Abstract
Background
One-dimensional (1-D) electrophoretic data obtained using the cDNA-AFLP method have attracted great interest for the identification of differentially expressed transcript-derived fragments (TDFs). However, high-throughput analysis of the cDNA-AFLP data is currently limited by the need for labor-intensive visual evaluation of multiple electropherograms. We would like to have high-throughput ways of identifying such TDFs.
Results
We describe a method, GOGOT, which automatically detects the differentially expressed TDFs in a set of time-course electropherograms. Analysis by GOGOT is conducted as follows: correction of fragment lengths of TDFs, alignment of identical TDFs across different electropherograms, normalization of peak heights, and identification of differentially expressed TDFs using a special statistic. The output of the analysis is a highly reduced list of differentially expressed TDFs. Visual evaluation confirmed that the peak alignment was performed perfectly for the TDFs by virtue of the correction of peak fragment lengths before alignment in step 1. The validity of the automated ranking of TDFs by the special statistic was confirmed by the visual evaluation of a third party.
Conclusion
GOGOT is useful for the automated detection of differentially expressed TDFs from cDNA-AFLP temporal electrophoretic data. The current algorithm may be applied to other electrophoretic data and temporal microarray data.
Background
Expression analysis based on comparison of one-dimensional (1-D) electrophoretic patterns is one of the few genome-wide approaches that don't require sequence information. There are a few methods such as differential display [1], amplified fragment length polymorphism (AFLP) [2], and its variants like cDNA-AFLP, an AFLP-derived technique for RNA fingerprinting [3]. The cDNA-AFLP method and related techniques such as HiCEP have been widely used for gene discovery and monitoring temporal expression changes of transcript-derived fragments (TDFs) by comparing sets of time-course electropherograms [4-15]. However, inaccurate DNA fragment sizing often interferes with high-throughput analysis.
A major source of incorrect estimation of fragment lengths is the use of wrong size marker peaks when the true peaks are masked by intense peaks nearby [12,16]. Such electropherograms are locally expanded or compressed and the deviation from the true electropherogram reaches a maximum around the length of the wrong marker peak. Although a previous normalization strategy for HiCEP (a cDNA-AFLP-based expression profiling technique) data analysis [12] can correct this kind of inappropriate fragment sizing, slight variations in the lengths of identical TDFs across different electropherograms still remain. Even if the variations of individual TDFs across electropherograms are very small (e.g., within 1 bp), cumulative errors of fragment sizing interfere with accurate alignment of identical TDFs and make visual evaluation troublesome. Therefore, the minimization of variations of identical TDFs is a prerequisite for accurate alignment and easy visual evaluation.
The purpose of the present study is the identification of differentially expressed TDFs from HiCEP time-course data using a method (called "GOGOT") proposed here. This is essentially the purpose of microarray analysis. However, the bottleneck is the construction of an expression matrix of TDFs (rows) per time points (columns) of HiCEP electrophoretic data due to the problem of imperfect alignment, though most microarray analysis uses such matrices as given data (e.g., [17,18]). GOGOT constructs an expression matrix consisting of valid TDFs whose alignment accuracies are objectively high and ranks TDFs according to their degrees of differential expression using a special statistic. The performance of GOGOT is demonstrated by analyzing a large set of HiCEP time-course data obtained from mouse embryonic stem (ES) cells.
Results and discussion
A total of 256 primer combinations (16 MspI-NN primers combined with 16 NN-MseI primers; N = {A, C, G, T}) of HiCEP time-course data (mouse embryonic stem cells at 0, 12, 24, 48, and 96 h after adding stimulation for differentiation) was analyzed. HiCEP samples were technically duplicated and thus designated as 0h-1, 0h-2, 12h-1, 12h-2, 24h-1, 24h-2, 48h-1, 48h-2, 96h-1, and 96h-2. The data were preprocessed by a method which corrects fragment sizing errors caused by the mis-selection of size marker peaks [12].
The remaining slight variations in the lengths of subjectively identical TDFs across different electropherograms would be sufficient to make visual evaluation tedious and tends to cause imperfect alignment of the same peaks across different electropherograms (such as shown in Fig. 1a). In this work, we applied the current method to each of the 256 sets (primer combinations).
Figure 1.
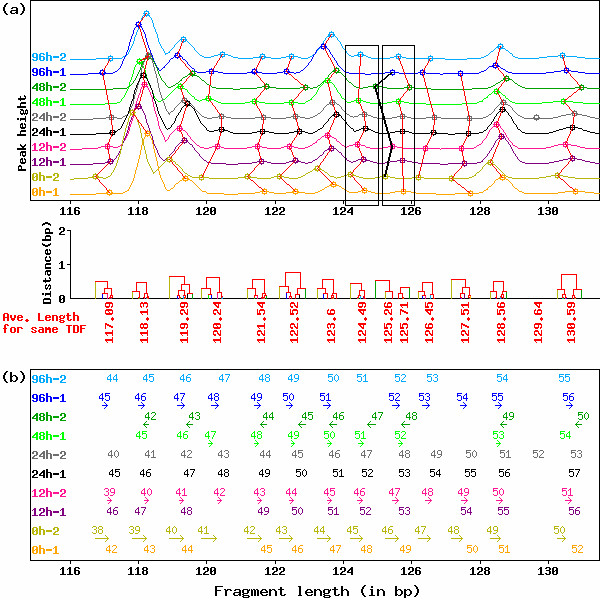
Typical example of HiCEP electropherograms before normalization of peak fragment lengths by GOGOTnormL. (a) Peak alignment of HiCEP electrophoretic data without GOGOTnormL normalization (upper) and the dendrograms obtained from complete-linkage clustering of the peak alignment (lower). Peaks connected by red lines and black bold lines are regarded as identical TDFs by the clustering-based peak alignment technique. Note that peak alignment subjectively failed in the range (124–126 bp) and that visual evaluation is also difficult because of the high variation in fragment lengths for individual TDFs. (b) Values of correction terms calculated by GOGOTnormL. For each serially numbered peak, directions and magnitudes are represented as arrows.
