Abstract
Background
The overproduction of recombinant proteins in host cells often leads to their misfolding and aggregation. Previous attempts to increase the solubility of recombinant proteins by co-overproduction of individual chaperones were only partially successful. We now assessed the effects of combined overproduction of the functionally cooperating chaperone network of the E. coli cytosol on the solubility of recombinant proteins.
Results
A two-step procedure was found to show the strongest enhancement of solubility. In a first step, the four chaperone systems GroEL/GroES, DnaK/DnaJ/GrpE, ClpB and the small HSPs IbpA/IbpB, were coordinately co-overproduced with recombinant proteins to optimize de novo folding. In a second step, protein biosynthesis was inhibited to permit chaperone mediated refolding of misfolded and aggregated proteins in vivo. This novel strategy increased the solubility of 70% of 64 different heterologous proteins tested up to 42-fold.
Conclusion
The engineered E. coli strains and the two-step procedure presented here led to a remarkable increase in the solubility of a various recombinant proteins and should be applicable to a wide range of target proteins produced in biotechnology.
Background
Chaperones assist the folding of newly synthesized proteins to the native state and provide a quality control system that refolds misfolded and aggregated proteins. In the E. coli cytosol, the folding of newly synthesized proteins is assisted by the ribosome-associated Trigger Factor, the DnaK system (DnaK with its DnaJ and GrpE cochaperones; KJE), and the GroEL system (GroEL with its GroES cochaperone; ELS) [1-4]. KJE and ELS also prevent aggregation and promote refolding of preexisting proteins which lost their native conformation e.g. due to thermal denaturation or intrinsic instability [5,6]. KJE furthermore cooperates with ClpB to solubilize aggregated proteins and refold them to the native state in a concerted action with ELS [7-9]. Moreover, the small heat shock proteins (sHSP) of E. coli, IbpA and IbpB (IbpAB), intercalate into protein aggregates and thereby facilitate the KJE/ClpB dependent disaggregation and refolding [10-12].
The overproduction of recombinant proteins in host cells often leads to their misfolding and aggregation [13-17]. Folding problems of overproduced client proteins can be caused by limitations in the chaperone capacity of the host cells. Several attempts were made to increase the yields of correctly folded, and hence soluble, recombinant proteins by the co-overproduction of individual chaperones in producing cells, however only with a limited success. For example, the co-overproduction of ELS increases the solubility of human ORP150, human lysozyme, p50csk protein tyrosine kinase, phosphomannose isomerase and fusion protein PreS2-S'-β-galactosidase and maize protoporphyrinogen IX oxidase [18-23]. The co-overproduction of KJE increases the solubility of endostatin, human ORP150, transglutaminase and PreS2-S'-β-galactosidase [21,24,25].
To assess the full potential of the cellular network of molecular chaperones for the production of soluble recombinant proteins we performed a systematic analysis of the combined power of the major cytosolic chaperone systems of E. coli, KJE, ELS, ClpB and IbpAB. We did not include Trigger Factor in our analyses since it acts through a 1:1 association with ribosomes and is already present in three-fold molar excess over ribosomes in wild type cells [2].
Results and Discussion
We first investigated a combination of the two major chaperone systems with folding activity, KJE and ELS, and the co-operating chaperone with disaggregating activity, ClpB. To allow for regulated overproduction of chaperones we generated a set of compatible plasmids, which (i) differ in copy number (pSC101, 3–4 copies/cell; p15A, 20–30 copies/cell), (ii) express chaperone genes under control of a strong (PA1) or weaker (Plac) promoter combined with the lacO1 operator (PA1/lacO1) [26], and (iii) encode the lacIq repressor gene. This set of plasmids was designed to allow for IPTG-inducible expression of chaperone genes in different combinations (Fig. 1, combinations 1 to 5), and to produce chaperones at the stoichiometries, which are optimal for their folding activity [27,28].
Figure 1.
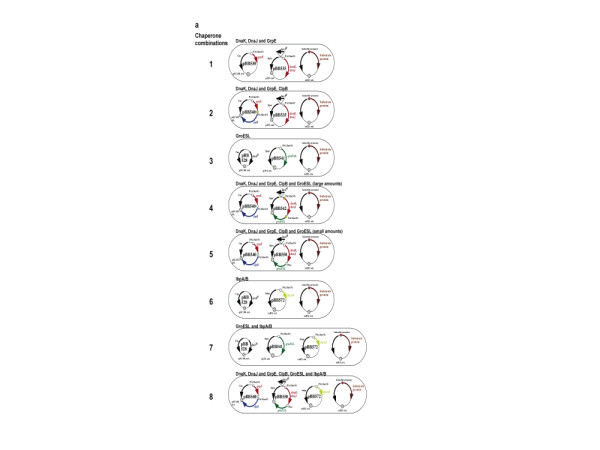
Design of E. coli host cells for the simultaneous overproduction of recombinant target proteins and major chaperone systems. Host cells for recombinant protein expression containing combinations (1 to 8) of two or three chaperone expression plasmids. Each combination of plasmids includes one plasmid carrying the lacIQ gene to allow for IPTG controlled chaperone expression.
