Abstract
Trinucleotide repeat diseases, such as Huntington's disease, are caused by the expansion of trinucleotide repeats above a threshold of about 35 repeats. Once expanded, the repeats are unstable and tend to expand further both in somatic cells and during transmission, resulting in a more severe disease phenotype. Flap endonuclease 1 (Fen1), has an endonuclease activity specific for 5' flap structures and is involved in Okazaki fragment processing and base excision repair. Fen1 also plays an important role in preventing instability of CAG/CTG trinucleotide repeat sequences, as the expansion frequency of CAG/CTG repeats is increased in FEN1 mutants in vitro and in yeast cells defective for the yeast homolog, RAD27. Here we have tested whether one copy of yeast FEN1 is enough to maintain CAG/CTG tract stability in diploid yeast cells. We found that CAG/CTG repeats are stable in RAD27 +/− cells if the tract is 70 repeats long and exhibit a slightly increased expansion frequency if the tract is 85 or 130 repeats long. However for CAG-155 tracts, the repeat expansion frequency in RAD27 +/− cells is significantly higher than in RAD27 +/+ cells. This data indicates that cells containing longer CAG/CTG repeats need more Fen1 protein to maintain tract stability and that maintenance of long CAG/CTG repeats is particularly sensitive to Fen1 levels. Our results may explain the relatively small effects seen in the Huntington's disease (HD) FEN1 +/− heterozygous mice and myotonic dystrophy type 1 (DM1) FEN1 +/− heterozygous mice, and suggest that inefficient flap processing by Fen1 could play a role in the continued expansions seen in humans with trinucleotide repeat expansion diseases.
Keywords: RAD27, Trinucleotide repeat, Huntington's disease, Myotonic dystrophy, DNA replication, DNA repair
1. Introduction
Thirteen inherited genetic diseases have been found to be caused by CAG/CTG trinucleotide repeat expansions, including Huntington's disease (HD), myotonic dystrophy type 1 (DM1), and several spinocerebellar ataxias (SCAs) (Cleary and Pearson, 2003; Pearson et al., 2005). Thenormal gene loci typically have 6 to 34 copies of the CAG/CTG repeats. When the copies exceed a threshold of about 35 repeats they become unstable and are prone to further expansions both somatically and in the following generations (Siianova and Mirkin, 2001; Cleary and Pearson, 2003; Pearson et al., 2005). The disease-causing repeat size ranges from 36 to 121 in Huntington's disease, and 80 to over 1000 in myotonic dystrophy (Siianova and Mirkin, 2001; Cleary and Pearson, 2003; Pearson et al., 2005).
The mechanisms of trinucleotide repeat expansion have been studied in different organisms, including E. coli, yeast, mice and mammalian cells (Siianova and Mirkin, 2001; Lenzmeier and Freudenreich, 2003). One of the models to explain the expansions of trinucleotide repeats is that expansions occur due to incorrect Okazaki fragment processing (Gordenin et al., 1997; Freudenreich et al., 1998; Schweitzer and Livingston, 1998; Lenzmeier and Freudenreich, 2003). Fen1, the human homolog of yeast Rad27, is a structure-specific endo/exonuclease that cleaves the 5' flap created by strand displacement during Okazaki fragment maturation (Rossi et al., 2006). In vitro, Fen1 prefers to cleave a double flap substrate containing a 5' flap with a one nucleotide 3' overhang (Rossi et al., 2006). Secondary structures formed by trinucleotide repeats on the 5' flap can inhibit Fen1 cleavage, leading to ligation of the unprocessed flap and repeat sequence expansion (Spiro et al., 1999; Henricksen et al., 2000). The inhibition is length dependent: the longer the repeats are, the more inefficient the flap processing (Henricksen et al., 2000).
Fen1 is an evolutionarily conserved nuclease and has been isolated and characterized from many organisms, from archaebacteria to humans (Greene et al., 1999). Human Fen1 protein shares 61% identity and 79% similarity with Saccharomyces cerevisiae Rad27 protein (Shen et al., 1998). Recent work has shown that expression of human Fen1 can correct a number of mutant phenotypes in rad27 deletion (rad27Δ) yeast cells, including sensitivity to DNA-damaging agents, high rates of recombination and mutation, and synthetic lethality with double-strand-break repair mutants (Hansen et al., 2000).
