Abstract
Phosphoinositide lipids function as both signaling molecules and as compartment-specific localization signals for phosphoinositide-binding proteins. In recent years, both phosphoinositides and phosphoinositide-binding proteins have been reported to display a restricted, rather than a uniform, distribution across intracellular membranes. Here, we examine recent data documenting the restricted distribution of both phosphoinositides and phosphoinositide-binding proteins and examine how phosphoinositide-binding proteins might engage multiple binding partners to achieve these restricted localizations, effectively acting as detectors of coincident localization signals.
Introduction
Phosphoinositides are a family of stereochemically distinct phosphorylated derivatives of the minor membrane lipid phosphatidylinositol (PtdIns; Figure 1). Phosphoinositides function both as signaling molecules and as localization cues, allowing for the recruitment of phosphoinositide-binding proteins to phosphoinositide-containing membranes [1]. Phosphoinositide-binding proteins commonly contain modular phosphoinositide-binding domains, such as pleckstrin homology (PH); phox-homology (PX); Fab1, YotB, Vac1, EEA1 (FYVE) and epsin amino-terminal homology (ENTH) domains [1–4] (Figure 2). This review will not attempt to catalogue the lipid-binding capabilities of these domains – for such information, readers are referred to excellent discussions of this subject elsewhere [1–4]. Here, we explore mechanisms that allow phosphoinositides and phosphoinositide-binding proteins to adopt a restricted distribution over cellular membranes.
Figure 1.
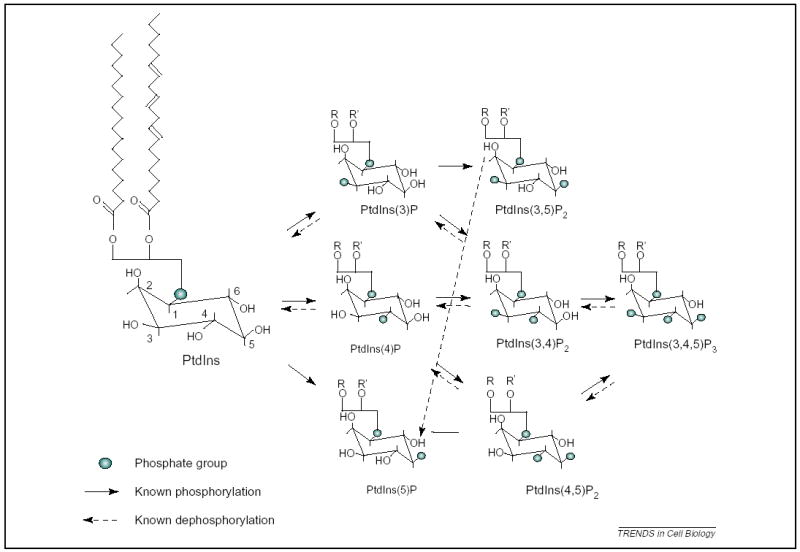
Phosphoinositide species. Phosphatidylinositol (PtdIns) consists of a D-myo-inositol 1-phosphate headgroup attached, almost exclusively, to 1-stearoyl, 2-arachanonyl, 3-phosphoglycerol as depicted. A variety of phosphoinositide kinases and phosphatases act to generate various phosphorylated derivatives, as indicated.
Figure 2.
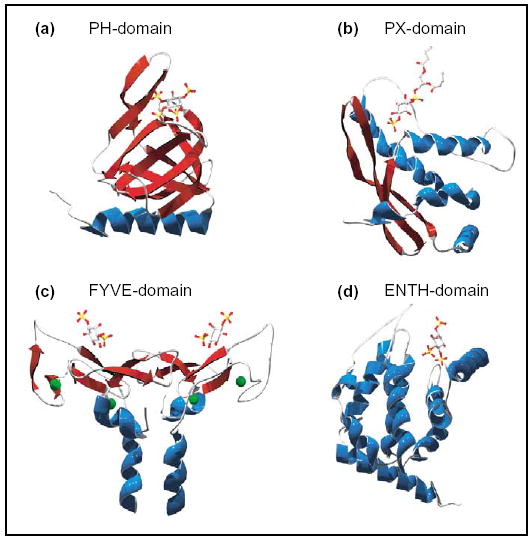
Phosphoinositide-binding domains. Structural examples of modular phosphoinositide-binding domains in complex with phosphoinositide-headgroup ligands and oriented such that the membrane interaction surface is top-most. Alpha-helices rendered in blue, beta-sheets in red. (a) Grp1 PH domain in complex with Ins(1,3,4,5)P4 [70]. (b) p40phox PX domain in complex with di-C4-PtdIns(3)P [71]. (c) Homodimeric EEA1 FYVE-domain (truncated to residues 1335–1411) in complex with Ins(1,3) P2. Green spheres represent Zn2+ ions [72]. (d) Epsin-1 ENTH-domain in complex with Ins(1,4,5)P3 [73]. Structures were manipulated in Deepview 3.7 and rendered using POV-ray 3.6.
Phosphoinositides as targeting modules
Phosphatidylinositol comprises ~8% of the total cellular phospholipid content [5]. Intracellular membranes are enriched in various phosphoinositides, present at levels of between 0.01–5% of that observed for PtdIns itself [6]. Phosphoinositides are not uniformly distributed among intracellular membranes but appear enriched upon different organelles (Figure 3); for example, PtdIns(4,5)P2 is enriched upon the inner leaflet of the plasma membrane and present upon membranes of the Golgi, whereas PtdIns(3)P appears restricted to membranes of the endosomal system, and PtdIns(4)P appears present upon the Golgi and plasma membrane [1,4,7]. Accounting for the proportional area of individual membranes, it has been estimated that the more abundant PtdIns(3)P, PtdIns(4)P and PtdIns(4,5)P2 lipids comprise ~4–5% of the total lipid content on the cytosolic face of their host organelles [4]. In this manner, phosphoinositides act as compartment-specific recognition signals for cytosolic binding partners and assist in the targeting of peripheral membrane proteins to these membranes.