Host cell were transformed with plasmids in 5 combinations. Combination 1 (pBB530 and pBB535) for overproduction of KJE; combination 2 (pBB540 and pBB535) for overproduction of KJE and ClpB; combination 3 (pBB528 and pBB541) for overproduction of ELS; combination 4 (pBB540 and pBB542) for overproduction of KJE, ClpB and high amounts of ELS; combination 5 (pBB540 and 550) for overproduction of KJE with ClpB and lower amounts of ELS (Fig. 1). Continuous growth of these cells in medium containing 100 μM IPTG resulted in an increase of DnaK (18–22 fold), ClpB (15–18 fold), GroEL (30 fold, combination 4; 5 fold, combination 5) over wild type levels (Fig. 2), without causing apparent growth defects (data not shown). Host cells containing different combinations of these plasmids were subsequently transformed with plasmids expressing 50 different recombinant genes of prokaryotic and eukaryotic origin (Table 1) from IPTG-controlled promoters. The encoded proteins include monomers and oligomers, cytosolic, membrane bound and secreted proteins, full-length, fragmented and fused proteins (fusion to GST, Trx, DsbA or NusA) [see Additional file 1], with molecular weights ranging between 7.5 and 118 kDa. Proteins were hexa-histidine tagged to allow affinity purification of the soluble fractions.
Figure 2.
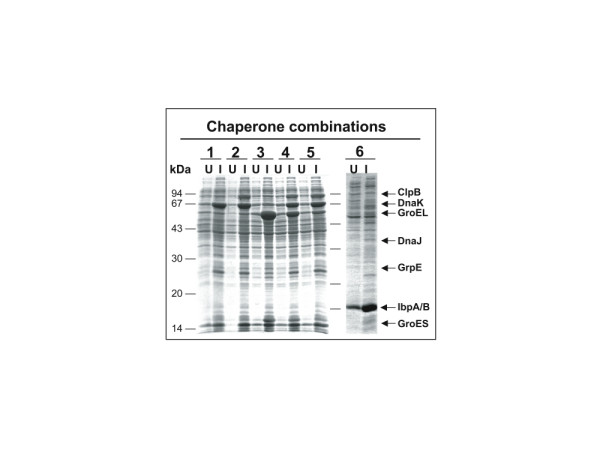
Chaperone overproduction upon IPTG induction in E. coli cells carrying plasmid combinations 1 to 6. Lysates of non-induced cells (U) and IPTG-induced cells (I) grown overnight at 20°C were separated by SDS-PAGE and Coomassie-stained. The various plasmid combinations present in the tested cells are indicated.
Table 1.
List of proteins co-expressed without and with chaperone combinations 1 to 5 and effects of chaperones on yields of purified, soluble target proteins, applying either the one-step or two-step procedure.
| Protein | MW (kDa) | Increased solubility by | |||
| one step procedure | two step procedure | ||||
| Yield increase | Best chaperone combination | Yield increase | Best chaperone combination | ||
| 1Fringe | 43 | ∞ | 4 | 3 | 4 |
| 2Ap | 54 | ∞ | 4 | 3 | 3 |
| 22f21 | 52 | ∞ | 3 | 3 | 4 |
| LZip | 41 | ∞ | 1 | n.d. | n.d. |
| Oskar3 | 118 | ∞ | 2/4 | 2.5 | 2 |
| 2c18 | 50 | 3 | 4 | 8 | 4 |
| 2Tep4 | 68 | 3.5 | 4 | 13 | 4 |
| BtKe | 55 | 7 | 3 | 42 | 4 |
| PhK | 29 | 3 | 4 | 7 | 5 |
| Tep1 | 8 | 3 | 5 | 6 | 1 |
| Susy | 86 | 2.5 | 3 | 5 | 5 |
| Xklp3A+Xklp3B | 20+ 16 | 2.5 | 4 | 3.5 | 4 |
| B1R | 59 | 3.5 | 3 | 3.5 | 3 |
| E8R2 | 85 | 5.5 | 3 | 5.5 | 3 |
| GTR1 | 35 | 3 | 4 | 3 | 4 |
| HbpH | 9 | 3.5 | 4 | 3.5 | 4 |
| Rolled | 43 | 4.5 | 3 | 4.5 | 3 |
| Xklp3A4 | 44 | 2.5 | 5 | 2.5 | 5 |
| Xklp3B3 | 70 | 2.5 | 4 | 2.5 | 4 |
| Mash+ Susy | 90+ 86 | 3 | 4 | 3 | 4 |
| 1Tep3 | 47 | 4 | 3 | n.d. | n.d. |
| 1Tep4 | 45 | 3.5 | 5 | n.d. | n.d. |
| BtKc | 64 | 3 | 5 | n.d. | n.d. |
| E8R1 | 58 | 5 | 5 | n.d. | n.d. |
| MaxF | 7.5 | 3 | 4 | n.d. | n.d. |
| Tev prot. | 30 | 3.5 | 3 | n.d. | n.d. |
| Xklp3A1 | 62 | 1* | - | ∞ | 2 |
| Xklp3B1 | 40 | 1* | - | ∞ | 5 |