In S. cerevisiae, the expansion and the contraction of CAG/CTG repeats is dramatically increased in a rad27Δ strain compared to wild-type (Freudenreich et al., 1998; Schweitzer and Livingston, 1998; Spiro et al., 1999), consistent with the idea that defective flap processing can lead to sequence expansions in vivo. CTG repeat expansions are also observed when human or yeast FEN1 mutants defective in nuclease activity are used in an in vitro assay for flap processing (Liu and Bambara, 2003; Liu et al., 2004). In mammals, the loss of FEN1 is embryonic lethal. In order to investigate the role of flap processing in repeat instability in mammals, Spiro and McMurray used mice heterozygous for FEN1 and harboring an expanded CAG-120 repeat within the human HD gene (Spiro and McMurray, 2003). Expansion of CAG repeats was increased in the progeny of the FEN1 heterozygous male mice, showing a shift from about equal expansions and contractions to five-fold more expansions than contractions. However no differences in somatic cell repeat instability were observed compared to wild-type mice (Spiro and McMurray, 2003). Recently another group observed that a (CAG/CTG)n (104≤n≥110) repeat at the DM1 locus did not exhibit increased instability either somatically or intergenerationally in FEN1 +/− heterozygous mice (van den Broek et al., 2006). These results raise the question of whether one copy of FEN1 was sufficient to prevent most repeat expansions in the HD and DM1 mouse models. We wished to address this question in order to gain further insight into whether defective flap processing by Fen1 may play a role in expansion of trinucleotide repeats in humans.
2. Materials and methods
2.1 Yeast strains and yeast mating
All the yeast haploid strains containing CAG-70 or 155 repeat tracts, cloned on a yeast artificial chromosome (YAC), were the VPS105 background (MATα ade2 can1 leu2-3,112 Δtrp1 ade3 Δura3 lys2-801 amber). All the yeast haploid strains containing CAG-85 and 130 repeat tracts, cloned on yeast chromosome II, were the YPH500 background (MATα ura3-52, lys2-801 amber, ade2-101 ochre, trp1-Δ63, his3-Δ200, leu2-Δ1). In order to create diploid yeast cells, a MATα strain containing a chromosome harboring an expanded CAG repeat tract (CAG-70, 85, 130 or 155) was mated with an isogenic MATa strain at 30°C for over night. Cells were plated for single colonies onto either YEPD for RAD27 +/+ or selective plates for RAD27 +/− and rad27 −/−. Single isolated colonies were checked for diploid status in two ways: (1) a mating type test that can distinguish diploid and haploid (MATa or MATα) cells (Sprague, 1991) and (2) By PCR for RAD27 +/− and rad27 −/− strains, which have different markers at each RAD27 locus resulting in different size PCR products which can be distinguished by gel electrophoresis.
2.2 CAG stability assay
To test CAG repeat expansion and contraction frequencies, 10 independent colonies with correct tract length (CAG-70, 85, 130 or 155) were chosen as starting colonies from either RAD27 + haploid cells, RAD27 +/+, rad27 −/−, or heterozygous RAD27 +/− diploid cells, and grown in liquid media for 16 hours at 30°C. These 10 individual cultures were mixed together and plated for single colonies using proper dilutions. Mixing 10 individual cultures was done to make sure we sampled multiple independent events and to avoid “jackpot” effects of a single early expansion or contraction event which could make up a significant fraction of a culture and bias the observed frequency. About 50 daughter colonies were then tested for repeat contractions or expansions by colony PCR. The assay was repeated a minimum of three times for each tract length in each strain background (Table S1). For CAG-70 and 155 which are on the YAC, repeat sequences were amplified by primers CTG rev2 (CCCAGGCCTCCAGTTTGC) and T720 (TAATACGACTCACTATAGGG). For CAG-85 and 130 on chromosome II, repeat sequences were amplified by primers CTG rev (GTGGAGGATGGAACACGG) and T720. Reactions were run as described previously (Callahan et al., 2003).
In order to check if loss of heterozygosity occurs in RAD27 +/− cells during the course of the experiment, the presence of both RAD27 and rad27Δ alleles were assessed independently by PCR for three sets of putative RAD27 +/− daughter colonies from the stability assay. All 152 daughter colonies tested retained both alleles, indicating that loss of heterozygosity was not occurring or was very rare.