Figure 3.
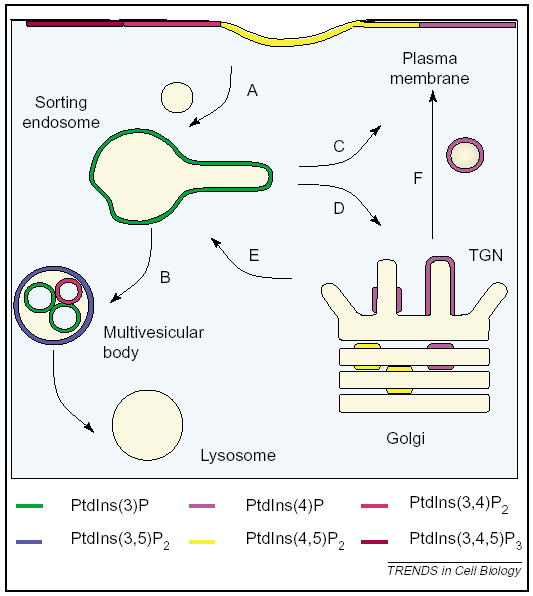
Cartoon depicting intracellular membranes in the endocytic and biosynthetic pathways and their hypothesized phosphoinositide content. These organelles control the trafficking of cargo to a variety of subcellular localizations. Organelle names given in type, trafficking pathways given by letters. A, internalization from the plasma membrane; B, degradative sorting to the lysosome; C, recycling from endosomes to the plasma membrane; D, retrieval of cargo from endosomes back to the trans-Golgi network (TGN); E, delivery of cargo from the TGN to endosomes; F, secretion of cargo from the TGN to the plasma membrane.
Avidity-mediated targeting
Phosphoinositide-binding domains commonly bind phosphoinositides with low affinities, favoring a highly plastic system whereby phosphoinositide-binding proteins can continually sample the membrane environment, searching out and becoming enriched at sites of phosphoinositide presence [1]. Weak binding is often enhanced through avidity effects (for example, as induced by oligomerization) and the ability to engage substrates in addition to the phosphoinositide lipid. A classic example of avidity-based targeting is observed for dynamin, a PH-domain-containing GTPase that regulates vesicle scission from membranes. In isolation, the dynamin PH domain interacts only weakly with membranes, but forced dimerization of dynamin PH domains greatly enhances the strength of membrane interaction [8]. In other words, the ability of a protein complex to engage multiply with the lipid surface enhances the avidity of that complex for such surfaces. Avidity-mediated enhancement in binding strength is due not only to the additive affinities of individual monomers but is assisted by a spatial constraint in that individual dissociated monomers remain in close proximity to, and thus have a higher probability of re-binding, their substrate. Avidity effects can be likened to the ability of Velcroe™ to create a high-strength bond despite the relatively weak strength of the interaction between individual hooks and loops, and such effects are thought to drive productive association of many peripheral membrane proteins with surfaces containing both lipid and non-lipid binding sites.
Plasma membrane microdomains of phosphoinositides
Early reports, employing a GFP-tagged PtdIns(4,5)P2-binding PLCδ1-PH domain, demonstrated an apparently uniform distribution of this lipid within the inner leaflet of the plasma membrane [9]. However, recent advances in fluorescent imaging, and the electron-microscopical (EM) examination of lipid-binding probes upon pre-fixed tissues [10], have led us to question just how ‘uniform’ the distribution of PtdIns(4,5)P2 actually is. Using high-resolution microscopy, Czech and colleagues demonstrated that the PLCdelta;1-PH domain decorated stable patches upon the plasma membrane of 3T3-L1 adipocytes [11]. Examination of concanavalin-A (a membrane-binding lectin) and BODIPY-HPC (a lipid dye) enrichment within these microdomains, demonstrated that, in contrast to previous reports [12], the enrichment of PLCdelta;1-PH at these sites was greater than that provided for by local membrane folding, suggesting that mechanisms existed to restrict the localization of PtdIns(4,5)P2 to microdomains at the plasma membrane. Similarly, using the PLCdelta;1-PH domain, or antisera specific for PtdIns(4,5)P2, PtdIns(4,5)P2 microdomains have been detected in the plasma membrane of PC12 cells [13], in COS cells and in neuronal growth cones [14]. Use of a GST-tagged PLCdelta;1-PH domain upon pre-fixed 1321N1 human astrocytoma cells demonstrated that, at EM resolution, this probe decorated lamellipodia-like profiles at the plasma membrane [15], consistent with the body of evidence implicating PtdIns(4,5)P2 in the control of actin dynamics [16–18]. Evidence therefore suggests the presence of plasma membrane microdomains of PtdIns(4,5)P2, but what function could these patches have?
Interestingly, clathrin and polymerized actin were found immediately beneath these patches of PtdIns(4,5) P2 [11], and it has recently been shown that these patches promote actin polymerization through the actin-regulatory protein neural Wiskott–Aldrich syndrome protein (N-WASP) [19]. N-WASP itself binds PtdIns(4,5)P2 in a highly cooperative mechanism by means of a lysine-rich domain [20], allowing it to respond in a switch-like manner to increases in PtdIns(4,5)P2 density. Normally, N-WASP inhibits Arp2/3-dependent actin polymerization, but this inhibition can be relieved through its binding to PtdIns(4,5)P2, suggesting that restricted localization of PtdIns(4,5)P2 to high-concentration patches might function to locally activate the Arp2/3 complex and regulate actin dynamics in the vicinity of these domains.
