Abstract
C/EBPδ (CCAAT/enhancer-binding protein δ) is a member of the C/EBP family of nuclear proteins that function in the control of cell growth, survival, differentiation and apoptosis. We previously demonstrated that C/EBPδ gene transcription is highly induced in G0 growth-arrested mammary epithelial cells but the C/EBPδ protein exhibits a t1/2 of only ∼120 min. The goal of the present study was to investigate the role of C/EBPδ modification by ubiquitin and C/EBPδ proteasome-mediated degradation. Structural and mutational analyses demonstrate that an intact leucine zipper is required for C/EBPδ ubiquitination; however, the leucine zipper does not provide lysine residues for ubiquitin conjugation. C/EBPδ ubiquitination is not required for proteasome-mediated C/EBPδ degradation and the presence of ubiquitin does not increase C/EBPδ degradation by the proteasome. Instead, the leucine zipper stabilizes the C/EBPδ protein by forming homodimers that are poor substrates for proteasome degradation. To investigate the cellular conditions associated with C/EBPδ ubiquitination we treated G0 growth-arrested mammary epithelial cells with DNA-damage- and oxidative-stress-inducing agents and found that C/EBPδ ubiquitination is induced in response to H2O2. However, C/EBPδ protein stability is not influenced by H2O2 treatment. In conclusion, our results demonstrate that proteasome-mediated protein degradation of C/EBPδ is ubiquitin-independent.
Keywords: CCAAT/enhancer-binding protein δ (C/EBPδ), H2O2, leucine zipper, oxidative stress, proteasome, ubiquitin
Abbreviations: BCA, bicinchoninic acid; C/EBP, CCAAT/enhancer-binding protein; CHOP10, C/EBP-homologous protein 10; DTT, dithiothreitol; HA, haemagglutinin; LZ domain, leucine zipper domain; Me-Ub, methylated ubiquitin; MMS, methyl methanesulfonate; NP40, Nonidet P40; ODC, ornithine decarboxylase; RD, regulatory domain; SUMO, small ubiquitin-related modifier; TAD, transactivation domain; TRAF-6, tumour-necrosis-factor-receptor-associated factor-6; UbR7, ubiquitin with all seven lysine residues mutated to arginine residues
INTRODUCTION
C/EBPs (CCAAT/enhancer-binding proteins) are a family of highly conserved bZIP (basic leucine zipper)-type transcription factors containing a TAD (transactivation domain), an RD (regulatory domain), a basic DBD (DNA-binding domain) and an LZ (leucine zipper) dimerization domain. Six C/EBP proteins have been identified including C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ, C/EBPϵ and C/EBPζ [CHOP10 (C/EBP-homologous protein 10)] [1,2]. The LZ domains of C/EBPs are highly conserved, containing four or five leucine residues in a heptad repeat permitting C/EBP proteins to form homo- or hetero-dimers [3]. C/EBP homo- and hetero-dimers bind to a relatively well-conserved consensus binding site (A[TTGCGCAA]T) in gene promoters [4]. In addition to dimerization between C/EBP family members, C/EBPs can also interact with other leucine zipper-containing proteins such as c-Fos, c-Jun and ATF (activating transcription factor) family members [5,6]. C/EBPs function in a wide range of cellular processes including differentiation, proliferation, inflammation, intermediary metabolism and apoptosis [1,2].
We previously reported that C/EBPδ gene expression is rapidly and persistently induced in human and mouse mammary epithelial cells in response to serum and growth factor withdrawal, contact inhibition and IL-6 (interleukin-6) family cytokine treatment and that C/EBPδ gene expression is reduced in primary human breast cancer [7–10]. We investigated the regulation of C/EBPδ and found, using nuclear run-on assays, that C/EBPδ gene transcription was induced ∼6-fold in G0 growth-arrested mammary epithelial cells compared with growing cells [11]. We further showed that the C/EBPδ mRNA and protein exhibited relatively short half-lives (∼45 and ∼120 min) in G0 growth-arrested human and mouse mammary epithelial cells [9,12]. The significant induction of C/EBPδ gene transcription coupled with the rapid turnover of C/EBPδ gene products during G0 growth arrest was somewhat unexpected as biochemical activity is dramatically reduced in G0 growth-arrested cells [13]. We further showed that antisense-mediated reduction in C/EBPδ levels delays entry into G0 growth arrest and that ectopic overexpression induces apoptosis [14]. Taken together, these results suggest that C/EBPδ plays an important role in mammary epithelial cell G0 growth arrest and that the cellular C/EBPδ content is tightly controlled.