The Fisher's exact test was used to analyze whether the total number of contractions or expansions in RAD27 +/−, rad27 −/− or RAD27 + strains was significantly different from that in the RAD27 +/+ strain. Data from each repeated experiment were combined together for the statistical analysis using a 2 × 2 contingency table. Analysis using a one-tailed non-parametric Mann-Whitney test was also done, and values reported as significant by the Fisher's exact test were also significant using this method. The means +/− standard error that are plotted in Fig. 1 are averaged across at least three repetitions of each experiment (Table S1).
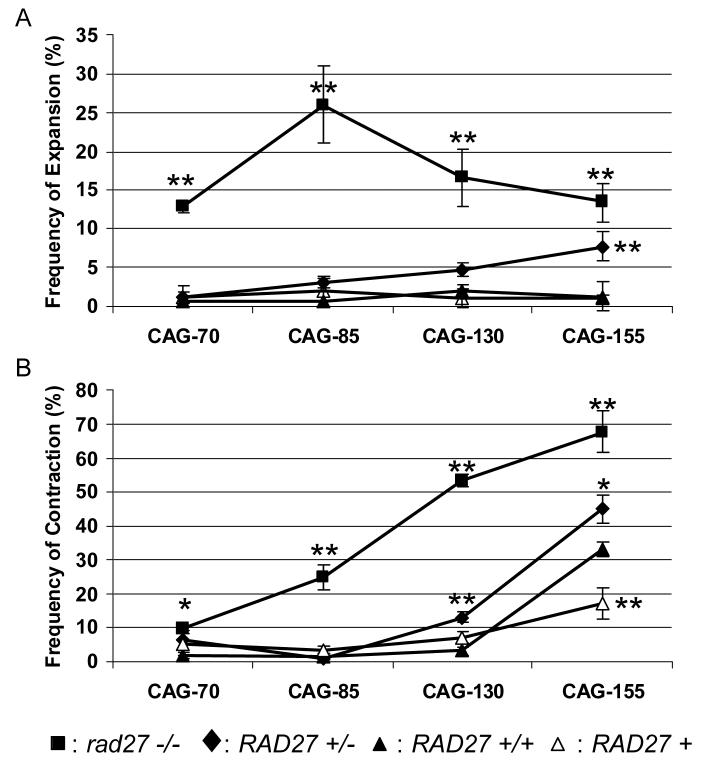
Figure 1.
Frequency of CAG repeat expansion (A) and contraction (B) in RAD27 + (Δ), RAD27 +/+ (▲), rad27 −/− (■) and heterozygous RAD27 +/− (◆) strains. 148 to 312 colonies were evaluated for each tract length of each strain in at least three independent experiments. We note that the frequency of expansions for the rad27 −/− CAG-130 and 155 strains in (A) could be an underestimate due to inefficient PCR amplification of tracts longer than about 180 repeats in this strain background. **, P<0.01; *, P<0.05 compared to RAD27 +/+ value, by Fisher's Exact Test. Standard error is shown.
2.3 Western blot analysis
Yeast protein was purified as described (Knop et al., 1999) with the following changes: 5ml cells (OD 2-3) were collected and resuspended in 1ml ice-cold water. The cell suspension was mixed with 150μl 1.85M NaOH and placed on ice for 10 min. 150μl 55% TCA was added and the mixture was incubated for 10 min on ice. Cells were pelleted for 5 min at 14000 rpm at 4°C. The supernatant was removed and the cell pellet resuspended in 150 μl 2X SDS-sample buffer (200mM DTT, 4% SDS, 100mM Tris-HCl pH 6.8, 20% glycerol and 30 μl/ml bromphenol blue). In order to neutralize the remaining traces of the TCA, 3-5 μl of 1.5M Tris-HCl, pH 8.8 was added. Proteins were separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to a nitrocellulose membrane (Amersham Biosciences). Rad27 protein was detected by anti-Rad27 antibody (Santa Cruz Biotechnology, Inc.). Actin was detected by anti-actin (Chemicon International). Signals were detected with an ECL detection reagent (Millipore) according to the manufacturer. Blots were stripped by washing with 1x TBST (10mM Tris-HCl pH 7.4; 150mM NaCl; 0.1% Tween20) and incubating for 30 min at 50°C with freshly made strip-buffer (100mM 2-mercaptoethanol; 2% SDS; 62.5mM Tris-HCl, pH 6.7). After stripping, blots were washed twice for 10 min in TBS-T.