To both improve the alignment and make visual evaluation less difficult, we adopted a method (called GOGOT) for the HiCEP expression analysis. Briefly, the procedure consists of four steps: (1) normalization of peak fragment lengths, (2) peak alignment, (3) normalization of peak heights, and (4) identification of differentially expressed TDFs. In our experience, step 1 is peculiarly important for easy visual evaluation, especially when the number of electropherograms being compared is increased, regardless of successful or unsuccessful peak alignment [12,19].
In this paper, peak alignment for HiCEP electrophoretic data (Step 2) is performed using an algorithm based on complete linkage hierarchical clustering [20], though algorithms based on dynamic programming (DP) have widely been used for the purpose [21-25]. Perhaps a sophisticated DP-based method could perform accurate alignment such as shown in Fig. 2 for electropherograms in Fig. 1 without step 1. Nevertheless, the results of peak alignment for normalized electropherograms such as shown in Fig. 2 obtained from our two-step process (step 1 and 2) were satisfactory and those visual evaluations were very easy. The advantageous characteristics of our two-step approach over conventional DP-based methods [21-25] may be (i) easy visual evaluation by virtue of step 1 and (ii) easy traceability of why peaks are merged into a single TDF by virtue of a simple clustering-based method at step 2 (for details, see Methods). In general, labor-intensive visual evaluation of the electropherograms imposes bottlenecks on high-throughput expression analysis by electrophoretic methods including cDNA-AFLP [23]. Although there is currently no convincing rationale for choosing between the different methods, our two-step approach may eventually increase throughput. We demonstrate the feasibility of GOGOT in the rest of this section.
Figure 2.
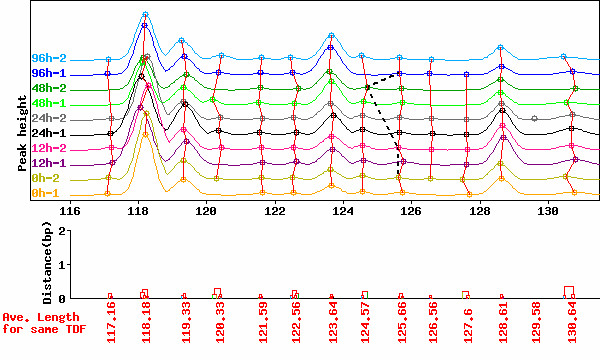
Normalized peak fragment lengths in HiCEP electropherograms in Fig. 1. Note that individual TDFs are represented by tight clusters and all peaks in the cluster are of course correctly aligned. The alignment connected by black bold lines in Fig. 1a is represented by black dashed lines and sectioned when peak alignment is reapplied to the normalized electropherograms.
Normalization of peak fragment lengths and its effect to peak alignment (Step 1 and 2)
Let be the ith TDF (i = 1,..., nk) in electropherogram Pk (k = 1,..., m; in this case m = 10). A TDF F is characterized by its length L (in bp), peak height H, and area A (in arbitrary units). The input data can be represented as 256 sets of (Li, Hi, Ai)k. Each electropherogram can be approximated by a Gaussian kernel using the input data [21-24]. The approximate electropherogram P(t), which is composed of n fragments Fi (i = 1,..., n), at length t (in bp) is given by
, where σi = Ai/(Hi), the standard deviation of a Gaussian curve approximated to the shape of the Fi.
As demonstrated in Fig. 1a, the direct application of clustering-based peak alignment (step 2) to HiCEP electrophoretic data does not work well because of the variation in the lengths of subjectively identical TDFs across electropherograms. In our opinion the four peaks aligned with black bold lines do not originate from identical TDFs and should be merged into the neighboring TDFs so that the peaks in each black box are aligned as identical TDFs. Furthermore, variant peaks across electropherograms which can make visual evaluation tedious still remain even if identical peak alignment could be performed by a sophisticated DP-based algorithm.
To this end, we first developed a method (called GOGOTnormL), which corrects peak fragment lengths L across electropherograms, so that the corrected lengths L' for subjectively identical TDFs are close to each other. The output data is represented as (L', H, A)k, where L' is defined as L' = L - c and the correction term c is calculated using a moving window approach (see Methods).
Fig. 1b shows the directions ("←" for c > 0 and "→" for c < 0) and magnitudes (|c|, represented by the length of the arrow) for the correction of peak fragment lengths L. In this figure, arrows are assigned to fragments having |c| > 0.10, and GOGOTnormL correction is mainly performed for one electropherogram (P48h-2) to the short side and four electropherograms (P0h-2, P12h-2, P48h-1, and P96h-1) to the long side. Although the other lanes (P0h-1, P12h-1, P24h-1, P24h-2, and P96h-2) in the figure are of course slightly corrected (the respective average correction terms were 0.05, -0.04, 0.06, 0.06, and 0.01), they are used as references.
Fig. 2 shows the result of Fig. 1 after normalizing by GOGOTnormL. Visually, the electropherograms are normalized nearly perfectly. The average correlation coefficients among the ten electrophoretic patterns in the range shown in the figure before and after normalization are 0.79 and 0.91, respectively. The alignment consisting of the four questionable peaks discussed above (shown as black dashed lines in Fig. 2) disappears when clustering-based peak alignment (see Methods) is applied to the normalized electropherograms. Nevertheless, objective evaluation of the peak alignment for the electropherograms after GOGOTnormL normalization compared to that before normalization is difficult in practice and the goodness of peak alignment is judged by subjective visual evaluation. Although we believe the strategy of correcting for peak fragment lengths before peak alignment can increase the accuracy of peak alignment and make visual evaluation of aligned peak sets easier, some researchers might not agree.
GOGOTnormL can be regarded as an image warping method for adjusting different mobilities among corresponding peaks. There are some image warping methods for 1-D electrophoretic data produced by various experimental technologies such as single-stranded conformational polymorphism (SSCP) or pulsed-field gel electrophoresis (PFGE) [19,25-27]. However, the comparison between these methods and the GOGOTnormL is difficult in practice because of (i) different frameworks such as input data format, (ii) the subjectivity caused by visual evaluation of normalized electropherograms and aligned peaks, and (iii) a multitude of adjustable parameters.