Results from our laboratory have demonstrated that the C/EBPδ protein is degraded by the proteasome but the biochemical mechanisms, specifically the role of ubiquitination in C/EBPδ protein degradation, is not well understood [12]. Ubiquitin was the first member described in the ubiquitin family of modifying proteins that includes SUMO1 (small ubiquitin-related modifier 1)–SUMO4, NEDD8, Apg8 and Apg12 [15,16]. Target protein ubiquitination is catalysed by a three-step sequential reaction mechanism that involves ATP-dependent ubiquitin activation (E1), transfer of the activated ubiquitin to a conjugating enzyme (E2) and ubiquitin transfer to the target protein substrate by a ubiquitin ligase (E3) [17]. The ubiquitin protein contains seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63) and each can be a target for the subsequent ubiquitin moieties to form polyubiquitin chains [18]. The best-characterized role of ubiquitination is in targeting proteins for degradation by the proteasome [19]. The ubiquitin–proteasome proteolytic pathway is one of the two major cellular pathways that function in protein degradation [19]. Ubiquitin-mediated proteasome degradation is associated with the addition of a Lys48-linked polyubiquitin chain of at least four ubiquitins to the target protein [20,21].
Despite the well-documented association between ubiquitination and proteasome degradation, an increasing number of proteins exhibit ubiquitin-independent proteasome degradation [22,23]. For example, ODC (ornithine decarboxylase), a key protein that functions in cell proliferation, is degraded by a ubi-quitin-independent, proteasome-mediated mechanism [24,25]. Ubiquitin-independent, proteasome-mediated degradation of ODC depends on proteasome targeting by antizyme, a member of the antizyme gene family, which is highly conserved from yeast to humans [26,27]. The cyclin-dependent kinase inhibitor p21 is another key cell-cycle-regulatory protein that exhibits ubiquitin-independent, proteasome-mediated degradation [28,29]. The degradation of p21 differs from ODC in that the p21 N-terminus binds to the C8 α subunit of the proteasome to mediate p21 ubiquitin-independent, proteasome-mediated degradation [28,29]. Other proteins such as c-Jun and calmodulin also exhibit ubiquitin-independent, proteasome-mediated degradation [30,31].
In addition to proteasome-mediated degradation, ubiquitination also influences protein function [16,32]. For example, mono- and di-ubiquitination have been associated with membrane protein endocytosis and protein sorting [33,34]. In addition, Lys63-linked polyubiquitin has been linked to DNA repair, transcriptional control and protein kinase activation [35–37].
The overall goal of the present study was to investigate the structural requirements and functional consequences of C/EBPδ ubiquitination in the context of proteasome degradation and to explore the role of C/EBPδ ubiquitination in the cellular response to stress. Structural analyses demonstrated that C/EBPδ is modified by ubiquitin, and that an intact LZ domain is required for efficient ubiquitination. Stability experiments demonstrate that the LZ domain stabilizes the C/EBPδ protein by forming homodimers and that proteasome degradation of C/EBPδ is independent of ubiquitin. We further demonstrate that H2O2 treatment increases C/EBPδ ubiquitination in a dose-dependent manner, suggesting that ubiquitination may influence C/EBPδ transcriptional activation function or protein–protein interactions under oxidative stress conditions.