3. Results
3.1 The effect of RAD27 haploinsufficiency on CAG repeat expansions and contractions is tract-length dependent
In yeast, cells can grow with either haploid or diploid chromosome content. In order to create heterozygous RAD27 +/− diploid yeast cells, a rad27Δ MATα strain containing a chromosome harboring an expanded CAG repeat tract (CAG-70, 85, 130 or 155) was mated with an isogenic wild-type MATa strain. For the cells containing CAG-85 or 130, the repeats were cloned on yeast chromosome II (Freudenreich et al., 1998), and for CAG-70 and 155, the repeats were cloned on a yeast artificial chromosome (YAC) (Callahan et al., 2003). In all strains, the repeat tract is oriented such that the CAG sequence is on the lagging strand template and the CTG sequence on the nascent lagging strand (i.e. the more expansion-prone orientation, referred to as orientation I in Kang et al., 1995 and Freudenreich et al., 1997). Based on available data on the location of replication origins (Nenguke et al., 2003) and also the expansion bias seen at the human disease loci, this orientation is likely to be equivalent to that found at the HD locus. The heterozygous yeast is similar to the HD and DM1 mouse models used (Spiro and McMurray, 2003; van den Broek et al., 2006), since each diploid yeast cell contains two copies of each yeast chromosome, one copy of the yeast FEN1 gene, and one expanded CAG repeat tract. To test CAG repeat expansion and contraction frequencies, stability assays were performed on RAD27 +/+, rad27 −/−, or heterozygous RAD27+/− diploid cells containing repeat tracts of different lengths (CAG-70, 85,130 or 155). The stability assay was repeated at least three times for each strain for a total of at least 148 colonies tested per strain (Table S1). For the RAD27 +/− cells, it was verified that loss of heterozygosity did not occur in the course of the experiment (0/152 colonies checked; see Methods 2.2).
For CAG-70 heterozygous RAD27 +/− diploids, the frequency of repeat expansion is similar to that of RAD27 +/+ cells (Fig. 1A; Table 1). The frequency of repeat expansion increased slightly as the repeat length increased to CAG-85 or 130 (∼2 to 4-fold), although the increase was not enough to be statistically significant. However, when the repeat tract length reached CAG-155, the RAD27 +/− heterozygote exhibited a significantly greater frequency of expansions than the RAD27 +/+ wild-type diploid control (P<0.006). The increase in CAG-155 expansions in the RAD27 +/− heterozygote was not as great as the expansion frequency observed in the absence of Rad27 protein, but rather showed an intermediate phenotype (Fig. 1A; Table 1). These results indicate that one copy of RAD27 in a diploid cell is sufficient to maintain the shorter CAG-70 repeat tracts to the same degree as in a wild-type cell, but that expansion of longer CAG/CTG repeat tracts is sensitive to RAD27 dose.
Table 1.
Frequency of Expansions and Contractions
| Expansion (%) |
Contraction (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| CAG-70 | CAG-85 | CAG-130 | CAG-155 | CAG-70 | CAG-85 | CAG-130 | CAG-155 | |
| rad27 −/− | 12.8** | 25.6** | 16.6** | 13.4** | 9.6* | 24.4** | 53.4** | 67.8** |
| RAD27 +/− | 1.3 | 2.6 | 4.7 | 7.7** | 6.4 | 1.3 | 12.8** | 45.2* |
| RAD27 +/+ | 0.6 | 0.6 | 1.9 | 1.3 | 1.9 | 1.6 | 3.2 | 33.3 |
| RAD27 + | 1.3 | 1.9 | 1.3 | 0.5 | 5.1 | 3.2 | 7.1 | 16.8** |
P<0.01
P<0.05 compared to RAD27 +/+ value, by Fisher's Exact Test.
A similar pattern was observed for the frequency of repeat contractions (Fig. 1B; Table 1). CAG-70 and CAG-85 RAD27 +/− heterozygotes exhibited a low level of contractions that was similar to the RAD27 +/+ strains. For CAG-130 and 155, a significant increase of contraction frequency in the RAD27 +/− heterozygotes compared to RAD27 +/+ wild-type diploids was observed (Fig. 1B; Table 1).