The effectiveness of GOGOTnormL (Step 1) depends on the choice of the parameters T (the number of consecutive fragments as a "window" for the normalization; see equation 1 in the Methods section) and D (the empirically estimated maximum variation in the lengths of subjectively identical TDFs). The magnitude (|c|) of the correction term c for making the corrected length L' tends to decrease when T is large or D is small. In this case GOGOTnormL is ineffective since L' approaches L. On the other hand, when T is small or D is large GOGOTnormL is likely to produce erroneous results such as Li' > Li+1' (the size relationship must always be Li' <Li+1' regardless of normalization). Indeed, we observed such unfavourable cases when for example T = 3 and D = 20. Although we conservatively give T = 8 as the minimum number for which the size discrepancy disappears for all 256 sets of the HiCEP data (and D = 2 bp empirically), it is variable for each of the 256 sets used here and the other datasets. Similarly, the effectiveness of peak alignment via clustering (Step 2) depends on the choice of the parameter u which specifies the maximum difference of lengths for the aligned fragments. The parameter value u = 2 was chosen to specify the maximum variation among fragment lengths originating from TDFs determined (by eye) to be identical. Determination of parameter settings is ultimately the subjective decision of the researcher.
Normalization of peak heights (Step 3)
Since cDNA-AFLP analysis handles small volumes of samples, problems during electrophoretic analysis such as over- or under-loading of samples cause variations in the overall peak heights H among different samples (or runs). Accordingly, normalization is essential when comparing cDNA-AFLP electrophoretic data.
One simple approach is to assume that the average peak height of all the reported TDFs among samples is approximately the same [12,28]. It is formulated as = constant for a set of electropherograms Pk (k = 1,..., m). However, this approach sometimes fails because it includes two kinds of questionable peaks [22]. One is peaks near a preset detection limit, resulting in some peaks being detected and others not (for example, two peaks at 217 bp and four at 223.5 bp in Fig. 3). The other is peaks incorrectly identified as either broad peaks or two overlapping peaks because of the similar appearance of these two types.
Figure 3.
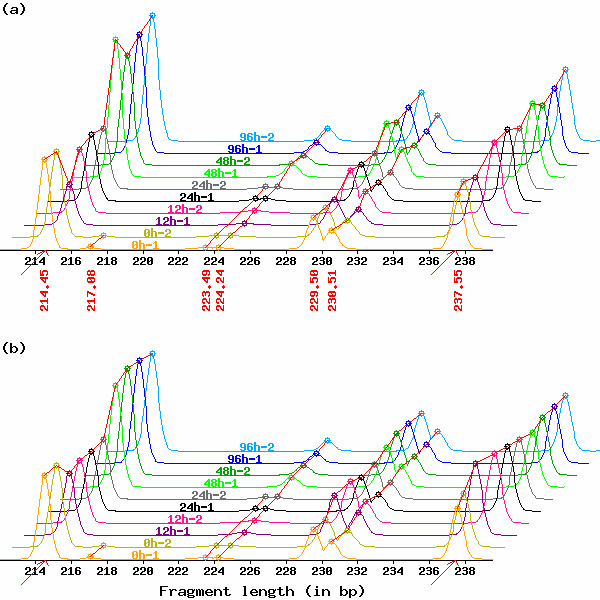
Effect of peak height normalization by GOGOTnormH. Electropherograms when peak height normalizations are performed using (a) all the reported TDFs (a conventional method used in [12, 28]) and (b) a subset of the selected TDFs (GOGOTnormH).
We selected TDFs (or peaks) satisfying the following three conditions for peak height normalization: (1) peaks corresponding to an identical TDF exist in all samples, (2) they are not close to neighboring TDFs, and (3) the quality scores [12] assigned to each peak are high (for details, see the Methods section). We performed the normalization by adjusting the average peak heights of the selected TDFs among samples (we call the procedure GOGOTnormH for convenience) by assuming that only a minority of the selected TDFs display temporal expression changes. In general, the more selected TDFs you use for normalization, the more reliable the analysis. If you relax the standards for choosing TDFs, however, you compromise the reliability of the selected TDFs and the resulting peak alignment is less accurate. It's a tradeoff.
Peak height after normalization (H') is defined as H' = H × N, where N is a normalization factor. GOGOTnormH outputs (L', H', A)k from the input dataset (L', H, A)k (k = 1,..., m). Fig. 3 demonstrates the effect of GOGOTnormH (peak height normalizations are performed using all the reported TDFs in Fig. 3a and a subset of the selected TDFs in Fig. 3b). In Fig. 3, two TDFs indicated by red arrows satisfy the three conditions above. Since two electropherograms in each time point (e.g., 12h-1 and 12h-2) are the technical replicates, we can measure the power of GOGOTnormH (Fig. 3b) with the conventional method [12,28] (Fig. 3a) in light of the reproducibility in peak heights H' between the replicates. In comparison with electropherograms normalized using the conventional method [12,28] (Fig. 3a), we observed higher reproducibility in peak heights H' between replicate experiments in the normalized electropherograms (Fig. 3b): Peak heights H' in 12h-1 (and 48h-2) after conventional normalization are consistently lower than those in 12h-2 (and 48h-1) in electropherograms.
We next show the statistics about ratios of peak heights between replicate experiments (Fig. 4). In the analysis of 256 sets (primer combinations) of HiCEP data, a total of 10,624 TDFs were used for peak height normalization and each TDF has five time points (0 h, 12 h, 24 h, 48 h, and 96 h). We observed a smaller dispersion for 53,120 (10,624 × 5) expression ratios after GOGOTnormH normalization. For example, there were 75.5% (or 94.8%) of 53,120 ratios satisfying ≤ 1.2 (or 1.5) fold difference after GOGOTnormH normalization, compared to 59.3% (or 89.5%) after normalization using the conventional method [12,28]. These results demonstrate the validity of our strategy at least for peak height normalization of the 10,624 TDFs. The minimization of differences between technical replicates is, of course, one quality criterion and it remains unclear how efficiently both algorithms reveal genuine temporal expression changes. The comparison of our GOGOTnormH and other conventional methods on real data which contain genuinely induced/suppressed TDFs is one of the next important task.
