel JP, Ehresmann C. Secondary and topographic structure of ribosomal RNA 16S of Escherichia coli. C. R. Seances Acad. Sci. D. 1980;291:937–940. [PubMed] [Google Scholar]
- 41.Zwieb C, Glotz C, Brimacombe R. Secondary structure comparisons between small subunit ribosomal RNA molecules from six different species. Nucleic Acids Res. 1981;9:3621–3640. doi: 10.1093/nar/9.15.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noller HF, Woese CR. Secondary structure of 16S ribosomal RNA. Science. 1981;212:403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- 43.Branlant C, Krol A, Machatt MA, Pouyet J, Ebel JP, Edwards K, Kossel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA.Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981;9:4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glotz C, Zwieb C, Brimacombe R, Edwards K, Kossel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981;9:3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gutell RR, Woese CR. Higher order structural elements in ribosomal RNAs: pseudo-knots and the use of noncanonical pairs. Proc. Natl. Acad. Sci. USA. 1990;87:663–667. doi: 10.1073/pnas.87.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haselman T, Camp DG, Fox GE. Phylogenetic evidence for tertiary interactions in 16S-like ribosomal RNA. Nucleic Acids Res. 1989;17:2215–2221. doi: 10.1093/nar/17.6.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haselman T, Gutell RR, Jurka J, Fox GE. Additional Watson-Crick interactions suggest a structural core in large subunit ribosomal RNA. J. Biomol. Struct. Dyn. 1989;7:181–186. doi: 10.1080/07391102.1989.10507759. [DOI] [PubMed] [Google Scholar]
- 48.Gutell RR, Gray MW, Schnare MN. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res. 1993;21:3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larsen N. Higher order interactions in 23S rRNA. Proc. Natl. Acad. Sci. USA. 1992;89:5044–5048. doi: 10.1073/pnas.89.11.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelles L, Fang BL, Volckaert G, Vandenberghe A, DeWachter R. Nucleotide sequence of a crustacean 18S ribosomal RNA gene and secondary structure of eukaryotic small subunit ribosomal RNAs. Nucleic Acids Res. 1984;12:8749–8768. doi: 10.1093/nar/12.23.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huysmans E, DeWachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1986;14:r73–r118. doi: 10.1093/nar/14.suppl.r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dams E, Hendriks L, VandePeer Peer, Neefs JM, Smits G, Vandenbempt I, DeWachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1988;16:r87–r173. doi: 10.1093/nar/16.suppl.r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neefs JM, VandePeer Y, De Rijk P, Goris A, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1991;19:1987–2015. doi: 10.1093/nar/19.suppl.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neefs JM, VandePeer Y, Hendriks L, DeWachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1990;18:2237–2317. doi: 10.1093/nar/18.suppl.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeRijk P, Neefs JM, Van de Peer Y, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1992;20:2075–2089. doi: 10.1093/nar/20.suppl.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neefs JM, VandePeer Y, De Rijk P, Chapelle S, De Wachter R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.VandePeer Peer, Nicolai S, DeRijk P, DeWachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1996;24:86–91. doi: 10.1093/nar/24.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.VandePeer Peer, VandenBroeck Broeck, DeRijk P, DeWachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1994;22:3488–3494. doi: 10.1093/nar/22.17.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16S ribosomal RNA and 30S ribosomal subunits using primer extension. J. Mol. Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 60.Gutell RR, Lee JC, Cannone JJ. The accuracy of ribosomal RNA comparative structure models. Curr. Opin. Struct. Biol. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 61.Wuyts J, DeRijk P, VandePeer Y, Pison G, Rousseeuw P, DeWachter R. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res. 2000;28:4698–4708. doi: 10.1093/nar/28.23.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 63.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank. A computer-based archival file for macromolecular structures. Eur. J. Biochem. 1977;80:319–324. doi: 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- 64.Studnicka GM, Rahn GM, Cummings IW, Salser WA. Computer method for predicting the secondary structure of single-stranded RNA. Nucleic Acids Res. 1978;5:3365–3387. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.VandePeer Peer, Neefs JM, DeRijk P, DeWachter R. Reconstructing evolution from eukaryotic small-ribosomal-subunit RNA sequences: calibration of the molecular clock. J. Mol. Evol. 1993;37:221–232. doi: 10.1007/BF02407359. [DOI] [PubMed] [Google Scholar]
- 66.VandePeer Y, Van der Auwera G, De Wachter R. The evolution of stramenopiles and alveolates as derived by “substitution rate calibration” of small ribosomal subunit RNA. J. Mol. Evol. 1996;42:201–210. doi: 10.1007/BF02198846. [DOI] [PubMed] [Google Scholar]
- 67.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jukes TH, Osawa S, Muto A, Lehman N. Evolution of anticodons: variations in the genetic code. Cold Spring Harb. Symp. Quant. Biol. 1987;52:769–776. doi: 10.1101/sqb.1987.052.01.086. [DOI] [PubMed] [Google Scholar]
- 69.Ban N, Nissen P, Hansen J, Capel M, Moore PB, Steitz TA. Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature. 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- 70.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 71.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 72.Clemons WM, Jr, May JL, Wimberly BT, McCutcheon JP, Capel MS, Ramakrishnan V. Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 73.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Dounda Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 74.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 75.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 76.Yonath A. Ribosomal crystallography: peptide bond formation, chaperone assistance and antibiotics activity. Mol. Cells. 2005;20:1–16. [PubMed] [Google Scholar]
- 77.Hopwood DA. Soil to genomics: the streptomyces chromosome. Annu. Rev. Genet. 2006;40:1–23. doi: 10.1146/annurev.genet.40.110405.090639. [DOI] [PubMed] [Google Scholar]
- 78.Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5S Ribosomal RNA Database. Nucleic Acids Res. 2002;30:176–178. doi: 10.1093/nar/30.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.VandePeer Y, Jansen J, DeRijk P, DeWachter R. Database on the structure of small ribosomal submit RNA. Nucleic Acids Res. 1997;25:111–116. doi: 10.1093/nar/25.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]