3.2 The frequency of contraction is cell type dependent for CAG-155
To determine whether CAG/CTG repeat tract stability is affected by the haploid or diploid status of the cells, CAG repeat instability was compared in wild-type diploid cells and wild-type haploid cells. For both cell types only one expanded CAG repeat tract was present. For CAG-70, 85 and 130, no significant difference in CAG repeat instability between wild-type haploid (RAD27 +) and wild-type diploid (RAD27 +/+) strains was observed (Fig. 1). Similarly for CAG-155, the wild-type diploid expansion frequency (1.3%) is approximately equal to the wild-type haploid frequency (0.5%). Interestingly, CAG-155 contractions were more frequent in wild-type diploid cells compared to wild-type haploids (33.3% vs 16.8%) (Fig. 1B; Table 1). The CAG-155 tract is carried on a YAC which is present in only one copy in both the haploid and diploid cells used. Therefore the increase cannot be explained by increased recombination between homologs.
3.3 Yeast Fen1 levels are not upregulated in RAD27 +/− heterozygote cells
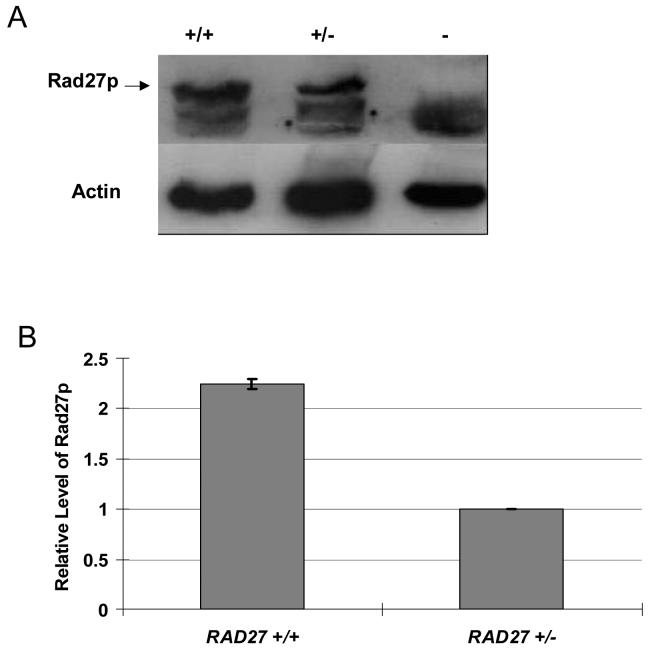
The phenotypic difference in repeat instability between the RAD27 +/+ diploid cells and RAD27 +/− diploid cells strongly suggests that Rad27 protein is not upregulated in the heterozygote. In order to directly determine Rad27 protein levels, protein was extracted from each cell type and detected using an antibody to yeast Rad27 protein (Fig. 2A). Quantification compared to a control protein (actin) confirmed that Rad27 protein levels in the heterozygote were half that detected in RAD27 +/+ diploids (Fig. 2B), indicating that Rad27p is not upregulated in RAD27 +/− cells.
Figure 2.
Detection of Rad27 protein levels by Western blot. (A) Protein extracts were separated on an SDS polyacrylamide gel, blotted, and probed with anti-Rad27 antibody. The same blot was stripped and re-probed with anti-actin antibody. Results for RAD27 +/+, RAD27 +/− heterozygotes and rad27 − strains are shown. (B) The relative level of Rad27p was first normalized by the amount of Actin in the same blot by comparing the band density. Quantification of band density was performed by Image J (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/). The normalized Rad27p level in RAD27 +/+ cells was compared to that in the RAD27 +/− cells to get the relative level of Rad27p. The experiment was done twice. Standard error is shown.