Figure 4.
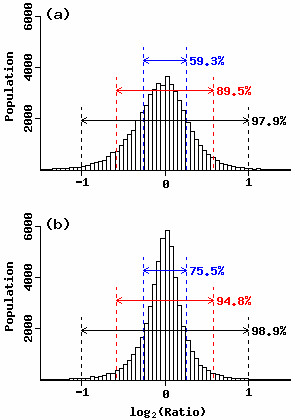
Distribution of peak height ratios between replicate experiments. Ratios are calculated using peak heights when the normalizations are performed using (a) all the reported TDFs (a conventional method used in [12, 28]) and (b) a subset of the selected TDFs (GOGOTnormH). Dashed lines in blue, red, and black indicate 1.2-, 1.5-, and 2.0-fold differences in peak heights, respectively.
Although we here calculated the normalization factor N using the average peak height of the selected TDFs (GOGOTnormH) to compare the effect of different sets of TDFs, there are many other possible approaches for calculating the normalization factor N such as the median, the trimmed mean [29], Tukey's one-step biweight method [30], and so on. Further improvement in the choice of those methods as well as the selection of valid TDFs remains to be studied.
Identification of differentially expressed TDFs (Step 4)
We have an expression matrix (called the "HiCEP expression matrix") consisting of 10,624 TDFs and ten temporal samples obtained from HiCEP electrophoretic data. We searched through these 10,624 TDFs looking for differentially expressed TDFs, though there are many others in the original data (such as the four TDFs in the range 222–232 bp shown in Fig. 3). This is because the 10,624 TDFs have two advantages: (1) they have high reproducibility between replicate experiments (Fig. 4) and (2) their annotation is potentially easy by virtue of the above three conditions used in GOGOTnormH normalization.
We used a special statistic called GOGOTstat (equation 4) for the detection of differentially expressed TDFs (see Methods). The statistic gives a nonnegative score, with the value 0 for TDFs expressed uniformly in all the interrogated samples and a high score for differential expression. Of course, there are many ways to rank TDFs according to the degrees of their differential expression. For example, a t-like statistic (equation 5) obtained by substituting the standard deviation of peak heights in replicate experiments for the ratio in equation 4 can be considered. However, such a t-like statistic often gives a high score (rank) to questionable TDFs whose overall peak heights are quite low (Additional file 1). This high score is mainly caused by a low value for the denominator in the t-like statistic. We do not give high scores to these questionable TDFs for the following two reasons. One is they have high relative error (low signal-to-noise ratio). In general, relative error increases for low peak heights when the peak height approaches the background level [31]. The reliability of such candidates is thus implausible [32]. The other reason is the disagreement with visual evaluation (Additional file 2 and Fig. 5). Unlike in microarray analysis, the true significance of the candidate patterns obtained from high-dimensional electrophoretic data such as HiCEP or Differential Display must be confirmed visually. It is important to develop a score metric compatible with visual evaluation. Our statistic (GOGOTstat) is a straightforward application of this idea.
Figure 5.
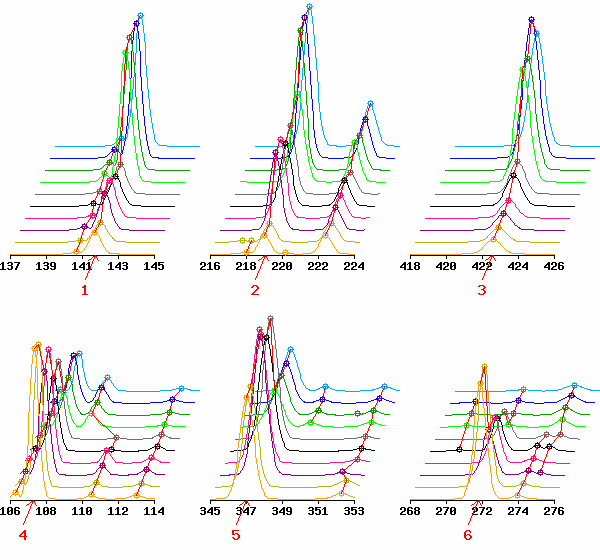
Expression patterns of top six TDFs listed in Table 2. Local (8 bp of range) electropherograms including the top-ranked TDFs indicated by red arrows are shown. Numbers below arrows indicate the ranks of the TDFs. Each electropherogram is shown in common scaling.
Table 1 lists expression data (peak heights) and the statistics for the top ten TDFs. As expected, there is a wide range of peak heights across time points and high reproducibility between the replicates for each time point. Visual evaluation of those local electrophoretic patterns also demonstrates that peak alignment is correctly performed (see Fig. 5). The expression data (peak heights) and two statistics (GOGOTstat and t-like statistic) for all 10,624 TDFs in the HiCEP expression matrix are available in Additional file 1.
Table 1.
Expression data for top ten TDFs ranked by GOGOTstat. Statistic: score of GOGOTstat. The values of peak heights after GOGOTnormH normalization are shown.
| Rank | Peak height | Statistic | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H0h-1 | H0h-2 | H12h-1 | H12h-2 | H24h-1 | H24h-2 | H48h-1 | H48h-2 | H96h-1 | H96h-2 | ||
| 1 | 134 | 121 | 228 | 236 | 183 | 186 | 811 | 828 | 843 | 817 | 134.7 |
| 2 | 115 | 115 | 483 | 489 | 388 | 425 | 554 | 870 | 881 | 873 | 134.0 |
| 3 | 91 | 88 | 105 | 112 | 189 | 201 | 704 | 698 | 868 | 706 | 129.0 |
| 4 | 938 | 894 | 650 | 710 | 455 | 485 | 214 | 237 | 295 | 233 | 123.7 |
| 5 | 636 | 627 | 865 | 835 | 712 | 756 | 231 | 247 | 248 | 261 | 116.8 |
| 6 | 719 | 752 | 285 | 282 | 203 | 172 | 16 | 12 | 16 | 10 | 116.6 |
| 7 | 803 | 811 | 320 | 332 | 293 | 299 | 188 | 203 | 212 | 180 | 114.9 |
| 8 | 141 | 153 | 335 | 353 | 342 | 338 | 743 | 763 | 646 | 774 | 112.7 |
| 9 | 643 | 627 | 600 | 634 | 560 | 527 | 90 | 84 | 65 | 72 | 106.5 |
| 10 | 684 | 704 | 650 | 665 | 572 | 666 | 149 | 133 | 132 | 136 | 104.4 |
Since the detection of differentially expressed TDFs is based on the HiCEP expression matrix, we can estimate the false discovery rate (FDR), defined as the expected proportion of false positives among true differentially expressed TDFs [33]. The random permutation test suggests that tens of top-ranked TDFs have statistical significance at low (5–10%) FDR (see Table 2). Visual evaluation confirmed the validity of the differential expression patterns for the top 100 TDFs and the non-differential patterns for the last 100 TDFs (data not shown except for the top six TDFs).