Ordered lipids and phosphoinositide microdomains
The partitioning of lipids into ‘ordered’ or ‘disordered’ phases, due to the abilities of their acyl chains to pack together [21,22], has long been considered a mechanism of compartmentalizing membranes. Regions of ordered lipids often contain cholesterol, sphingolipids and glyco-sylphosphatidylinositol-anchored proteins and have become known as ‘rafts’. PtdIns(4,5)P2-rich patches at the plasma membrane have been shown to contain cholesterol and ordered-lipids [20] and were found to colocalize with raft-enriched, actin-regulatory, GAP-43-like proteins such as GAP43, MARCKS (myristoylated alanine-rich C kinase substrate) and CAP23 [14]. Thus, the presence of ordered and disordered membrane microdomains might induce lateral segregation of phosphoinositide lipids within cellular membranes, providing a possible mechanism for microdomain biogenesis.
However, before leaving with the notion that the presence of PtdIns(4,5)P2 microdomains is universally accepted, it would be prudent to note some counterarguments. Just as phosphoinositides can recruit proteins to membranes, some proteins are able to laterally recruit phosphoinositides within the bilayer; for example, GAP-43-like proteins, and possibly the PLCdelta;1-PH domain itself, seem able to cluster PtdIns(4,5)P2 upon liposomes in vitro [23,24]. Indeed, triton X-100, a detergent commonly used in immunofluorescence protocols, can even induce PtdIns(4,5)P2 clustering in vivo [25]. So, while a body of evidence exists describing plasma membrane patches of PtdIns(4,5)P2, these data must also come with a caution; for, like Schrödinger’s cat, the very tools used to examine phosphoinositides might induce lipid microdomains themselves.
Restricted localization of phosphoinositides upon internal membranes
As well as the plasma membrane patches of PtdIns(4,5)P2 described above, evidence exists for restricted localization of phosphoinositides upon internal membranes. At EM resolution, a multimerized-FYVE domain from Hrs (2xFYVEhrs), detected PtdIns(3)P exclusively upon endosomal membranes and the inner vesicles of multivesicular bodies [26]. Further work, using Alexa488- and biotinylated-2xFYVEhrs probes at confocal resolution, demonstrated the existence of PtdIns(3)P microdomains upon the surface of artificially enlarged endosomes and large endosomes in HepG2 cells. These microdomains contained some PtdIns(3)P-binding proteins, including EEA1 (early endosomal antigen-1), but excluded others, such as Hrs [27]. Thus, the lipid species upon the cytosolic face of endosomes seem able to adopt a non-uniform distribution and phosphoinositide-binding proteins themselves can adopt a restricted distribution across these membranes.
Creating non-uniform phosphoinositide distributions
How might these microdomains form? One can imagine how barriers must exist to prevent lateral diffusion of these lipids. If phosphoinositide-binding proteins can cluster phosphoinositides within the bilayer [23,24], perhaps recruitment and oligomerization of phosphoinositide-binding proteins themselves will stabilize phosphoinositides within membranes. Alternatively, spatial regulation of phosphoinositide-kinase and -phosphatase activities might also contribute towards microdomain formation.
Activation of kinases
The PtdIns 3-kinase complex mVps34/p150 is an effector of Rab5 (see Glossary) [28], an endosomally localized GTPase. Rab5 activation directs cytosolic mVps34 to specific sites upon endosomes, leading to the localized production of PtdIns(3)P and the recruitment of PtdIns(3)P-binding proteins, such as EEA1 (see Box 1), in a GTP–Rab5-dependent manner [29,30]. GTP–Rab5 can also recruit a complex of Rabaptin5–Rabex5. Rabex5 (a Rab5 activator) can enhance GTP loading upon Rab5, and its recruitment is thought to stabilize GTP–Rab5 at specific sites upon endosomes [30,31]. Thus, stabilization of GTP–Rab5 upon endosomes, coupled to the recruitment of PtdIns 3-kinases to Rab5 domains might contribute to the formation of PtdIns(3)P-enriched microdomains upon endosomes.
Box 1. Proteins that can bind phosphoinositides
Cvt13p/Cvt20p: Two PX-domain-containing proteins that operate in the yeast cytosol-to-vacuole trafficking pathway. Cvt13p is also known as Snx4p.
Centaurins: PH-domain-containing Arf-GTPase-activating proteins that deactivate Arfs by promoting GTP hydrolysis.
Disabled-1, Disabled-2: Mammalian adaptor proteins containing a phosphoinositide- and cargo-binding phosphotyrosine-binding domain.
Dynamin: A PH-domain-containing GTPase intrinsically linked with the scission of vesicles from membranes.
EEA1: Early endosomal antigen-1. A FYVE-domain-containing protein that binds GTP–Rab5 and controls endosome fusion.
FAPP-1, FAPP-2: Four-phosphate adaptor protein-1, four-phosphate adaptor protein-2. PH-domain-containing adaptor proteins that regulate secretory cargo export from the TGN.
GAP-43 like proteins (GAP-43, CAP-23 and MARCKS): Raft-enriched protein kinase C substrates that bind and localize with PtdIns(4,5)P2.
Hrs: Hepatocyte-growth-factor-regulated tyrosine kinase substrate. An FYVE-domain-containing protein that is a key regulator of degradative sorting within the endosomal system.
Myotubularins: GRAM- and FYVE-domain-containing phosphoinositide 3-phosphatases.
N-WASP: Neuronal Wiskott–Aldrich syndrome protein. Interacts with the Arp2/3 complex to initiate actin polymerization and can bind PtdIns(4,5)P2 through an uncharacterized basic domain.
Oligophrenins: PH-domain-containing GTPase-activating proteins that act upon Rho-family GTPases.
OSBP: Oxysterol-binding protein. A Golgi-resident PH-domain-containing protein implicated in vesicle trafficking, signal transduction and lipid metabolism.
Phospholipase-C: A PH-domain-containing phospholipid lipase that hydrolyses PtdIns(4,5)P2 to Ins(1,4,5)P3 and diacylglycerol.
Phospholipase-D: A PX- and PH-domain-containing phospholipid lipase that hydrolyses phosphatidylcholine to phosphatidic acid.
Sorting nexins: A family of PX-domain-containing proteins thought to regulate endosomal trafficking decisions.