4. Discussion
Our data indicate that maintenance of long CAG/CTG repeats is particularly sensitive to Fen1 levels. Comparing the RAD27 +/− diploids with RAD27 +/+ diploids, both contain one expanded CAG repeat tract in the genome and two copies of each natural yeast chromosome, but in the RAD27 +/− cells the total amount of Rad27p is half the level found in RAD27 +/+ cells. This situation is not a problem under normal circumstances. However, 5' flaps containing a CTG repeat (as in our case) pose a special challenge since they can form a stable hairpin structure that inhibits Fen1 cleavage (Spiro et al., 1999; Henricksen et al., 2000). In this case, flap equilibration can usually eventually result in correct processing by Fen1 (Liu et al., 2004), followed by DNA ligation and no repeat expansion. However, when Fen1 concentration is limiting, the unprocessed flaps could equilibrate into a variety of intermediate structures, some of which will be ligated by DNA ligase to cause a sequence expansion. This model is supported by in vitro data that shows that there is a competition between Fen1 and DNA ligase at the flap, with increasing amounts of ligase resulting in ligation of the unprocessed flaps to produce expansions (Henricksen et al., 2002). Similarly, limiting amount of Fen1 could also favor flap ligation to produce expansions.
In the RAD27 +/− heterozygote cells we studied, the same amount of Rad27 protein is present in cells containing the shorter or longer repeat tracts, yet only the longer repeat tracts were prone to expansions and contractions. One explanation is that there are more Okazaki fragments with CTG repeats on the 5' flap in cells containing the long repeat tracts. In eukaryotes, Okazaki fragments are 135-145 nucleotides long (Cleary and Pearson, 2005). Assuming that the average length of Okazaki fragments is ∼ 140 nucleotides, in cells containing shorter repeats (CAG-70 and 85) there will be no more than 2 Okazaki fragments containing CTG repeats on a 5' flap during one replication cycle. However, in CAG-130 or 155 cells, 3 or 4 Okazaki fragments will have CTG repeats located on 5' flaps waiting for Rad27p cleavage, increasing the probability that ligation will result before flap cleavage to cause a sequence expansion. Equilibrating flaps could also allow hairpin formation on the template strand opposite the unligated nick, which combined with extension of the 3' end of the next Okazaki fragment over the template hairpin, could lead to a contraction. Another explanation is that the longer repeat is more likely to incur DNA lesions that require Rad27 for repair. It has been shown that rad27Δ cells show an increased fragility of CAG repeats compared to wild type cells which is exacerbated as tract length increases (Callahan et al., 2003). Faulty repair could lead to either expansions or contractions.
A novel finding from this study was that wild-type diploid cells have a significantly higher contraction frequency than haploid cells, but again only for the long CAG-155 tract. Because of the experimental setup, both cell types had only one copy of the chromosome with the expanded CAG repeat, therefore the difference between haploids and diploids could not be due to recombinational repair between CAG repeats on homologs. It has been shown that non-homologous end-joining is much less efficient in diploid cells than in haploid cells (Astrom et al., 1999; Lee et al., 1999). Possibly, the repair choice for CAG repeats could shift towards a more contraction-prone repair pathway, such as single-strand annealing, in diploid cells. The chance of utilizing such a pathway could depend on the level of lesions in the cell, or could also be influenced by a specific structure or lesion formed more frequently at CAG-155 tracts. One candidate for this structure is a stalled replication fork (Freudenreich and Lahiri, 2004).
The sensitivity to repeat length we observed could explain the results seen in the FEN1 +/− mice with 120 CAG repeats at the HD locus (Spiro and McMurray, 2003). Full levels of Fen1 are apparently required for proper maintenance of 120 repeats in the developing male germ cells, indicating that significant flap processing is occurring in this cell type during either premeiotic germ cell divisions or postmeiotic gap repair (Kovtun and McMurray, 2001; Spiro and McMurray, 2003). However, in the somatic cells studied that are not undergoing rapid division or differentiation, one copy of FEN1 appears sufficient to maintain a CAG-120 tract without expansions (Spiro and McMurray, 2003). One copy was also sufficient to maintain a CTG-110 repeat at the DM1 locus (van den Broek et al., 2006). Our results suggest that a FEN1 +/− HD or DM1 mouse model with longer repeats might show a more dramatic phenotype. Similarly, a FEN1 heterozygote individual with a trinucleotide repeat in the long-normal range might accumulate a trinucleotide repeat expansion in a germ cell, even though general genome stability is not affected.