| 2Fringe | 55 | 1 | - | 2 | 4 |
| Xklp3A5 | 35 | 1 | - | 19 | 4 |
| 2Tep3 | 70 | 1 | - | 11 | 2 |
| BtKp | 55 | 1 | - | 28 | 4 |
| Mash | 91 | 1 | - | 3 | 2 |
| PPAT | 22 | 1 | - | 3 | 4 |
| Eg5 | 95 | 1 | - | 1 | - |
| F10L | 72 | 1 | - | 1 | - |
| Tep2 | 11 | 1 | - | 1 | - |
| 1AP | 52 | 1 | - | n.d. | n.d. |
| Chip | 64 | 1 | - | n.d. | n.d. |
| dLMO | 37 | 1 | - | n.d. | n.d. |
| Pex5P | 38 | 1 | - | n.d. | n.d. |
| Endostatin | 42 | 1* | - | 1* | - |
| Kringle | 30 | 1* | - | 1* | - |
| Lzip2 | 37 | 1* | - | 1* | - |
| UCP1 | 33 | 1* | - | 1* | - |
| Tlc4 | 57 | 1* | - | n.d. | n.d. |
| Xklp3A2 | 38 | 1* | - | n.d. | n.d. |
| Xklp3A3 | 72 | 1* | - | n.d. | n.d. |
| Xklp3B1 | 35 | 1* | - | n.d. | n.d. |
| Xpot1 | 108 | 1* | - | n.d. | n.d. |
The increase in solubility of target proteins relative to control cells without co-overexpression of chaperones (set as 1) is given in the column "yield increase". The chaperone combinations for optimal yield of each target protein in the one-step and two-step procedures are indicated. Proteins that exhibited solubility exclusively after chaperone co-overexpression are indicated by "∞". The symbol "*" indicates that no soluble protein was purified from cells with or without chaperone co-overproduction. n.d., not determined. The same protein name can appear more than one time in the list because different constructs (domains or fusions) have been used for its expression.
26 of the 50 target proteins tested showed an increase in the final yield of purified, soluble protein upon co-overproduction of chaperones (Table 1; see Fig. 3a for examples). Of these 26 proteins, 21 showed a 2.5- to 5.5-fold increase in solubility, and 5 proteins (e.g. Lzip and Oskar3) became soluble only upon chaperone co-overproduction, allowing its purification under native conditions. For 16 of the 26 target proteins the highest increase in solubility was achieved by co-overproduction of all three KJE, ELS and ClpB chaperone systems in combination 4 or 5, with a higher success rate (11 of 26) for combination 4, which produces higher levels of ELS. For 8 of the 26 proteins the highest increase in solubility was obtained with combination 3, which overproduces ELS alone, and in only one case each the combination 1 (KJE alone) or 2 (KJE and ClpB) yielded the highest degree of solubility of the substrate protein. Taken together, these results show that chaperone co-overproduction is successful in about 50% of the proteins tested, with KJE, ClpB and ELS being the most successful combination. We did not observe any influence of the protein fusion tags, which have been fused to some of the recombinant proteins, on the efficiency of chaperone mediated solubility increase [see Additional file 1]. Furthermore, identical beneficial effects of chaperone co-production were observed when selected recombinant proteins (Btke, 2Tep4, Susy, Oskar1, TEV, Xklp3A/B) were produced in larger culture volumes (1–10 L) with final yields in the range of 2–20 mg/L [29].
Figure 3.