Positive-feedback control of phosphoinositide synthesis can also occur at the plasma membrane. Type-I PtdIns(4)P 5-kinase-α can be activated by both GTP–Arf6 and phosphatidic acid [16], and its lipid product, PtdIns(4,5)P2, can activate phospholipase D to produce phosphatidic acid [32]. Thus, it can be seen that PtdIns(4)P 5-kinase-α activation might be augmented through PtdIns(4,5)P2-mediated activation of phospholipase D [1], leading to a restricted burst of PtdIns(4,5)P2 production. Thus, feed-forward control of kinase activity might contribute towards non-uniform phosphoinositide localization across intracellular membranes.
Activation of phosphatases
Phosphoinositide phosphatases can also shape the phosphoinositide distribution across membranes. PTEN (phosphatase and tensin homolog deleted on chromosome 10), dephosphorylates PtdIns(3,4,5)P3 to generate PtdIns(4,5) P2 [33]. In Dictyostelium discoideum, PtdIns(3,4,5)P3 is generated at the leading edge of the cell in response to chemoattractants [34], where it defines the direction of migration. This definition is enhanced through recruitment of PTEN to the posterior edge of the cell [34], demonstrating that restricted localization of phosphatases can shape the phosphoinositide profile across a single membrane. Indeed, like kinases, phosphatases are subject to feed-forward activity control. PTEN can be allo-sterically activated through the binding of its lipid product, PtdIns(4,5)P2, enhancing its ability to dephosphorylate PtdIns(3,4,5)P3 [35]. Similarly, myotubularin-1, -3 and -6, which are PtdIns(3,5)P2 3-phosphatases, are allosterically activated by their lipid product, PtdIns(5)P [36], suggesting that membranes containing allosteric activators provide a suitable environment for phosphatase activity and that phosphoinositide gradients might be sharpened through feed-forward control of kinase and phosphatase activation.
Communication through phosphoinositide metabolism
The finding that some phosphoinositide-binding modules can bind to multiple phosphoinositides raises rather intriguing points: either a homo-oligomeric complex will localize to membranes enriched in all these lipids, or phosphoinositide metabolism might allow for communication between individual phosphoinositide-containing regions. For example, the PX-domain of SNX5 binds PtdIns(3)P and PtdIns(3,4)P2. In resting cells, SNX5 resides upon PtdIns(3)P-enriched endosomes but, after epidermal growth factor (EGF) stimulation, translocates to the PtdIns(3,4)P2-enriched plasma membrane [37], suggesting that its localization can be modulated by phosphoinositide metabolism. Indeed, PtdIns(3,4)P2 has also been detected upon the internal vesicles of multivesicular bodies (MVBs), raising the possibility that conversion of PtdIns(3)P to PtdIns(3,4)P2 might regulate some facet of MVB biogenesis (Figure 3) [38]. Additionally, PIKfyve, a PtdIns(3)P 5-kinase that phosphorylates PtdIns(3)P to form PtdIns(3,5)P2, localizes to endosomes through its PtdIns(3)P-binding FYVE domain [39]. It can be hypothesized that localized PIKfyve activity will create a microdomain of PtdIns(3,5)P2 upon endosomes, excluding PtdIns(3)P-binding proteins and recruiting those able to bind PtdIns(3,5)P2. The recent description of endosomally localized PtdIns(3,5)P2-binding proteins, such as Svp1p [40], Ent3p [41], Ent5p [42] and mVps24 [43], will hopefully allow visualization of the relationship between endosomal PtdIns(3)P and PtdIns(3,5)P2 and reveal whether biological processivity can occur between domains containing these lipids.
Coincidence detection to restrict protein distribution
It is becoming clear that phosphoinositide-binding proteins themselves can adopt a restricted distribution across cellular membranes. A genome-wide analysis of the 33 yeast PH domains revealed that, of those that could bind phosphoinositides, most did so weakly and promiscuously, yet still localized to membranes in a phosphoinositide-dependent manner. Indeed, PH domains with similar phosphoinositide-binding specificities could localize differentially in cells, suggesting that factors other than phosphoinositides can restrict the localization of PH-domain-containing proteins [44]. A similar picture is seen with the genome-wide screen of the 15 yeast PX domains. While all bar one bound PtdIns(3)P, most did so with such low affinity that membrane localization was thought to require the engagement of other factors to enhance the strength of membrane binding [45]. Indeed, restricted localization of the PX-domain-containing yeast proteins Cvt13p and Cvt20p to the pre-autophagosome required Apg14p expression, suggesting that Apg14p modulated the localization of these phosphoinositide-binding proteins [46]. Furthermore, while both EEA1 and Hrs contain PtdIns(3)P-binding FYVE domains [47], the endogenous proteins do not overlap perfectly upon endosomes [27,48]. Hrs localizes to a clathrin-containing bilayered coat upon endosomes in a manner requiring its FYVE domain and coiled-coil region [48], whereas EEA1 localizes to distinct regions in a manner requiring both its FYVE domain and its Rab5-binding domain [27,29]. So, engagement of ligands other than phosphoinositides might be a common mechanism employed by a variety of phosphoinositide-binding modules to enhance avidity for surfaces and to impose a restriction upon their localization.
Coincidence detection of small GTPases and phosphoinositides
The distribution of small monomeric GTPases among cellular membranes can, in many cases, act to restrict the localization of many phosphoinositide-binding proteins. For example, the FAPPs (four-phosphate-adaptor proteins) localize to the trans-Golgi network (TGN) in a manner requiring binding to both PtdIns(4)P and Arf1 [49]. Oxysterol-binding-protein (OSBP), another Golgi resident, has a PH domain that binds PtdIns(4)P, PtdIns(4,5)P2 and Arf1 [50], and the adaptor-protein (AP) complex AP-1 localizes to the TGN in a manner requiring both Arf1 and binding to PtdIns(4)P [51–53]. Thus, Arf1 and 4-phosphoinositides appear to provide a combinatorial targeting mechanism for Golgi localization (Figure 4a).
Figure 4.