Table 2.
Numbers of TDFs called significant at various thresholds.
| Statistic | Randomized | Observed | FDR |
|---|---|---|---|
| 13.4 | 1017.8 | 2037 | 50% |
| 20.2 | 481.9 | 1202 | 40% |
| 30.0 | 163.2 | 543 | 30% |
| 40.1 | 53.9 | 270 | 20% |
| 57.2 | 8.3 | 83 | 10% |
| 76.5 | 1.4 | 28 | 5% |
Statistic: score of GOGOTstat satisfying given FDRs.
Randomized: average number called significant by analyzing 1,000 randomly permutated HiCEP expression matrices.
Observed: number called significant by analysing the original HiCEP expression matrix of 10,624 TDFs and 10 samples.
The FDR is defined as the percentage of falsely significant TDFs compared to the TDFs called significant.
Note that there must be other differentially expressed TDFs (i.e., true-negative TDFs) which are not included in the dataset (i.e., the 10,624 TDFs in the expression matrix) because they do not satisfy the above three conditions used in Step 3. For example, we cannot identify differentially expressed TDFs if peaks constituting an identical TDF are not detected due to the effect of a preset detection limit, with the current settings used for the selection of the 10,624 TDFs. Of course, they should be detected if they are genuine. However, as also discussed in the selection of the 10,624 TDFs, the more true-negatives you want to detect, the more tedious visual inspections you have to do. It's a tradeoff.
In practice one may want to detect the differentially expressed TDFs having no data at time point k due to reasons such as the above, though the current analysis does not analyze those TDFs. One way to deal with them would be to let Hk = constant (e.g., the predetermined value of the peak detection threshold). Of course, there are many possible ways to analyze those TDFs. Further improvement of GOGOTstat to make it universal remains to be done.
Conclusion
We propose an integrated strategy (called GOGOT) for identifying differentially expressed TDFs from HiCEP time-course data. As with mass spectrometry data, there remains the problem that the same peaks across electropherograms are not perfectly aligned in general [34,35]. We demonstrated that the application of GOGOTnormL (step 1) dramatically contributes to successful peak alignment via clustering (step 2) and facilitates visual evaluation (Figs. 1 and 2). This enabled us to construct a HiCEP expression matrix consisting of 10,624 valid TDFs and ten samples from a total of 256 sets of ten HiCEP electrophoretic data samples. Normalization of peak heights in the matrix using GOGOTnormH increased reproducibility between replicate experiments (Figs. 3 and 4). These results facilitate the use of various analysis methods for the identification of differentially expressed genes in microarray data.
We used a simple statistic (GOGOTstat at step 4) to rank TDFs according to the degree of their temporal expression change. Researchers involved in HiCEP analysis were very satisfied with the ranking of the differentially expressed TDFs (Fig. 5). Although the current statistic was developed for analyzing HiCEP data, it can also be applied to microarray data since the input data (expression matrix) is the same. As future work, it would be interesting to evaluate the potential of the statistic in the analysis of microarray data.
The current method GOGOT can be regarded as a method for analyzing 1-D electrophoretic data. There are a number of methods for analyzing 1-D electrophoretic data produced by various experimental technologies [19,21-29]. Compared to them, GOGOT can be positioned a method specialized for cDNA-AFLP data analysis. The fully automated GOGOT procedure dramatically increased the throughput of data analysis (approximately, 20–30 times). We also verified the power of the strategy using other HiCEP datasets (data not shown). In addition to cDNA-AFLP data, the algorithm should be easily applicable to other one-dimensional electrophoretic data such as Differential Display or AFLP. GOGOT can be a powerful tool for detecting differentially expressed TDFs from multiple one-dimensional electrophograms.
Methods
Data
Samples were prepared from mouse embryonic stem (ES) cells at 0, 12, 24, 48, and 96 h after removal of leukemia inhibitory factor (LIF) from the culture medium. The samples subjected to HiCEP reaction were technically duplicated (i.e., the replicates were from the same samples). We thus designated each sample as 0h-1, 0h-2, 12h-1, 12h-2, 24h-1, 24h-2, 48h-1, 48h-2, 96h-1, and 96h-2. The HiCEP reaction was performed according to a previous report [8]. Most of the steps are the same as in standard AFLP [2] except for (i) the primers, whose GC content is 55% to 60% and (ii) the annealing temperature (71.5°C) in selective PCR [8].
cDNA prepared from mRNA extracted from each sample were digested with two 4-bp-cutting endonucleases (MspI combined with MseI) and ligated with the corresponding adaptors. The resulting HiCEP templates, 5'-MspI-MseI-3' fragments, were amplified using fluorescently labeled primers. In total, 256 primer combinations (16 MspI-NN primers combined with 16 NN-MseI primers; N = {A, T, G, C}) were used in the HiCEP analysis. The details of the protocol of the HiCEP reaction are described elsewhere [8].