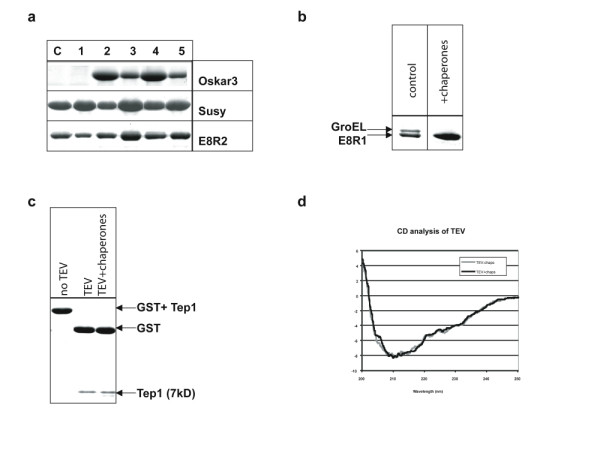
Effects of the chaperone co-expression on the yields of soluble target proteins. (a) Coomassie-stained SDS-PAGE showing target proteins purified by metal affinity chromatography either from control cells (C; without chaperone overexpression) or from different host cells overexpressing chaperone combinations 1 to 5 (indicated by the numbers above the lanes). The proteins Oskar3, Susy, and E8R2 were purified from cells grown with the one-step protocol. (b) Chaperone over-expression (combination 5) in host cells prevents the co-elution of the target protein E8R1 together with GroEL. (c) Comparison of the TEV activity using TEV protease purified from wild type cells and chaperone over-expressing cells (combination 3). (d) CD analysis of the TEV protein purified from wild type cells without (gray line) and with chaperone overexpression (combination 3, black line).
We tested for several recombinant proteins whether their solubility reflects the folding to the native state. TEV protease purified from the soluble fractions of cells with and without co-overproduction of ELS was active in cleaving a GST-Tep1 domain fusion protein (Fig. 3c) and showed highly similar circular dichroism spectra (Fig. 3d). For the target protein E8R1 the soluble material purified from cells without chaperone co-overproduction showed co-purification of significant amounts of EL, in contrast to the E8R1 protein purified from ELS/KJE/ClpB overproducing cells which appears EL-free (Fig. 3b), indicative of correct folding. These results suggest that the overproduction of chaperones not only results in improved solubility of the recombinant proteins tested but also enhances the acquisition of the native state.
To further improve the solubility of recombinant proteins we considered that it may be of advantage to allow chaperone-assisted folding in the absence of ongoing protein biosynthesis, thereby preventing the continuous generation of novel aggregation-prone proteins. In one set of experiments we prevented further synthesis of the plasmid-encoded target and chaperone proteins by withdrawal of IPTG. In another set of experiments synthesis of all cellular proteins was blocked by the addition of chloramphenicol (or tetracycline yielding similar results, data not shown) to the culture medium after removal of IPTG.
In 19 out of 34 tested recombinant proteins the two-step procedure resulted in higher solubility yields as compared to the one-step procedure (Table 1). Out of these, two proteins required the two-step procedure for any solubilization, and six proteins showed a chaperone-mediated solubilization only with the two-step procedure. For 13 of the 19 proteins the chaperone combinations 4 or 5 (ELS, KJE, ClpB) resulted in the highest yields of soluble recombinant protein (Table 1). The two-step procedure thus is clearly superior over the one-step procedure in the production of soluble recombinant protein.
As an example Fig. 4a shows the data obtained for Btke, a domain of the Brutons tyrosine kinase from human, applied to all 5 chaperone combinations (as well as 3 further combinations 6–8 described below). Even in control cells lacking chaperone expression plasmids, the two-step procedure resulted in a slight (3-fold) increase in Btke solubility as compared to the one-step procedure. This is in agreement with the previous observation that in wild type cells, the recovery of soluble recombinant proteins from aggregates can be improved by inhibition of protein biosynthesis [30]. The total amount of Btke produced under chaperone-coexpressing conditions was similar for all 5 strains tested. The one-step procedure resulted in an increased solubility of Btke only for chaperone combinations 3 and 5. In contrast, the two-step procedure, with a two-hour folding period in the presence of chloramphenicol, resulted in increased solubility for all five chaperone combinations, as compared to the control without chaperone co-overproduction. The most striking case was chaperone combination 4 (ELS, KJE, ClpB) in which the solubility of Btke, as compared to the control cells without chaperone co-overproduction, was 42-fold higher for the two-step procedure; in contrast, the one-step procedure (combination 3) yields only a 7-fold increase (Table 1). Interestingly, the optimal chaperone combination differs between the one-step procedure (ELS; combination 3) and the two-step procedure (KJE, ClpB, ELS; combination 4) (Fig. 4a). This switch in chaperone utilization probably reflects the fact that during the (re)folding period in step 2, aggregated Btke is solubilized in a process that requires the concerted action of KJE and ClpB.
Figure 4.