EXPERIMENTAL
Plasmid construction
The cDNA encoding C/EBPδ was cloned into PCDNA3.1/V5-His-TOPO TA expression vector (Invitrogen, Carlsbad, CA, U.S.A.). ΔLZ(1–233), ΔDBLZ(1–185), AD(1–102), ΔAD(102–268) and DBLZ(171–268) expression plasmids were constructed by PCR amplification of the corresponding cDNA sequences and cloned into the same vector. Deletion mutants and lysine to arginine mutants were generated by site-directed mutagenesis. Amino acids 103–185 were deleted in ΔRD, 186–213 were deleted in ΔNLS, and 224–249 were deleted in ΔL1–3. HA-Ub (ubiquitin) expression plasmid was a gift from Dr Xiong Yue (Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, U.S.A.). UbR7 (ubiquitin with all seven lysine residues mutated to arginine residues) coding sequence was amplified from CS2-UbR7, which was a gift from Dr Michele Pagano (Department of Pathology, New York University School of Medicine, New York, NY, U.S.A.), using a forward primer bearing a HA (haemagglutinin) tag coding sequence, and cloned into PCDNA3 vector, to obtain HA-UbR7 expression vector. All constructs were verified by DNA sequencing analysis.
In vitro ubiquitination/degradation assay
C/EBPδ fusion proteins were expressed in vitro using the TNT Quick Coupled in vitro transcription/translation system (Promega, Madison, WI, U.S.A.). In vitro ubiquitination assays were carried out at 37°C in 30 μg of HeLa cell S100 extract (Biomol, Plymouth Meeting, PA, U.S.A.) with addition of 2 μl of in vitro translation products, 5 mM ATP, 2.5 mM Mg2+, 1 mM DTT (dithiothreitol), 5 μg of ubiquitin (Biomol), 2.5 μM ubiquitin aldehyde (Biomol) and 100 μM MG-132 (the proteasome inhibitor carbobenzoxy-L-leucyl-L-leucyl-L-leucinal; Calbiochem, San Diego, CA, U.S.A.). Samples were taken at the indicated time points, and mixed with an equal volume of 2× Laemmli sample buffer [62.5 mM Tris/HCl, pH 6.8, 25% (v/v) glycerol, 2% (w/v) SDS, 0.01% Bromophenol Blue and 5% (v/v) 2-mercaptoethanol], and heated at 95°C for 5 min. Samples were then analysed by Western blot using mouse monoclonal anti-V5 antibody (Invitrogen). In vitro degradation assays were carried out in a similar way except that ubiquitin aldehyde and the proteasome inhibitor MG-132 were not added unless indicated. Samples of 5 μl were taken at the indicated time points, mixed with 5 μl of 2× Laemmli sample buffer and heated at 95°C for 5 min. Samples were then analysed by Western blot using mouse monoclonal anti-V5 antibody. Where indicated, 100 μM MG-132 was pre-incubated with the reaction mixture on ice for 1 h before addition of in vitro transcription/translation products. Me-Ub (methylated ubiquitin; 10 μg) was used to replace ubiquitin where indicated. Western blot results were quantified using the software ImageJ 1.34 [NIH (National Institutes of Health); Bethesda, MD, U.S.A.], and data were plotted using Microcal Origin 5.0.
Cell culture and transfection
HC11 cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% (v/v) foetal bovine serum, 10 ng/ml epidermal growth factor, 10 μg/ml insulin, 50 units/ml penicillin, 50 μg/ml streptomycin and 500 ng/ml fungazone at 37°C under 5% CO2 in a humidified incubator. Transient transfections were performed using Lipofectamine™ transfection reagent and Plus reagent (Invitrogen) according to the manufacturer's instructions.