The PCR products were denatured and loaded on an ABI Prism 310 (Applied Biosystems) for capillary gel electrophoresis. The digitized images were noise-reduced and baseline-corrected by the GeneScan software (Applied Biotech). After noise reduction and baseline correction, the software quantifies each TDF F by fragment length L (in bp), peak height H, and area A (in arbitrary units) in the size calibration range (35–700 bp in this analysis). Accordingly, the data obtained from a HiCEP electropherogram consists of a collection of TDFs and each TDF (or peak) (i = 1,..., nk) in electropherogram Pk (k = 1,..., m; m = 10 in this case) is characterized by (Li, Hi, Ai)k, where the peaks are originally numbered with respect to their sizes.
To correct fragment sizing errors caused by the mis-selection of size marker peaks, the preprocessing method of Kadota et al. [12] was adopted. Accordingly, the variation in the lengths of subjectively identical TDFs in the input data was small (± 1 bp at most; see Fig. 1).
Normalization of peak fragment lengths (Step 1; GOGOTnormL)
The purpose of step 1 using GOGOTnormL is to correct peak fragment lengths L across electropherograms so that the corrected lengths L' for subjectively identical TDFs are close to each other. The output data for the m electropherograms (k = 1,..., m) to be compared is represented as (L', H, A)k. The normalization is performed for each electropherogram Pk (k = {1,..., m}) using a moving window approach. Briefly, the procedure is as follows:
Step 1-1. Determination of the window (range) in the target electropherogram
Each electropherogram can be approximated by a Gaussian kernel using the input data (L, H, A). The approximate electropherogram P(t), which is composed of n fragments Fi (i = 1,..., n), at length t (in bp) is given by
, where σi = Ai/(Hi), the standard deviation of the Gaussian curve approximated to the shape of the Fi.
A total of (ntarget - T + 1) ranges are defined from a target electropherogram Ptarget (target = {1,..., m}), where ntarget is the number of fragments in Ptarget. The ith (i = 1,..., ntarget - T + 1) range consists of T fragments Fa (a = i,..., i+T-1). In this analysis, we let T = 8 though other numbers are of course possible. For example, the first range consists of eight fragments F1, F2,..., F8 and the (ntarget - T + 1)th range consists of . The ith range in the target electropherogram, (starti-endi bp)target, is given by:
| (1) |
Step 1–2. Selection of the reference for each range
The selection of the reference electropherogram (a kind of typical electrophoretic pattern) for the normalization of the ith range (starti-endi bp)target is performed according to Kadota et al. [12]. Briefly, quality scores Q() at fragment lengths in electropherogram Pk (k = {1,..., m}) are estimated by a windowing calculation of local average correlation coefficients between Pk and the other electropherograms (for details, see [12]). A high (or low) score for electropherogram Pk indicates a high (or low) level of relative similarity between the electropherogram and the others at around length . The reference to the ith range (starti-endi bp)target is the electropherogram Pk (k = {1,..., m}) satisfying both (i) the number of peaks satisfying starti ≤ ≤ endi is T/2 or more and (ii) the average quality score is the maximum.
Step 1–3. Normalization of the target electropherogram
For the ith (i = 1,..., ntarget - T + 1) range (starti-endi bp), GOGOTnormL searches for the sub-electropherogram Ptarget [starti+xi, endi+xi] (in the target electropherogram) that is most similar to the reference Preference [starti, endi] around the range. The most similar sub-electropherogram is the one that achieves the highest correlation coefficient ri between Preference [starti, endi] and the various candidates Ptarget [starti+x, endi+x] (-D <= x <= +D). xi is the x at the highest correlation coefficient ri. We set D = 2, as a maximal realistic displacement.
If we use a moving window of T fragments, most fragments (ntarget-2T+2 fragments) (i = T, T+1,..., ntarget - T+1) have T values of xa and ra (a = i-T+1, i-T+2,..., i).
Li' (Li in the target electropherogram after normalization) is obtained from Li' = Li - ci. The correction term ci is calculated by using xa and ra (a = i-T+1, i-T+2,..., i):
| (2) |
where w(ra) is the tricube weight function of ra, namely w(ra) = 1 - (1 - ra3)3.
Peak alignment via clustering (Step 2)
The major difficulty in one-dimensional (1-D) electrophoretic data (including HiCEP) analysis is the alignment of multiple peak sets. In order to solve this problem, we here use complete-linkage hierarchical clustering, since the strategy was successfully applied to mass spectrometry data by Tibshirani et al. [20]. To our knowledge, it is the first time clustering-based peak alignment has been used for 1-D electrophoretic data analysis.
The absolute difference in fragment lengths between two peaks is used as the distance. To guarantee that each cluster represents individual TDFs, two clusters are merged only when all the peaks in the two clusters are derived from different samples. Since we analyze ten samples, the maximum peak number in each cluster is thus ten. The dendrogram is cut off at height u bp. In this study, we set u = 2, implying that every peak in the cluster is at most 2 bp from any other peak in that same cluster by virtue of the use of complete-linkage.
As previously mentioned by Tibshirani et al. [20], the idea for peak alignment is that tight clusters should represent identical TDFs. The use of clustering-based peak alignment combined with the correction of peak fragment lengths by GOGOTnormL was successful (Figs. 1 and 2).
Normalization of peak heights (Step 3; GOGOTnormH)
As shown in Fig. 3a, the conventional normalization strategy [12,28], in which average peak heights of all the reported TDFs among samples are adjusted, sometimes does not work well. We assert the reason is the use of all the reported TDFs in individual samples. GOGOTnormH selects TDFs satisfying the following three conditions:
(i) peaks corresponding to identical TDFs are exhibited by all samples. The idea is essentially the same as that of Fushiki et al. [35]: peaks exhibited by only a few samples may just be noise, but peaks exhibited by many subjects appear to be true TDFs.
(ii) the neighboring TDFs are not so close. Suppose that (a) individual TDFs in a set of electropherograms are numbered with respect to their average sizes, (b) there are m (10 in this case) peaks in the ith TDF (i = 1,..., nTDF) by condition (i), and (c) the ten peaks corresponding to the ith TDF (i = 1,..., nTDF) in electropherograms Pk (k = 1,..., m) are characterized by length , peak height , area , and standard deviation . The width of the ith TDF is defined as
Finally, the TDFs satisfying following conditions are selected:
• Case i = 1,
• Case i = 2,..., nTDF -1,
and
• Case i = nTDF,
(iii) all of the quality scores Q() assigned for the ten peaks corresponding to the ith TDF are 0.7 or more. The score provides an objective goodness for the estimation of the fragment lengths and our previous study suggests the value of 0.7 is the minimum necessary for the accurate alignment of valid TDFs across electropherograms [12].