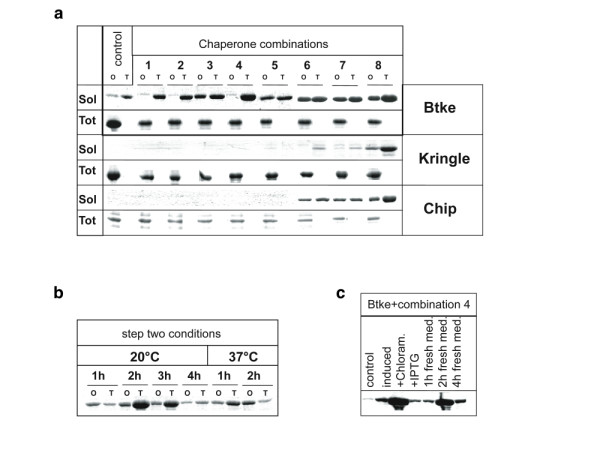
A two step procedure for successive periods of protein synthesis and folding. (a) Affinity chromatography purified proteins were separated by SDS-PAGE and stained with SimplyBlue (Invitrogen). Soluble recombinant proteins Btke, Kringle and Chip (Sol) recovered after chaperone-induced folding in BL21(DE3) control cells and cells co-expressing the eight different chaperone combinations described in Fig. 1. Cells were subjected to the one-step (O) or two-step (T) protocols. The total amounts of recombinant protein (Tot) expressed in control and chaperone co-overproducing cells were evaluated after purification of the protein under denaturating conditions. (b, c) Optimization of the (re)folding conditions in step two for Btke using chaperone combination 4. (b) After overnight culturing at 20°C the cells were pelleted, resuspended in fresh medium and cultured for 1 h, 2 h, 3 h, and 4 h at 20°C, or 1 h and 2 h at 37°C in the presence of 200 μg/ml chloramphenicol. For each time point the one-step (O) and two-step (T) procedures were compared. (c) Purified Btke from uninduced cells (control) or cells induced with IPTG according to the one-step procedure (induced) were compared with cells subjected to the two-step procedure with varying conditions: IPTG-induced overnight cultures were pelleted and further grown in fresh media without addition (growth for 1, 2, 4 h) or with addition of chloramphenicol or IPTG (growth for 2 h).
Several parameters were found to affect the solubilization efficiency. A two hour incubation of the cells in chloramphenicol containing media at 20°C was optimal for high yields of soluble Btke (Fig. 4b). Longer incubation times or higher temperature lowered the yield, probably because Btke is not entirely stable and is degraded in vivo as indicated by the decreased total amounts of Btke found in these cells. Furthermore, significant solubility of Btke (26-fold) could be achieved when transcription of the target gene was decreased by the removal of the inducer IPTG during the folding period, instead of inhibiting protein biosynthesis by chloramphenicol (Fig. 4c).
Although the chaperone combinations described so far showed remarkable effects on the solubility of 34 out of 50 recombinant proteins we aimed at a further optimization of the procedure. Recent findings indicate that the activity of ClpB in disaggregation and refolding of heat denatured proteins is facilitated by the presence of the bacterial sHSPs, IbpA and IbpB (IbpAB) [11]. We tested the effects of co-expression of the ibpAB genes (from a high copy number plasmid under pPA1/lacO1 control; pBB572) either alone (combination 6), or with ELS (combination 7) or ELS, KJE, ClpB (combination 8) (Fig. 1).
The co-overproduction of IbpAB (approximately 8–10 fold compared to wild type levels, Fig. 2) resulted in significant further improvement of the solubility of recombinant proteins (Table 2). Remarkably, IbpAB co-overproduction was beneficial even for some of the proteins, which remained completely insoluble with any of the previously tested 5 chaperone combinations. Fig. 4a shows two examples (Chip, Kringle) of such previously insoluble proteins. Out of the 23 proteins tested, the co-overproduction of IbpAB resulted in an increased solubility of 17 proteins in the one-step procedure (7 proteins with combination 6; 3 proteins with combination 7; 7 proteins with combination 8). Using the two-step procedure the yields of 3 additional soluble proteins was increased (Table 2). In sum, 20 out of 23 proteins tested showed increased solubility, and in 12 cases the chaperone co-expression was the only possibility to obtain any soluble protein.
Table 2.