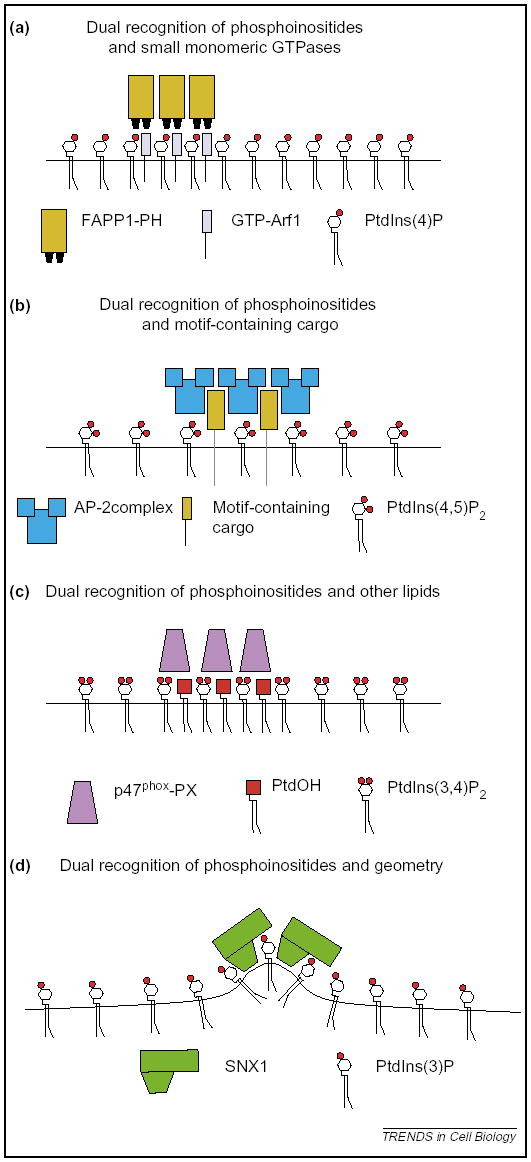
Coincidence detection to restrict localization. It is predicted that the following mechanisms will contribute towards the restricted localization of phosphoinositide-binding proteins across membrane. (a) Detection of phosphoinositides and small monomeric GTPases. Exemplified by the Arf1 and PtdIns(4)P-dependent localization of FAPP1 to the TGN. (b) Detection of phosphoinositides and cargo. Exemplified by the cargo and PtdIns(4,5)P2-dependent localization of the adaptor protein AP-2 complex to the plasma membrane. (c) Detection of phosphoinositides and other membrane lipids. Exemplified by the hypothetical PtdIns(3,4)P2- and PtdOH-dependent localization of p47phox. (d) Detection of phosphoinositides and geometric cues. Exemplified by the 3′-phosphoinositide and curvature-dependent localization of SNX1 to endosomes.
Osh1p and Osh2p, yeast orthologs of OSBP, contain highly similar PH domains that can bind PtdIns(4)P. While Osh2p-PH detects both plasma-membrane- and Golgi-localized PtdIns(4)P, Osh1p-PH detects solely Golgi-resident PtdIns(4)P [54]. Selective recognition of Golgi-localized PtdIns(4)P by Osh1p-PH stemmed from a putative membrane-receptor-interacting site within the Osh1p-PH domain, absent from the Osh2p-PH domain, suggesting that this site confers a Golgi-localization cue. Furthermore, Cla4p, a yeast PAK-related protein kinase, localized to sites of polarized growth at the plasma membrane, in a manner requiring its PH domain to bind to PtdIns(4)P and its p21-binding domain to bind to plasma-membrane-localized Cdc42, a Rho-family GTPase [55]. Thus, Cla4p appears to be a coincidence detector of plasma-membrane-localized PtdIns(4)P and Cdc42, whereas Osh1p and the FAPPs appear to be coincidence detectors selecting for Golgi membranes enriched in PtdIns(4)P and Arf1.
Coincidence detection through cargo–phosphoinositide interactions
Assembly of adaptor-protein complexes, often in conjunction with clathrin, and in a manner dependent upon phosphoinositides, allows for concentration of cargo into nascent carriers, a key step in every membrane-trafficking process [56]. Recently, it has been shown that proper assembly of the AP-2 complex requires coincident binding of both motif-containing cargo and PtdIns(4,5)P2 and that these interactions are mutually stabilizing [57]. These data demonstrate that cargo itself can initiate the assembly of the AP-2 complex and explain why this complex assembles exclusively at the plasma membrane – this is where the PtdIns(4,5)P2 is. Indeed, the crystal structures of the adaptor proteins disabled-1 and disabled-2 show that both phosphoinositide-binding and peptide-binding sites upon their PH-domain-like phosphotyrosine-binding domain were simultaneously occupied [58], suggesting that these, and possibly other adaptor-proteins or coat complexes, might localize to membranes through coincident detection of cargo and phosphoinositides (Figure 4b).
Coincidence detection through binding to multiple lipids
Lipids themselves might provide another mechanism to restrict protein distribution. p47phox, a component of the NADPH oxidase complex, has a PX domain that binds PtdIns(3,4)P2. However, a distinct site within its PX domain is able to bind to acidic lipids such as phosphatidic acid (PtdOH) [59]. Additionally, the EEA1-FYVE domain can bind both PtdIns(3)P and PtdSer [60], again through distinct sites, suggesting that these probes will be enriched upon membranes containing both lipids, and highlighting the fact that phosphoinositide-binding domains, once considered monogamous, can actually contain multiple lipid-binding sites that might refine their localization across membranes differentially enriched in these lipids (Figure 4c).