In vivo ubiquitination assay
HC11 cells were co-transfected with 4 μg of HA-Ub expression plasmid and 4 μg of C/EBPδ fusion protein constructs. At 16–24 h after transfection, cells were treated with 10 μM MG-132 for 2 h before harvest. Cell pellets were boiled for 10 min in TSD buffer (50 mM Tris/HCl, pH 7.5, 1% SDS and 0.5 mM DTT), and centrifuged at room temperature for 10 min. Protein concentration was determined using a BCA (bicinchoninic acid) protein assay kit (Pierce, Rockford, IL, U.S.A.). Precleared cell lysates containing equal amount of proteins (500 μg) were subjected to denaturing immunoprecipitation using 1 μg of rabbit anti-V5 antibody (Chemicon, Temecula, CA, U.S.A.) in 1 ml of TNN buffer [50 mM Tris/HCl, pH 7.5, 250 mM NaCl, 5 mM EDTA, 0.5% NP40 (Nonidet P40) and protease inhibitor cocktail] by rotating at 4°C overnight. Immune complexes were captured by 50 μl of 50% Protein A–agarose slurry by rotating at 4°C for 2 h, washed four to five times using TNN buffer, and boiled for 5 min in 40 μl of 2× Laemmli sample buffer. Samples were then analysed by Western blot using anti-HA antibody (Cell Signaling Technology, Danvers, MA, U.S.A.).
In vivo degradation assay
HC11 cells grown in 10 cm dishes were transfected with C/EBPδ expression plasmids, or co-transfected with HA-UbR7 expression vector. At 16–24 h after transfection, cells were split into eight to twelve 6 cm dishes. After a further 16–24 h, cells were treated with 100 μg/ml emetine (Sigma, St. Louis, MO, U.S.A.) and harvested at intervals of 2 h after addition of emetine. Where indicated, cells were pretreated with 10 μM MG-132 for 2 h before addition of emetine. Cell pellets were lysed in RIPA buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1% NP40, 0.5% deoxycholate and 0.1% SDS) with addition of Complete™ protease inhibitor cocktail. Protein concentration was determined by BCA assay. Proteins were assessed by Western blot. Western blot results were quantified using the software ImageJ 1.34 (NIH), normalized to β-actin levels, and data were plotted using Microcal Origin 5.0.
RESULTS AND DISCUSSION
C/EBPδ is ubiquitinated in vitro and in vivo
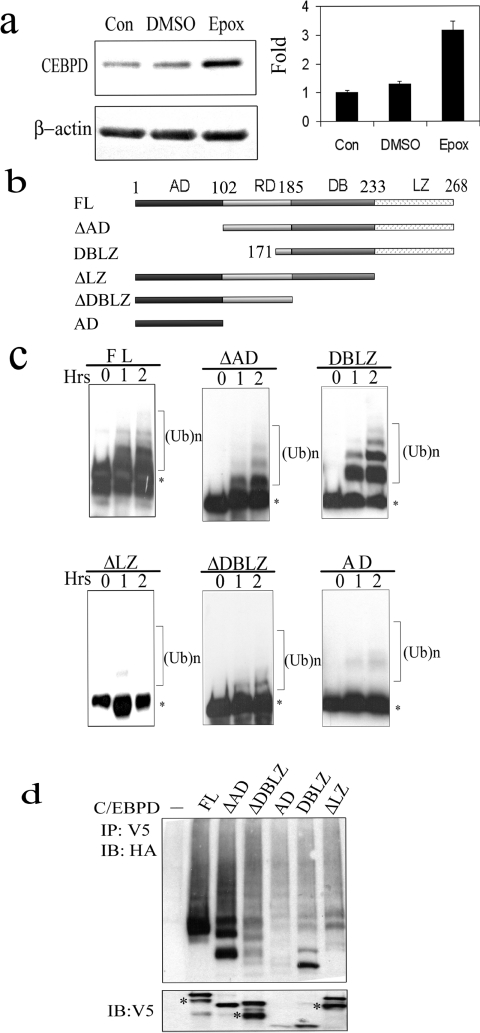
We previously demonstrated that the C/EBPδ protein half-life (t1/2) is relatively short (∼120 min) and that C/EBPδ protein degradation is blocked by the proteasome inhibitors MG132 and lactalysin [12]. In the present study, we confirm and extend these findings by demonstrating that C/EBPδ protein degradation is also inhibited by epoxomicin, a highly specific proteasome inhibitor (Figure 1a). Although these results demonstrate that C/EBPδ is degraded by the proteasome the role of ubiquitin conjugation in C/EBPδ protein degradation has not been reported. The goal of the present study was to investigate C/EBPδ ubiquitination and to determine the role of ubiquitination in C/EBPδ protein degradation and protein function. We first determined the role of C/EBPδ structural domains in C/EBPδ ubiquitination using a series of C/EBPδ full-length and domain-specific constructs (Figure 1b). In vitro transcribed and translated C/EBPδ constructs were incubated with HeLa S100 cell lysate (source of E1, E2 and E3 enzymes) and ubiquitination was assessed by Western blot. The results demonstrate that C/EBPδ constructs containing intact leucine zippers, i.e. full-length C/EBPδ (FL), ΔAD and DBLZ, all exhibit ubiquitination (Figure 1c). In contrast, C/EBPδ constructs lacking the LZ domain, i.e. ΔLZ, ΔDBLZ and AD, exhibited minimal ubiquitination.