Effects of IbpAB co-overproduction (combinations 6 to 8) on the yields of purified, soluble target proteins.
| Protein | MW (kDa) | Increased solubility by | |||
| one step procedure | two step procedure | ||||
| Yield increase | Best chaperone combination | Yield increase | Best chaperone combination | ||
| Ag1Ser | 30 | ∞ | 7 | 3 | 7 |
| Chip | 87 | ∞ | 8 | 4 | 8 |
| dLMO | 92 | ∞ | 6 | 3 | 6 |
| Isu2 | 46 | ∞ | 6 | 3 | 8 |
| Kringle | 27 | ∞ | 8 | 7 | 8 |
| Luc7 | 56 | ∞ | 8 | 4 | 8 |
| Oskar1 | 16 | ∞ | 8 | 3 | 8 |
| Ag3Ser | 33 | ∞ | 6 | 1 | - |
| Ag4Ser | 35 | ∞ | 8 | 1 | - |
| Icy1 | 45 | ∞ | 8 | 1 | - |
| Oskar4 | 70 | ∞ | 6 | 1 | - |
| 2c18 | 50 | 2.5 | 7 | 3.5 | 6 |
| E8R1 | 85 | 2.5 | 6 | 3 | 7 |
| Oskar2 | 115 | 1* | - | ∞ | 6 |
| Ag2Ser | 31 | 2.5 | 7 | 1 | - |
| Synapsin | 62 | 3 | 6 | 1 | - |
| 1Tep3 | 47 | 18 | 6 | 1 | - |
| 1Tep4 | 45 | 6 | 8 | 1 | - |
| 3x77 | 44 | 1 | - | 3 | 8 |
| Titin | 35 | 1 | - | 4 | 7 |
| Xklp3A3 | 72 | 1 | - | 1 | - |
| Msl1 | 66 | 1 | - | 1 | - |
| NF1 | 36 | 1* | - | 1* | - |
Increases in the amount of purified, soluble protein are given relative to the optimal solubility (set as 1) obtained either by the best of the combinations 1 to 6 tested before (Table 1) or to control cells (combination 4, data not shown). Symbols as in Table 1.
Conclusion
Taking all results together, the co-overproduction of the entire network of major cytosolic chaperones in E. coli cells, combined with a two-step procedure that allows for (re)folding of the recombinant proteins in the absence of ongoing de novo synthesis, resulted in an increased solubility of 70% of the 64 recombinant proteins tested. This efficiency is remarkable as most of the constructs used in this study encode proteins that are difficult to be produced in soluble form. The engineered E. coli strains and the two-step procedure presented here (patent application No. 10/500,883) should prove particularly useful for biotechnological applications.
Methods
Vector description
Plasmids (see Fig. 1a: pBB541, pBB542, pBB550, pBB535, pBB530, pBB540) expressing chaperone genes under the control of the IPTG-regulated promoter PA1/lacO-1 or Plac, and plasmid pBB528 constitutively expressing the LacIQ repressor were constructed as described earlier [28]. Plasmid pBB572 expressing the ibpA, ibpB operon under control of the PA1/lacO1 promoter was generated by PCR using primers ON1 (5'-CGGGATCCATATGCGTAACTTTGATTTATCCC-3') and ON4 (5'-GCTCTAGAGCTAGTTAGCTATTTAACGC-3') to amplify the ibpA, ibpB operon using chromosomal E. coli DNA as template. The PCR product was cut with BamHI and XbaI and cloned into plasmid pUHE212fdΔ12 [31]. Plasmids encoding various target proteins [see Additional file 1] were delivered from various laboratories to the Protein Expression Unit of the European Molecular Biology Laboratory Heidelberg.
Transformation procedure
Competent BL21 (DE3) cells (Novagen) were first transformed with a plasmid carrying the lacIQ gene (pBB535, pBB542, pBB550, pBB528) to allow for the controlled expression of chaperones. Subsequently, cells were transformed with appropriate plasmids for selective expression of eight different chaperone combinations and made competent (Fig. 1). Chaperone-overexpressing cells were finally transformed with a plasmid carrying the target gene. Cells were always grown in presence of appropriate antibiotics to ensure the maintenance of all plasmids.
Protein expression: One-step and two-step procedure
Single colonies from the transformed cells were used to inoculate 3 ml of LB medium supplemented with appropriate antibiotics. Liquid cultures were grown at 37°C until they reached an OD600 of 0.4 followed by transfer to 20°C. At an OD600 of 0.8 protein expression of plasmid encoded genes was induced by the addition of 0.1 mM IPTG and cells were further incubated overnight at 20°C. For the one-step procedure overnight cultures were pelleted, frozen and stored at -20°C until recombinant proteins were purified. For the two-step procedure overnight cell cultures were pelleted, the medium was removed and the cell pellet was resuspended in the same amount of fresh medium either supplemented with 200 μg/ml chloramphenicol or lacking IPTG. After 2 h incubation of the culture at 20°C the cells were harvested and target proteins were purified.