Coincidence detection through lipid and geometric cues
Coincidence detection need not arise solely through the recognition of dual protein–protein, or protein–lipid interactions: the recent identification of the BAR (Bin/ Amphiphysin/Rvs) domain as a membrane-binding domain able to sense membrane curvature [61] provides another example. BAR-domain-containing proteins often contain additional protein–lipid interaction domains, such as PH domains (in oligophrenins or centaurins) or PX domains (in sorting nexins). Endogenous sorting nexin-1 (SNX1) localizes to high-curvature membrane tubules emanating from endosomes. For this, SNX1 requires a functional PtdIns(3)P-binding PX domain and a functional curvature-sensing BAR domain [62]. Forced dimerization of the SNX1-PX domain did not significantly bring it to membranes [63], consistent with its low-affinity interaction with PtdIns(3)P in vitro [64], demonstrating that the SNX1-BAR domain functions as a membrane-binding domain within its own right and that membrane geometry can act as a localization cue (Figure 4d).
Thus, detection of signals other than phosphoinositides appears to be a mechanism employed by a variety of phosphoinositide-binding proteins to both restrict their distribution across membranes and enhance the strength of their interaction with membranes.
Regulating disassembly
If localization of phosphoinositide-binding proteins occurs through coincidence detection of phosphoinositide and non-phosphoinositide cues, then regulated abolition of a single cue might selectively delocalize these proteins. In many cases, localization requires coincident binding of both phosphoinositides and G-proteins. Thus, it can be imagined how the GTPase cycle could regulate the association and dissociation of phosphoinositide-binding proteins with membranes. Similarly, proteins recruited to dual-phosphoinositide and geometric cues could be selectively localized or delocalized through the alteration of membrane geometry, perhaps during vesicular budding or fusion, and proteins binding multiple lipids could be localized or delocalized through the metabolism of each lipid. Thus, employing coincidence detection might allow for more than just restricted localization of proteins: it might also allow for regulated disassembly of protein complexes at membranes.
Do we really know where phosphoinositides are?
The concept of coincidence detection raises a rather troubling question: if the localization of phosphoinositide-binding domains is determined by cues other than phosphoinositides, how accurately can they be used as reporters for these lipids [65]? For example, the Golgi localization of probes such as FAPP1 and OSBP is as dependent upon Arf1 as it is on PtdIns(4)P; hence PtdIns(4)P at other membranes will be invisible to these probes. While the Golgi/plasma membrane localization of Osh1p-PH and Osh2p-PH described depends upon PtdIns 4-kinases present upon these membranes [54], these PH domains actually bind a range of phosphoinositides in vitro [44] rather than being specific for PtdIns(4)P, suggesting that these probes might not report exclusively the presence of PtdIns(4)P. EM determination of PtdIns(4,5)P2 localization has demonstrated that significant pools of PtdIns(4,5)P2 exist upon Golgi and endosome membranes [15] that are not recognized by this probe in living cells [9]. In this light, data describing Double FYVE-containing protein-1, a PtdIns(3)P-binding protein that localizes to the Golgi [66], the presence of PtdIns(3)P at the plasma membrane [67] or the ER localization of PtdIns(3,4,5)P3 [68] lead us to question whether the tools we currently use to detect phosphoinositides might only ‘see’ a restricted population of these lipids and that phosphoinositides might be more widely distributed than previously thought.
Intermolecular coincidence recognition
Finally, it has emerged that ‘coincidence’ detection might occur at stages other than interactions with membranes. Screening for low-homology PH-domain fragments, Snyder and colleagues recently demonstrated the existence of intermolecular-PH domains: PH domains formed upon the interaction of two proteins containing half-PH domains. The interaction between PLCγ1 and the TrpC3 Ca2+ channel generates a functional PtdIns(4,5)P2-binding PH domain, retaining TrpC3 at the plasma membrane (Figure 5) [69]. These data raise the exciting possibility that many more phosphoinositide-binding domains than originally thought might exist, owing to the fact that they are formed only upon coincident interaction between two proteins, and are thus undetectable by conventional search algorithms. Future efforts ought to examine whether there exist split-ENTH, split-FYVE or split-PX domains as well as the split-PH domains described above. Thus, as well as enhancing the avidity of interaction, protein oligomerization might also enhance binding strength by creating entirely new lipid-binding domains.
Figure 5.
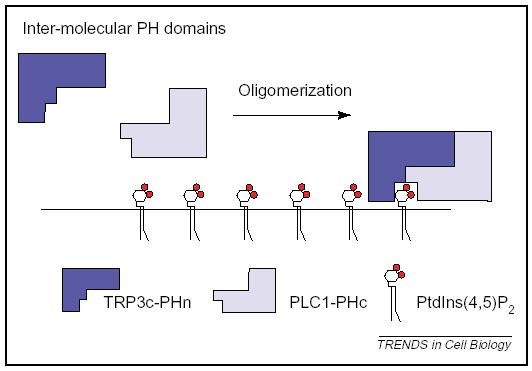
Intermolecular coincidence detection. Oligomerization of PLCγ1 and the TrpC3 Ca2+ channel creates a PtdIns(4,5)P2-binding PH domain from two half-PH domains.
Concluding remarks
Thus, while phosphoinositides commonly act as recruitment signals for peripheral membrane proteins, it is becoming clear that additional interactions, other than recognition of the phosphoinositide species, can restrict the localization of phosphoinositide-binding proteins upon cellular membranes. Future work should more fully characterize the micro-localization of both phosphoinositides and phosphoinositide-binding proteins and provide insight into the physiological role that this compartmentalization might bring.
Acknowledgments
We thank members of the Cullen laboratory for critical reading of the manuscript and for helpful discussion. J.C. is funded by a grant from Diabetes UK; work in the Cullen laboratory is supported by grants from the Biotechnology and Biological Sciences Research Council, the Medical Research Council and the Wellcome Trust.
Glossary
- Adaptor protein
Proteins or protein complexes that link cargo to vesicular carriers; for example the AP-1, -2, -3 and -4 adaptor protein complexes.
- Apg14p
A component of the yeast autophagic PI 3-kinase complex that localizes to the autophagosome.
- Arf1
ADP-ribosylation factor-1. A small monomeric GTPase that controls trafficking decisions at the Golgi and at endosomes.
- MVB
Multivesicular body. Endosomes containing internal vesicles upon which cargo that is sorted for degradation becomes incorporated.