Figure 1. C/EBPδ is degraded by the proteasome and ubiquitinated.
(a) Epoxomicin (Epox) inhibits C/EBPδ protein degradation. C/EBPδ protein levels were assessed in G0 growth-arrested HC11 cells at zero time [Control (Con), untreated] and after treatment with DMSO (vehicle) or 5 μM epoxomicin for 4 h. C/EBPδ protein levels were normalized to the β-actin control and plotted. (b) Schematic representation of C/EBPδ constructs. (c) In vitro ubiquitination of C/EBPδ. Asterisks denote the unmodified form of C/EBPδ proteins; the higher molecular mass species represent ubiquitinated C/EBPδ proteins. (d) In vivo ubiquitination of C/EBPδ. HC11 cells were co-transfected with an HA-Ub expression vector and specific C/EBPδ constructs. Co-transfection of HA-Ub with an empty vector control is designated as the ‘–’ lane. The bottom panel shows the protein levels of transfected C/EBPδ constructs. Asterisks denote the truncated proteins generated by using internal start codons.
To extend the C/EBPδ ubiquitination results from cell extracts to intact cells, HC11 cells were transfected with individual C/EBPδ expression plasmids plus an HA-tagged ubiquitin expression plasmid (HA-Ub). The results demonstrate that C/EBPδ constructs containing intact leucine zippers, particularly full-length C/EBPδ (FL) and ΔAD, exhibited significant levels of ubiquitination (Figure 1d). Modest levels of ubiquitination were also detected for the DBLZ construct. However, the ΔDBLZ and ΔLZ C/EBPδ constructs, which lack the LZ domain, exhibited less ubiquitination and the AD construct, which is highly unstable (see below), was minimally ubiquitinated. These results demonstrate that the C/EBPδ LZ domain plays a key role in the ubiquitination of C/EBPδ.
The LZ domain is required for C/EBPδ ubiquitination
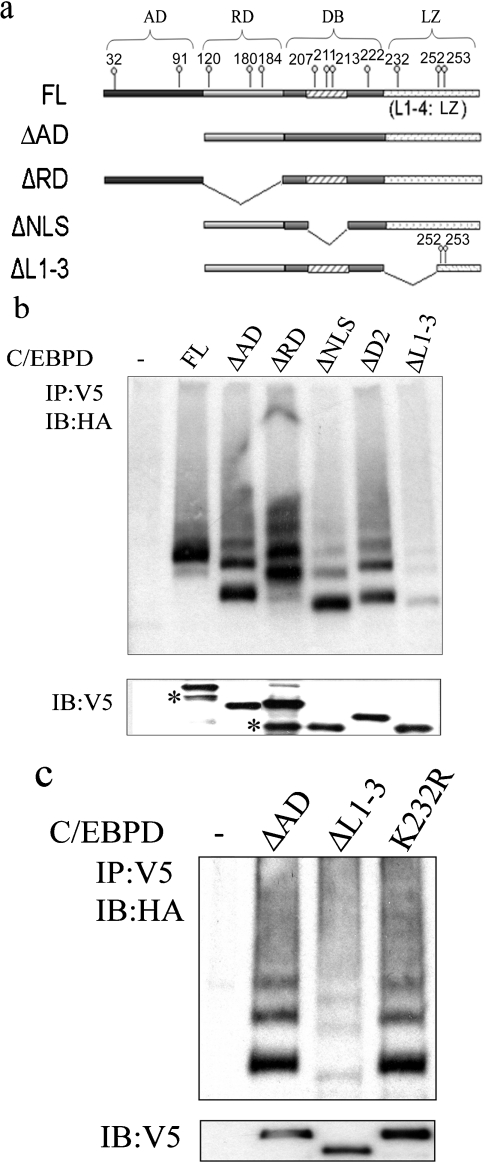
Proteins that are ubiquitinated are characterized by the presence of an E3 ligase recognition site and target lysine residue(s), or a free N-terminal NH2 group, for ubiquitin conjugation [38,39]. To determine the approximate location of these elements in the C/EBPδ protein we developed C/EBPδ deletion constructs (Figure 2a) and