The peak height after normalization (H') is obtained from H' = H × N, where N is a normalization factor. The normalization factor Nk for electropherogram Pk is calculated by
| (3) |
where is the average peak height for the selected TDFs in electropherogram Pk. After normalization, the average peak height in each electropherogram is adjusted to 100.
Identification of differentially expressed TDFs (Step 4; GOGOTstat)
The differentially expressed TDFs are detected from a total of 10,624 valid TDFs used for peak height normalization at step 3. Consider one expression vector H consisting of peak heights Hp-q at the qth replicate experiment (q = 1, 2,..., np) in time point p. To quantify the degree of differential expression, we define a statistic GOGOTstat as
| (4) |
where is the average of np normalized peak heights at time point p and Hp-q is the normalized peak height in the qth replicate experiment at time point p. Since each TDF has ten normalized peak heights in five time points (i.e., p = (0h, 12h, 24h, 48h, 96h)) and each time point has two replicates (i.e., np = 2), the generalized expression vector can be written as H = (H0h-1, H0h-2,..., H96h-2). For example, the statistic of the top-ranked TDF shown in Table 1 is calculated as follows:
The t-like statistic compared to our GOGOTstat statistic (see Results and discussion) is defined as
| (5) |
where σp is the standard deviation of np normalized peak heights at time point p.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
KK invented the method and wrote the paper. RA, YN, and MA provided critical comments and led the project.
Supplementary Material
Expression data for 10,624 valid TDFs. Peak heights after GOGOTnormH for 10,624 valid TDFs are provided. It also contains two statistics measured by GOGOTstat and a t-like statistic and their ranks.
Expression patterns of top six TDFs ranked by a t-like statistic. Legends are the same as given in Fig. 5.
Contributor Information
Koji Kadota, Email: kadota@iu.a.u-tokyo.ac.jp.
Ryoko Araki, Email: a_ryo@nirs.go.jp.
Yuji Nakai, Email: yunakai@iu.a.u-tokyo.ac.jp.
Masumi Abe, Email: abemasum@nirs.go.jp.
Acknowledgements
We would like to thank A. Nifuji for visual evaluation of the GOGOT results and J.J. Rodrigue for editing the English. This study was performed through Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science and Technology, the Japanese Government. This study was also supported by a Research Revolution 2002 on Innovative Development Project grant and a Grant-in Aid for Young Scientists (B) (19700273) to K. Kadota from the Ministry of Education, Culture, Sports, Science and Technology, the Japanese Government.
References
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;2:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;2:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem CW, van der Hoeven RS, de Bruijn SM, Vreuqdenhil D, Zabeau M, Visser RG. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 1996;2:745–753. doi: 10.1046/j.1365-313X.1996.9050745.x. [DOI] [PubMed] [Google Scholar]
- Donson J, Fang Y, Espiritu-Santo G, Xing W, Salazar A, Miyamoto S, Armendarez V, Volkmuth W. Comprehensive gene expression analysis by transcript profiling. Plant Mol Biol. 2002;2:75–97. doi: 10.1023/A:1013722224489. [DOI] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Cannoot B, Rombaut D, Vandepoele K, Rombauts S, Vanderhaeghen R, Inze D, Zabeau M. Quantitative cDNA-AFLP analysis for genome-wide expression studies. Mol Genet Genomics. 2003;2:173–179. doi: 10.1007/s00438-003-0830-6. [DOI] [PubMed] [Google Scholar]
- Yao YX, Li M, Liu Z, Hao YJ, Zhai H. A novel gene, screened by cDNA-AFLP approach, contributes to lowering the acidity of fruit in apple. Plant Physiol Biochem. 2007. in press . [DOI] [PubMed]
- Cnudde F, Hedatale V, de Jong H, Pierson ES, Rainey DY, Zabeau M, Weterings K, Gerats T, Peters JL. Changes in gene expression during male meiosis in Petunia hybrida. Chromosome Res. 2006;2:919–932. doi: 10.1007/s10577-006-1099-5. [DOI] [PubMed] [Google Scholar]
- Fukumura R, Takahashi H, Saito T, Tsutsumi Y, Fujimori A, Sato S, Tatsumi K, Araki R, Abe M. A sensitive transcriptome analysis method that can detect unknown transcripts. Nucleic Acids Res. 2003;2:e94. doi: 10.1093/nar/gng094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani Y, Suzuki K, Kondo K, Okumura K, Tamura T. Gene expression analysis using a modified HiCEP method applicable to prokaryotes: A study of the response of Rhodococcus to isoniazid and ethambutol. J Biotechnol. 2006;2:259–272. doi: 10.1016/j.jbiotec.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Umeda N, Tsutsumi Y, Fukumura R, Ohkaze H, Sujino M, van der Horst G, Yasui A, Inoue ST, Fujimori A, Ohhata T, Araki R, Abe M. Mouse dexamethasone-induced RAS protein 1 gene is expressed in a circadian rhythmic manner in the suprachiasmatic nucleus. Brain Res Mol Brain Res. 2003;2:1–6. doi: 10.1016/S0169-328X(02)00543-0. [DOI] [PubMed] [Google Scholar]
- Araki R, Takahashi H, Fukumura R, Sun F, Umeda N, Sujino M, Inoue SI, Saito T, Abe M. Restricted expression and photic induction of a novel mouse regulatory factor X 4 transcript in the suprachiasmatic nucleus. J Biol Chem. 2004;2:10237–10242. doi: 10.1074/jbc.M312761200. [DOI] [PubMed] [Google Scholar]