In humans, CAG/CTG expansions can occur in both dividing pre-meiotic germ cells and nondividing postmeiotic germ cells (Yoon et al., 2003) as well as in both proliferating and non-proliferating somatic cells (Sinden, 2001). Our data indicate that cells with larger expanded repeat tracts that are actively undergoing either replication or repair may be the most sensitive to Fen1 protein levels. The myotonic dystrophy locus, where CTG tracts larger than 150 repeats are often observed, exhibits instability in proliferating cell types that are presumably undergoing Fen1-dependent flap processing during replication (Fortune et al., 2000; Pearson et al., 2005). The SCA2 and SCA7 loci also expand to 200-300 repeats (Siianova and Mirkin, 2001). The majority of inherited Huntington's alleles are less than 150 repeats, however there is evidence that somatic expansions in affected human brain cells are common, with a significant percentage of cells containing CAG tracts greater than 150 repeats, and that these increases precede pathogenesis (Kennedy et al., 2003). These non-dividing neural cells could be prone to repeat expansion events during Fen1-dependent repair. Since brain cells are known to have reduced repair capacity, it is possible that the normal expression level of Fen1 in the affected cells is not sufficient to prevent expansions in a long CAG repeat tract.
In summary, we have shown that the balance between Fen1 levels and repeat maintenance is critical, implying that any condition that upsets this balance (more flaps because of DNA damage or longer repeat lengths; less Fen1 because of cell type, developmental window, or a mutated allele) could lead to repeat expansions. Overall, our data imply that inefficient flap processing by Fen1 is a mechanism that may be operating to generate CAG/CTG expansions in humans.
Supplementary Material
Acknowledgements
We would like to thank Sara Lewis for advice on statistical analysis. This work was supported by a National Institute of Health grant (GM63066) to C.H.F.
Abbreviations
- FEN1
Flap endonuclease 1
- Rad
Radiation sensitive
- HD
Huntington's disease
- DM1
Myotonic dystrophy type 1
- SDS
Sodium dodecyl sulfate
- PAGE
Polyacrylamide gel electrophoresis
- MAT
Mating type locus
- YAC
Yeast artificial chromosome
- SCA
Spinocerebellar ataxia
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Astrom SU, Okamura SM, Rine J. Yeast cell-type regulation of DNA repair. Nature. 1999;397:310. doi: 10.1038/16833. [DOI] [PubMed] [Google Scholar]
- Callahan JL, Andrews KJ, Zakian VA, Freudenreich CH. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol. Cell Biol. 2003;23:7849–7860. doi: 10.1128/MCB.23.21.7849-7860.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Pearson CE. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet Genome Res. 2003;100:25–55. doi: 10.1159/000072837. [DOI] [PubMed] [Google Scholar]
- Cleary JD, Pearson CE. Replication fork dynamics and dynamic mutations: the fork-shift model of repeat instability. Trends Genet. 2005;21:272–280. doi: 10.1016/j.tig.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Fortune MT, Vassilopoulos C, Coolbaugh MI, Siciliano MJ, Monckton DG. Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum. Mol. Genet. 2000;9:439–445. doi: 10.1093/hmg/9.3.439. [DOI] [PubMed] [Google Scholar]
- Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- Freudenreich CH, Lahiri M. Structure-forming CAG/CTG repeat sequences are sensitive to breakage in the absence of Mrc1 checkpoint function and Sphasecheckpoint signaling: implications for trinucleotide repeat expansion diseases. Cell Cycle. 2004;3:1370–1374. doi: 10.4161/cc.3.11.1246. [DOI] [PubMed] [Google Scholar]
- Freudenreich CH, Stavenhagen JB, Zakian VA. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell Biol. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordenin DA, Kunkel TA, Resnick MA. Repeat expansion--all in a flap? Nat. Genet. 1997;16:116–118. doi: 10.1038/ng0697-116. [DOI] [PubMed] [Google Scholar]
- Greene AL, Snipe JR, Gordenin DA, Resnick MA. Functional analysis of human FEN1 in Saccharomyces cerevisiae and its role in genome stability. Hum. Mol. Genet. 1999;8:2263–2273. doi: 10.1093/hmg/8.12.2263. [DOI] [PubMed] [Google Scholar]