these constructs were transfected into HC11 cells and ubiquitination was assessed. The results demonstrate that all constructs retaining an intact LZ domain exhibit evidence of single or multiple ubiquitin additions (Figure 2b). The exception is the ΔL1–3 construct, which lacks most of the LZ but contains Lys252 and Lys253. The ΔL1–3 construct exhibits minimal ubiquitination, suggesting that a full, intact LZ domain is required for optimal ubiquitination. Alternatively, the absence of Lys232 in the ΔL1–3 construct suggested the possibility that reduced ubiquitination of the ΔL1–3 construct may be due to loss of Lys232 ubiquitination target lysine residue. To test the functional role of Lys232 as a ubiquitin target and to further test the hypothesis that an intact LZ domain is required for efficient C/EBPδ ubiquitination we compared the ubiquitination profile of ΔAD [intact leucine zipper (Figure 2a)], ΔL1–3 [major portion of LZ deleted (Figure 2a)] and K232R [intact leucine zipper, Lys232 converted into arginine residue (R)]. The results demonstrated that ubiquitination of the K232R mutant was similar to ΔAD; however, ubiquitination of the ΔL1–3 mutant was significantly reduced (Figure 2c). These results suggest that the leucine zipper region deleted in the ΔL1–3 construct plays a key role in the C/EBPδ–ubiquitin machinery interaction, but does not appear to contain a target lysine residue.
Figure 2. The L1–3 region of the C/EBPδ leucine zipper is required for ubiquitination.
(a) Schematic representation of C/EBPδ constructs. (b) In vivo ubiquitination of C/EBPδ mutants containing lysine deletions. The ‘–’ lane designates co-transfection of HA-Ub plus empty vector control. The bottom panel shows the protein levels of transfected C/EBPδ constructs. Asterisks denote the truncated proteins generated by using internal start codons. (c) Lys232 is not the target lysine residue for C/EBPδ ubiquitination. Ubiquitination of ΔAD, ΔL1–3 and ΔL1–3K232R is presented (upper panel). The ‘–’ lane designates co-transfection of HA-Ub plus empty vector control. The bottom panel shows the protein levels of transfected C/EBPδ constructs.