Protein purification and evaluation
The frozen bacterial pellet was resuspended in 350 μl of 20 mM Tris HCl buffer (pH 8.0) containing 2 mM PMSF, 0.05% Triton X-100, 1 μg/ml DNase, 5 mM MgCl2 and 1 mg/ml lysozyme and incubated on ice for 30 min with periodic stirring. The suspension was sonicated in water for 5 min, an aliquot of 5 μl was withdrawn (homogenate), and the cell debris was pelleted in a minifuge. A 5 μl aliquot of the supernatant was stored and the residual lysate was added to 15 μl of pre-washed magnetic beads (Qiagen) and incubated further for 30 min under agitation before the supernatant was removed. Beads were washed for 30 min with 20 mM K-phosphate buffer (pH 7.8) containing 300 mM NaCl, 20 mM imidazole, 8% glycerol, 0.2%Triton X-100 and subsequently with PBS buffer plus 0.05% Triton X-100. Finally beads were boiled in 12 μl SDS sample buffer and the supernatant loaded on a SDS PAGE. Proteins were detected by Simply Blue Safestain staining (Invitrogen) and the gels were recorded using a Umax Astra 4000U scanner. Protein bands were analyzed using the public NIH Image 1.62f software. Alternatively, the protein was eluted from washed beads using 30 μl PBS buffer plus 0.5 M imidazole and its relative concentration was measured by determination of its absorbance at 280 nm. The folding status was evaluated by circular dichroism using a J-710 spectropolarimeter (Jasco). The proteolytic activity of TEV protease was measured by incubating 500 μg of a fusion protein (GST-Tep1) containing a TEV recognition sequence in its linker in the presence of 5 μg of the TEV protease in Tris-HCl buffer (pH 8), 1 mM EDTA, 1 mM DTT, for 2 hours at 30°C.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
A.dM., designed experiments, performed experiments, interpreted data and wrote the manuscript. E.D., A.M., T.T., and B.B., designed experiments, interpreted data and wrote the manuscript.
Supplementary Material
Supplemental tables 1a and 1b.
Contributor Information
Ario de Marco, Email: ario.demarco@ifom-ieo-campus.it.
Elke Deuerling, Email: e.deuerling@zmbh.uni-heidelberg.de.
Axel Mogk, Email: a.mogk@zmbh.uni-heidelberg.de.
Toshifumi Tomoyasu, Email: tomoyasu@bio.tokushima-u.ac.jp.
Bernd Bukau, Email: bukau@zmbh.uni-heidelberg.de.
References
- Bukau B, Deuerling E, Pfund C, Craig EA. Getting newly synthesized proteins into shape. Cell. 2000;101:119–122. doi: 10.1016/S0092-8674(00)80806-5. [DOI] [PubMed] [Google Scholar]
- Deuerling E, Bukau B. Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit Rev Biochem Mol Biol. 2004;39:261–277. doi: 10.1080/10409230490892496. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Buchner J, Schmidt M, Fuchs M, Jaenicke R, Rudolph R, Schmid FX, Kiefhaber T. GroE facilitates refolding of citrate synthase by suppressing aggregation. Biochemistry. 1991;30:1586–1591. doi: 10.1021/bi00220a020. [DOI] [PubMed] [Google Scholar]
- Schröder H, Langer T, Hartl FU, Bukau B. DnaK, DnaJ, GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloubinoff P, Mogk A, Peres Ben Zvi A, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Tomoyasu T, Goloubinoff P, Rüdiger S, Röder D, Langen H, Bukau B. Identification of thermolabile E. coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe YH, Motohashi K, Taguchi H, Yoshida M. Heat-inactivated proteins managed by DnaKJ-GrpE-ClpB chaperones are released as a chaperonin-recognizable non-native form. J Biol Chem. 2000;275:12388–12392. doi: 10.1074/jbc.275.17.12388. [DOI] [PubMed] [Google Scholar]
- Laskowska E, Wawrzynow A, Taylor A. IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie. 1996;78:117–122. doi: 10.1016/0300-9084(96)82643-5. [DOI] [PubMed] [Google Scholar]
- Mogk A, Deuerling E, Vorderwulbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50:585–595. doi: 10.1046/j.1365-2958.2003.03710.x. [DOI] [PubMed] [Google Scholar]
- Veinger L, Diamant S, Buchner J, Goloubinoff P. The small heat shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J Biol Chem. 1998;273:11032–11037. doi: 10.1074/jbc.273.18.11032. [DOI] [PubMed] [Google Scholar]
- Jaenicke R, Seckler R. Spontaneous versus assisted protein folding. In: Bukau B, editor. Molecular Chaperones and Folding Catalysts Regulation, Cellular Function and Mechanism. Amsterdam, Harwood Academic Publishers; 1999. pp. 407–436. [Google Scholar]