- Kadota K, Fukumura R, Rodrigue JJ, Araki R, Abe M. A normalization strategy applied to HiCEP (an AFLP-based expression profiling) analysis: Toward the strict alignment of valid fragments across electrophoretic patterns. BMC Bioinformatics. 2005;2:43. doi: 10.1186/1471-2105-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama S, Jakt ML, Suzuki M, Araki R, Hikichi T, Kishigami S, Ohta H, Van Thuan N, Mizutani E, Sakaide Y, Senda S, Tanaka S, Okada M, Miyake M, Abe M, Nishikawa SI, Shiota K, Wakayama T. Equivalency of nuclear transfer-derived embryonic stem cells to those derived from fertilized mouse blastcyst. Stem Cells. 2006;2:2023–2033. doi: 10.1634/stemcells.2005-0537. [DOI] [PubMed] [Google Scholar]
- Araki R, Nakahara M, Fukumura R, Takahashi H, Mori K, Umeda N, Sujino M, Inouye SI, Abe M. Identification of genes that express in response to light exposure and express rhythmically in a circadian manner in the mouse suprachiasmatic nucleus. Brain Res. 2006;2:9–18. doi: 10.1016/j.brainres.2006.04.096. [DOI] [PubMed] [Google Scholar]
- Araki R, Fukumura R, Sasaki N, Kasama Y, Suzuki N, Takahashi H, Tabata Y, Saito T, Abe M. More than 40,000 transcripts including novel and non-coding transcripts in mouse embryonic stem cells. Stem Cells. 2006;2:2522–2528. doi: 10.1634/stemcells.2006-0005. [DOI] [PubMed] [Google Scholar]
- Augustynowicz E, Gzyl A, Szenborn L, Banys D, Gniadek G, Slusarczyk J. Comparison of usefulness of randomly amplified polymorphic DNA and amplified-fragment length polymorphism techniques in epidemiological studies on nasopharyngeal carriage of non-typable Haemophilus influenzae. J Med Microbiol. 2003;2:1005–1014. doi: 10.1099/jmm.0.05341-0. [DOI] [PubMed] [Google Scholar]
- Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;2:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota K, Ye J, Nakai Y, Terada T, Shimizu K. ROKU: a novel method for identification of tissue-specific genes. BMC Bioinformatics. 2006;2:294. doi: 10.1186/1471-2105-7-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higasa K, Kukita Y, Baba S, Hayashi K. Software for machine-independent quantitative interpretation of SSCP in capillary array electrophoresis (QUISCA) Biotechniques. 2002;2:1342–1348. doi: 10.2144/02336bc05. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Soltys S, Shi G, Koong A, Le QT. Sample classification from protein mass spectrometry, by 'peak probability contrasts'. Bioinformatics. 2004;2:3034–3044. doi: 10.1093/bioinformatics/bth357. [DOI] [PubMed] [Google Scholar]
- Aittokallio T, Ojala P, Nevalainen TJ, Nevalainen O. Analysis of similarity of electrophoretic patterns in mRNA differential display. Electrophoresis. 2000;2:2947–2956. doi: 10.1002/1522-2683(20000801)21:14<2947::AID-ELPS2947>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Aittokallio T, Ojala P, Nevalainen TJ, Nevalainen O. Automated detection of differently expressed fragments in mRNA differential display. Electrophoresis. 2001;2:1935–1945. doi: 10.1002/1522-2683(200106)22:10<1935::AID-ELPS1935>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Aittokallio T, Pahikkala T, Ojala P, Nevalainen TJ, Nevalainen O. Electrophoretic signal comparison applied to mRNA differential display analysis. Biotechniques. 2003;2:116–122. doi: 10.2144/03341rr02. [DOI] [PubMed] [Google Scholar]
- Aittokallio T, Ojala P, Nevalainen TJ, Nevalainen OS. Automated pattern ranking in differential display data analysis. Methods Mol Biol. 2006;2:111–122. doi: 10.1385/1-59259-968-0:111. [DOI] [PubMed] [Google Scholar]
- Glasbey C, Vali L, Gustafsson J. A statistical model for unwarping of 1-D electrophoresis gels. Electrophoresis. 2005;2:4237–4242. doi: 10.1002/elps.200500365. [DOI] [PubMed] [Google Scholar]
- Drury HA, Green P, McCauley BK, Olson MV, Politte DG, Thomas LJ Jr. Spatial normalization of one-dimensional electrophoretic gel images. Genomics. 1990;2:119–126. doi: 10.1016/0888-7543(90)90233-K. [DOI] [PubMed] [Google Scholar]
- Bajla I, Hollander I, Fluch S, Burg K, Kollar M. An alternative method for electrophoretic gel image analysis in the GelMaster software. Comput Methods Programs Biomed. 2005;2:209–231. doi: 10.1016/j.cmpb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Hong Y, Chuah A. A format for databasing and comparison of AFLP fingerprint profiles. BMC Bioinformatics. 2003;2:7. doi: 10.1186/1471-2105-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Hettwer U, Koopmann B, Karlovsky P. Conversion of cDNA differential display results (DDRT-PCR) into quantitative transcription profiles. BMC Genomics. 2005;2:51. doi: 10.1186/1471-2164-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsis A, Andersson U, Bauren G, Ernfors P, Lonnerberg P, Montelius A, Oldin M, Pihlak A, Linnarsson S. Whole-genome expression profiling through fragment display and combinatorial gene identification. Nucleic Acids Res. 2004;2:e127. doi: 10.1093/nar/gnh126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;2(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;2:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statist Soc. 1995;2:289–300. [Google Scholar]
- Yasui Y, McLerran D, Adam BL, Winget M, Thornquist M, Feng Z. An automated peak identification/calibration procedure for high-dimensional protein measures from mass spectrometers. J Biomed Biotechnol. 2003;2:242–248. doi: 10.1155/S111072430320927X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushiki T, Fujisawa H, Eguchi S. Identification of biomarkers from mass spectrometry data using a "common" peak approach. BMC Bioinformatics. 2006;2:358. doi: 10.1186/1471-2105-7-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression data for 10,624 valid TDFs. Peak heights after GOGOTnormH for 10,624 valid TDFs are provided. It also contains two statistics measured by GOGOTstat and a t-like statistic and their ranks.
Expression patterns of top six TDFs ranked by a t-like statistic. Legends are the same as given in Fig. 5.