- NADPH oxidase
A superoxide-generating complex found in the plasma membrane of phagocytes with a microbiocidal function.
- PI 3-kinase
Phosphoinositide 3-kinase. A phosphoinositide kinase that phosphorylates phosphoinositides at the 3-position.
- Rab5
A small GTPase of the Rab family that localizes to endosomes, recruits a number of effectors to endosomes and regulates a variety of endocytic sorting processes.
- Rabaptin5
A Rab5 effector that localizes to endosomes and controls Rab5-dependent endosomal fusion.
- Rabex5
A Rab5 GDP–GTP exchange factor (GEF) that activates Rab5 by promoting GTP-exchange upon it.
- Rho-family GTPases
A family of small GTPases including Rho, Rac and Cdc42 that regulate actin dynamics.
References
- 1.Cullen PJ, et al. Modular phosphoinositide-binding domains –their role in signalling and membrane trafficking. Curr Biol. 2001;11:R882–R893. doi: 10.1016/s0960-9822(01)00523-1. [DOI] [PubMed] [Google Scholar]
- 2.Lemmon MA. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 3.Balla T. Inositol-lipid binding motifs: signal integrators through protein–lipid and protein–protein interactions. J Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- 4.Roth MG. Phosphoinositides in constitutive membrane traffic. Physiol Rev. 2004;84:699–730. doi: 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi T, et al. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- 6.Stephens L, et al. Phosphoinositide 3-kinases: regulation by cell-surface receptors and function of 3-phosphorylated lipids. In: Cockcroft S, editor. Biology of Phosphoinositides. Oxford University Press; 2000. pp. 32–108. [Google Scholar]
- 7.De Matteis MA, Godi A. PI-loting membrane traffic. Nat Cell Biol. 2004;6:487–492. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- 8.Klein DE, et al. The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide-binding. J Biol Chem. 1998;273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- 9.Stauffer TP, et al. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- 10.Irvine R. Inositol lipids: to PHix or not to PHix? Curr Biol. 2004;14:R308–R310. doi: 10.1016/j.cub.2004.03.051. [DOI] [PubMed] [Google Scholar]
- 11.Huang S, et al. Phosphatidylinositol-4,5-bisphosphate-rich plasma membrane patches organize active zones of endocytosis and ruffling in cultured adipocytes. Mol Cell Biol. 2004;24:9102–9123. doi: 10.1128/MCB.24.20.9102-9123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Rheenen J, Jalink K. Agonist-induced PIP(2) hydrolysis inhibits cortical actin dynamics: regulation at a global but not at a micrometer scale. Mol Biol Cell. 2002;13:3257–3267. doi: 10.1091/mbc.E02-04-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoyagi K, et al. The activation of exocytotic sites by the formation of phosphatidylinositol-4,5-bisphosphate microdomains at syntaxin clusters. J Biol Chem. 2005;280:17346–17352. doi: 10.1074/jbc.M413307200. [DOI] [PubMed] [Google Scholar]
- 14.Laux T, et al. GAP43, MARCKS, and CAP23 modulate PI(4,5) P(2) at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J Cell Biol. 2000;149:1455–1472. doi: 10.1083/jcb.149.7.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watt SA, et al. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem J. 2002;363:657–666. doi: 10.1042/0264-6021:3630657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honda A, et al. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 17.Tall EG, et al. Dynamics of phosphatidylinositol 4,5-bisphosphate in actin-rich structures. Curr Biol. 2000;10:743–746. doi: 10.1016/s0960-9822(00)00541-8. [DOI] [PubMed] [Google Scholar]
- 18.Martin TF. PI(4,5)P(2) regulation of surface membrane traffic. Curr Opin Cell Biol. 2001;13:493–499. doi: 10.1016/s0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- 19.Golub T, Caroni P. PI(4,5)P2-dependent microdomain assemblies capture microtubules to promote and control leading edge motility. J Cell Biol. 2005;169:151–165. doi: 10.1083/jcb.200407058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papayannopoulos V, et al. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol Cell. 2005;17:181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 21.Maxfield FR. Plasma membrane microdomains. Curr Opin Cell Biol. 2002;14:483–487. doi: 10.1016/s0955-0674(02)00351-4. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee S, Maxfield FR. Membrane domains. Annu Rev Cell Dev Biol. 2004;20:839–866. doi: 10.1146/annurev.cellbio.20.010403.095451. [DOI] [PubMed] [Google Scholar]
- 23.Gambhir A, et al. Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys J. 2004;86:2188–2207. doi: 10.1016/S0006-3495(04)74278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caroni P. New EMBO members’ review: actin cytoskeleton regulation through modulation of PI(4,5)P(2) rafts. EMBO J. 2001;20:4332–4336. doi: 10.1093/emboj/20.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Rheenen J, et al. PIP(2) signaling in lipid domains: a critical re-evaluation. EMBO J. 2005;24:1664–1673. doi: 10.1038/sj.emboj.7600655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillooly DJ, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillooly DJ, et al. Phosphatidylinositol 3-phosphate is found in microdomains of early endosomes. Histochem Cell Biol. 2003;120:445–453. doi: 10.1007/s00418-003-0591-7. [DOI] [PubMed] [Google Scholar]
- 28.Christoforidis S, et al. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 29.Lawe DC, et al. The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J Biol Chem. 2000;275:3699–3705. doi: 10.1074/jbc.275.5.3699. [DOI] [PubMed] [Google Scholar]
- 30.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 31.Lippe R, et al. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol Biol Cell. 2001;12:2219–2228. doi: 10.1091/mbc.12.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Divecha N, et al. Interaction of the type Ialpha PIPkinase with phospholipase D: a role for the local generation of phosphatidylinositol 4,5-bisphosphate in the regulation of PLD2 activity. EMBO J. 2000;19:5440–5449. doi: 10.1093/emboj/19.20.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stokoe D. PTEN. Curr Biol. 2001;11:R502. doi: 10.1016/s0960-9822(01)00303-7. [DOI] [PubMed] [Google Scholar]