- Kiefhaber T, Rudolph R, Kohler HH, Buchner J. Protein aggregation in vitro and in vivo: a quantitative model of the kinetic competition between folding and aggregation. BioTechnology. 1991;9:825–829. doi: 10.1038/nbt0991-825. [DOI] [PubMed] [Google Scholar]
- King J, Betts S. A green light for protein folding. Nature Biotechnol. 1999;17:637–638. doi: 10.1038/10848. [DOI] [PubMed] [Google Scholar]
- Schrodel A, de Marco A. Characterization of the aggregates formed during recombinant protein expression in bacteria. BMC Biochem. 2005;6:10. doi: 10.1186/1471-2091-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann J, Ventzki R, Schrodel A, de Marco A. Comparative analysis of protein aggregates by blue native electrophoresis and subsequent sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a three-dimensional geometry gel. Proteomics. 2005;5:2002–2009. doi: 10.1002/pmic.200401091. [DOI] [PubMed] [Google Scholar]
- Amrein KE, Takacs B, Stieger M, Molnos J, Flint NA, Burn P. Purification and characterization of recombinant human p50csk protein-tyrosine kinase from an Escherichia coli expression system overproducing the bacterial chaperones GroES and GroEL. Proc Natl Acad Sci U S A. 1995;92:1048–1052. doi: 10.1073/pnas.92.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale GE, Schonfeld HJ, Langen H, Stieger M. Increased solubility of trimethoprim-resistant type S1 DHFR from Staphylococcus aureus in Escherichia coli cells overproducing the chaperonins GroEL and GroES. Protein Eng. 1994;7:925–931. doi: 10.1093/protein/7.7.925. [DOI] [PubMed] [Google Scholar]
- de Marco A, Volrath S, Bruyere T, Law M, Fonne-Pfister R. Recombinant maize protoporphyrinogen IX oxidase expressed in Escherichia coli forms complexes with GroEL and DnaK chaperones. Protein Expr Purif. 2000;20:81–86. doi: 10.1006/prep.2000.1274. [DOI] [PubMed] [Google Scholar]
- Nishihara K, Kanemori M, Yanagi H, Yura T. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl Environ Microbiol. 2000;66:884–889. doi: 10.1128/AEM.66.3.884-889.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot AE, Goffin L, Payton MA, Wells TN, Bernard AR. In vivo and in vitro folding of a recombinant metalloenzyme, phosphomannose isomerase. Biochem J. 1996;318 ( Pt 2):437–442. doi: 10.1042/bj3180437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JG, Baneyx F. Protein folding in the cytoplasm of Escherichia coli: requirements for the DnaK-DnaJ-GrpE and GroEL-GroES molecular chaperone machines. Mol Microbiol. 1996;21:1185–1196. doi: 10.1046/j.1365-2958.1996.651436.x. [DOI] [PubMed] [Google Scholar]
- Thomas JG, Baneyx F. Protein misfolding and inclusion body formation in recombinant Escherichia coli cells overexpressing Heat-shock proteins. J Biol Chem. 1996;271:11141–7 Record 25 of 52 - MEDLINE (R) 1/96-12/96. doi: 10.1074/jbc.271.19.11141. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Kikuchi Y, Yasueda H. Overproduction of DnaJ in Escherichia coli improves in vivo solubility of the recombinant fish-derived transglutaminase. Biosci Biotechnol Biochem. 1998;62:1205–1210. doi: 10.1271/bbb.62.1205. [DOI] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Research. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Schröder H, Hesterkamp T, Schönfeld HJ, Bukau B. Substrate shuttling between the DnaK and GroEL systems indicates a chaperone network promoting protein folding. J Mol Biol. 1996;261:328–333. doi: 10.1006/jmbi.1996.0465. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol. 2001;40:397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- De Marco V, De Marco A, Goldie KN, Correia JJ, Hoenger A. Dimerization properties of a Xenopus laevis kinesin-II carboxy-terminal stalk fragment. EMBO Rep. 2003;4:717–722. doi: 10.1038/sj.embor.embor884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrio MM, Villaverde A. Protein aggregation as bacterial inclusion bodies is reversible. FEBS Lett. 2001;489:29–33. doi: 10.1016/S0014-5793(01)02073-7. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Valencia A, McMacken R, Sander C, Bukau B. The chaperone function of DnaK requires the coupling of ATPase activity with substrate binding through residue E171. EMBO J. 1994;13:1687–1695. doi: 10.1002/j.1460-2075.1994.tb06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental tables 1a and 1b.