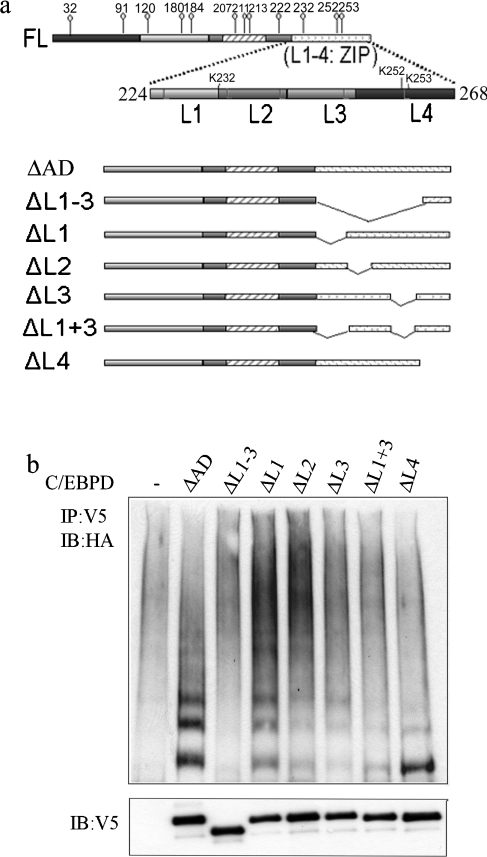
To further characterize the functional role of specific regions located within the C/EBPδ LZ domain, we assessed ubiquitination of C/EBPδ LZ domain deletion constructs in HC11 cells. The C/EBPδ LZ domain contains five evenly spaced leucine-containing amino acid sequences (regions L1–L4). The C/EBPδ LZ domain deletion constructs analysed included: ΔL1–3 (L1–3 is deleted, L4 is retained); ΔL1 (L1 is deleted, L2–4 retained); ΔL2 (L2 is deleted, L1, L3 and L4 are retained); ΔL3 (L3 is deleted, L1, L2 and L4 are retained); ΔL1+3 (L1, L3 are deleted, L2, L4 are retained); ΔL4 (L4 is deleted, L1–3 are retained) (Figure 3a). The ΔAD C/EBPδ construct containing an intact LZ domain (L1–L4) was used as a positive control. The ΔL1 construct exhibited polyubiquitination which paralleled the ΔAD positive control (Figure 3b). In contrast, ΔL1–3 did not exhibit evidence of ubiquitination (Figure 3b). The ΔL2, ΔL3 and ΔL1+3 constructs exhibited minimal ubiquitination. The ΔL4 construct exhibited a relatively strong band consistent with mono-ubiquitination. The capacity of individual C/EBPδ LZ domain deletion constructs to form functional zippers and homo- or hetero-dimers was assessed using Prosite analysis (http://www.expasy.org). The predictions indicated that C/EBPδ LZ domain deletion constructs with deletions of L1 or L4 would retain a functional leucine zipper; however, deletion of L2, L3, L1+3 and L1–3 would disrupt the leucine zipper. These predictions are consistent with the experimental results and indicate that significant disruptions in the LZ domain reduce C/EBPδ ubiquitination. This suggests that the intact LZ domain is required for efficient interaction between the substrate, C/EBPδ, and ubiquitinating enzymes. Our findings are consistent with previous investigations of the role of coiled-coiled domains in protein modification by the ubiquitin family proteins. For example, Yang et al. [40] reported that the coiled-coil domain of TRAF-6 (tumour-necrosis-factor-receptor-associated factor-6) mediates its interaction with Ubc13/Uev1A, a ubiquitin conjugating enzyme E2, and is required for TRAF-6 auto-ubiquitination. Kim et al. [41] also reported that the coiled-coil domain of PML (promyelocytic leukaemia)-RARα (retinoic acid receptor α) mediates its interaction with Ubc9, the SUMO conjugating enzyme E2, and is required for its modification by SUMO, the small ubiquitin-like modifier. The leucine zipper structure is a special coiled-coil protein domain. It is possible that the LZ domain functions in the recruitment of C/EBPδ to the ubiquitinating machinery. The proteins that facilitate the interaction between C/EBPδ and the ubiquitin machinery interactions and the enzymes that catalyse C/EBPδ ubiquitination are under investigation.
Figure 3. The intact LZ domain is required for C/EBPδ ubiquitination.
(a) Schematic representation of C/EBPδ partial leucine zipper deletion mutants. (b) In vivo ubiquitination of C/EBPδ partial leucine zipper deletion mutants. The bottom panel shows the protein levels of transfected C/EBPδ constructs.