- 34.Van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 35.Campbell RB, et al. Allosteric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2003;278:33617–33620. doi: 10.1074/jbc.C300296200. [DOI] [PubMed] [Google Scholar]
- 36.Schaletzky J, et al. Phosphatidylinositol-5-phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr Biol. 2003;13:504–509. doi: 10.1016/s0960-9822(03)00132-5. [DOI] [PubMed] [Google Scholar]
- 37.Merino-Trigo A, et al. Sorting nexin 5 is localized to a subdomain of the early endosomes and is recruited to the plasma membrane following EGF stimulation. J Cell Sci. 2004;117:6413–6422. doi: 10.1242/jcs.01561. [DOI] [PubMed] [Google Scholar]
- 38.Watt SA, et al. Detection of novel intracellular agonist responsive pools of phosphatidylinositol 3,4-bisphosphate using the TAPP1 pleckstrin homology domain in immunoelectron microscopy. Biochem J. 2004;377:653–663. doi: 10.1042/BJ20031397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sbrissa D, et al. Phosphatidylinositol 3-phosphate-interacting domains in PIKfyve. Binding specificity and role in PIKfyve Endomembrane localization. J Biol Chem. 2002;277:6073–6079. doi: 10.1074/jbc.M110194200. [DOI] [PubMed] [Google Scholar]
- 40.Dove SK, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friant S, et al. Ent3p Is a PtdIns(3,5)P2 effector required for protein sorting to the multivesicular body. Dev Cell. 2003;5:499–511. doi: 10.1016/s1534-5807(03)00238-7. [DOI] [PubMed] [Google Scholar]
- 42.Eugster A, et al. Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Mol Biol Cell. 2004;15:3031–3041. doi: 10.1091/mbc.E03-11-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitley P, et al. Identification of mammalian Vps24p as an effector of phosphatidylinositol 3,5-bisphosphate-dependent endosome compartmentalization. J Biol Chem. 2003;278:38786–38795. doi: 10.1074/jbc.M306864200. [DOI] [PubMed] [Google Scholar]
- 44.Yu JW, et al. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 45.Yu JW, Lemmon MA. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J Biol Chem. 2001;276:44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]
- 46.Nice DC, et al. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Misra S, et al. Recognizing phosphatidylinositol 3-phosphate. Cell. 2001;107:559–562. doi: 10.1016/s0092-8674(01)00594-3. [DOI] [PubMed] [Google Scholar]
- 48.Raiborg C, et al. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci. 2001;114:2255–2263. doi: 10.1242/jcs.114.12.2255. [DOI] [PubMed] [Google Scholar]
- 49.Godi A, et al. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 50.Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- 51.Wang YJ, et al. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- 52.Stamnes MA, Rothman JE. The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell. 1993;73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- 53.Traub LM, et al. Biochemical dissection of AP-1 recruitment onto Golgi membranes. J Cell Biol. 1993;123:561–573. doi: 10.1083/jcb.123.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roy A, Levine TP. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J Biol Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- 55.Wild AC, et al. The p21-activated protein kinase-related kinase Cla4 is a coincidence detector of signaling by Cdc42 and phosphatidylinositol 4-phosphate. J Biol Chem. 2004;279:17101–17110. doi: 10.1074/jbc.M314035200. [DOI] [PubMed] [Google Scholar]
- 56.Robinson MS. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Honing S, et al. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol Cell. 2005;18:519–531. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 58.Yun M, et al. Crystal structures of the Dab homology domains of mouse disabled 1 and 2. J Biol Chem. 2003;278:36572–36581. doi: 10.1074/jbc.M304384200. [DOI] [PubMed] [Google Scholar]
- 59.Karathanassis D, et al. Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 2002;21:5057–5068. doi: 10.1093/emboj/cdf519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kutateladze TG, et al. Multivalent mechanism of membrane insertion by the FYVE domain. J Biol Chem. 2004;279:3050–3057. doi: 10.1074/jbc.M309007200. [DOI] [PubMed] [Google Scholar]
- 61.Peter BJ, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 62.Carlton J, et al. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 63.Zhong Q, et al. Determinants of the endosomal localization of sorting Nexin 1. Mol Biol Cell. 2005;16:2049–2057. doi: 10.1091/mbc.E04-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cozier GE, et al. The phox homology (PX) domain-dependent, 3-phosphoinositide-mediated association of sorting nexin-1 with an early sorting endosomal compartment is required for its ability to regulate epidermal growth factor receptor degradation. J Biol Chem. 2002;277:48730–48736. doi: 10.1074/jbc.M206986200. [DOI] [PubMed] [Google Scholar]
- 65.Balla T, et al. How accurately can we image inositol lipids in living cells? Trends Pharmacol Sci. 2000;21:238–241. doi: 10.1016/s0165-6147(00)01500-5. [DOI] [PubMed] [Google Scholar]
- 66.Ridley SH, et al. FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and Golgi compartments. J Cell Sci. 2001;114:3991–4000. doi: 10.1242/jcs.114.22.3991. [DOI] [PubMed] [Google Scholar]
- 67.Maffucci T, et al. Insulin induces phosphatidylinositol-3-phosphate formation through TC10 activation. EMBO J. 2003;22:4178–4189. doi: 10.1093/emboj/cdg402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sato M, et al. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nat Cell Biol. 2003;5:1016–1022. doi: 10.1038/ncb1054. [DOI] [PubMed] [Google Scholar]
- 69.van Rossum DB, et al. Phospholipase Cgamma1 controls surface expression of TRPC3 through an intermolecular PH domain. Nature. 2005;434:99–104. doi: 10.1038/nature03340. [DOI] [PubMed] [Google Scholar]
- 70.Lietzke SE, et al. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell. 2000;6:385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- 71.Bravo J, et al. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- 72.Dumas JJ, et al. Multivalent endosome targeting by homodimeric EEA1. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 73.Ford MG, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]