- Hansen RJ, Friedberg EC, Reagan MS. Sensitivity of a S. cerevisiae RAD27 deletion mutant to DNA-damaging agents and in vivo complementation by the human FEN-1 gene. Mutat. Res. 2000;461:243–248. doi: 10.1016/s0921-8777(00)00056-2. [DOI] [PubMed] [Google Scholar]
- Henricksen LA, Tom S, Liu Y, Bambara RA. Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J. Biol. Chem. 2000;275:16420–16427. doi: 10.1074/jbc.M909635199. [DOI] [PubMed] [Google Scholar]
- Henricksen LA, Veeraraghavan J, Chafin DR, Bambara RA. DNA ligase I competes with FEN1 to expand repetitive DNA sequences in vitro. J. Biol. Chem. 2002;277:22361–22369. doi: 10.1074/jbc.M201765200. [DOI] [PubMed] [Google Scholar]
- Kang S, Jaworski A, Ohshima K, Wells RD. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- Kennedy L, Evans E, Chen CM, Craven L, Detloff PJ, Ennis M, Shelbourne PF. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- Lee SE, Paques F, Sylvan J, Haber JE. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 1999;9:767–770. doi: 10.1016/s0960-9822(99)80339-x. [DOI] [PubMed] [Google Scholar]
- Lenzmeier BA, Freudenreich CH. Trinucleotide repeat instability: a hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet Genome Res. 2003;100:7–24. doi: 10.1159/000072836. [DOI] [PubMed] [Google Scholar]
- Liu Y, Bambara RA. Analysis of human flap endonuclease 1 mutants reveals a mechanism to prevent triplet repeat expansion. J. Biol. Chem. 2003;278:13728–13739. doi: 10.1074/jbc.M212061200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang H, Veeraraghavan J, Bambara RA, Freudenreich CH. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol. Cell Biol. 2004;24:4049–4064. doi: 10.1128/MCB.24.9.4049-4064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenguke T, Aladjem MI, Gusella JF, Wexler NS, Arnheim N. Candidate DNA replication initiation regions at human trinucleotide repeat disease loci. Hum. Mol. Genet. 2003;12:1021–1028. doi: 10.1093/hmg/ddg111. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Edamura KN, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- Rossi ML, Purohit V, Brandt PD, Bambara RA. Lagging strand replication proteins in genome stability and DNA repair. Chem. Rev. 2006;106:453–473. doi: 10.1021/cr040497l. [DOI] [PubMed] [Google Scholar]
- Schweitzer JK, Livingston DM. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- Shen B, Qiu J, Hosfield D, Tainer JA. Flap endonuclease homologs in archaebacteria exist as independent proteins. Trends Biochem. Sci. 1998;23:171–173. doi: 10.1016/s0968-0004(98)01199-2. [DOI] [PubMed] [Google Scholar]
- Siianova E, Mirkin SM. Expansion of trinucleotide repeats. Mol. Biol. 2001;35:168–182. [PubMed] [Google Scholar]
- Sinden RR. Neurodegenerative diseases. Origins of instability. Nature. 2001;411:757–758. doi: 10.1038/35081234. [DOI] [PubMed] [Google Scholar]
- Spiro C, McMurray CT. Nuclease-deficient FEN-1 blocks Rad51/BRCA1-mediated repair and causes trinucleotide repeat instability. Mol. Cell. Biol. 2003;23:6063–6074. doi: 10.1128/MCB.23.17.6063-6074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro C, Pelletier R, Rolfsmeier ML, Dixon MJ, Lahue RS, Gupta G, Park MS, Chen X, Mariappan SV, McMurray CT. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell. 1999;4:1079–1085. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- Sprague GF., Jr. Assay of yeast mating reaction. Methods Enzymol. 1991;194:77–93. doi: 10.1016/0076-6879(91)94008-z. [DOI] [PubMed] [Google Scholar]
- van den Broek WJ, Nelen MR, van der Heijden GW, Wansink DG, Wieringa B. Fen1 does not control somatic hypermutability of the (CTG)(n)*(CAG)(n) repeat in a knock-in mouse model for DM1. FEBS Lett. 2006;580:5208–5214. doi: 10.1016/j.febslet.2006.08.059. [DOI] [PubMed] [Google Scholar]
- Yoon SR, Dubeau L, de Young M, Wexler NS, Arnheim N. Huntington disease expansion mutations in humans can occur before meiosis is completed. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8834–8838. doi: 10.1073/pnas.1331390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.