These results differ somewhat from those reported by Hattori et al. [42], in which the presence of an intact LZ domain suppressed ubiquitination of C/EBPγ (Ig/EBP) and C/EBPζ (CHOP10). Although it is unclear how the LZ domain might exhibit opposite functions within the C/EBP family members, the differences could be due to differences in cell-type-specific regulation of protein steady-state levels or due to structural differences between C/EBPγ and C/EBPζ and C/EBPδ. Both C/EBPγ and C/EBPζ lack transcriptional activation domains and typically function in transcriptional repression [42]. In contrast, C/EBPδ contains a TAD and C/EBPδ primarily functions as a transcriptional activator [43]. In addition, published reports indicate that C/EBP family members are degraded by divergent cellular mechanisms. For example, C/EBPβ is degraded by a calpain-dependent, proteasomeindependent manner [44]. The use of alternative mechanisms to degrade C/EBPs indicates that cells use multiple strategies to degrade C/EBPs. This may be due to differences in cell-specific transcriptional functions or different biological roles of individual C/EBP family members.
The presence of an intact LZ domain stabilizes the C/EBPδ protein
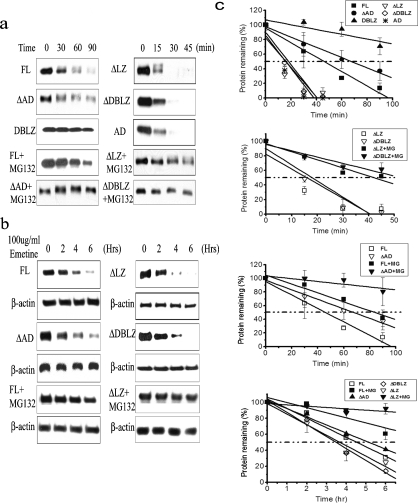
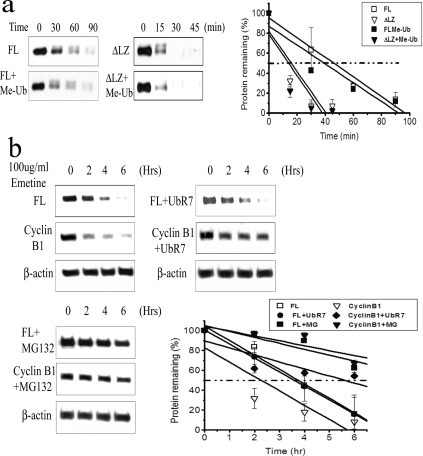
Ubiquitination is associated with protein degradation as well as alterations in protein function [16,32]. To determine the influence of the LZ domain on C/EBPδ protein degradation we compared the stability of C/EBPδ constructs containing intact LZ domains (FL, ΔAD and DBLZ) to C/EBPδ constructs lacking an intact LZ domain (ΔLZ, ΔDBLZ and AD) in vitro. The C/EBPδ LZ-containing constructs (FL, ΔAD and DBLZ) exhibited t1/2 values of ∼45 to >90 min (the experimental end point) (Figure 4a). In contrast, the C/EBPδ constructs lacking the LZ domain (ΔLZ, ΔDBLZ and AD) exhibited t1/2 values of less than 15 min (Figure 4a). The t1/2 values of all C/EBPδ constructs were significantly increased in the presence of MG-132, demonstrating that C/EBPδ protein constructs are degraded by a proteasome-dependent mechanism (Figure 4a).
Figure 4. The C/EBPδ protein is stabilized by the LZ domain.
(a) In vitro degradation of C/EBPδ. Two sets of time points were used for the two sets of C/EBPδ constructs with or without the LZ domain (0–90 and 0–45 min). (b) In vivo degradation of C/EBPδ. C/EBPδ protein contents were normalized to β-actin. (c) Western blot results from three independent experiments were quantified and plotted.
Having demonstrated that an intact LZ domain stabilizes C/EBPδ constructs incubated in HeLa cell lysates we next investigated the influence of the LZ domain on the stability of C/EBPδ constructs expressed in HC11 cells. The C/EBPδ FL- and ΔAD LZ-containing constructs exhibited t1/2 values of ∼320 min in HC11 cells (Figure 4b). In contrast, the C/EBPδ constructs lacking the LZ domain (ΔLZ and ΔDBLZ) exhibited in vivo t1/2s of ∼210 min (Figure 4b). The stability of the FL and ΔLZ constructs was significantly increased (>360 min, the experimental end point) by treatment of the HC11 cells with the proteasome inhibitor MG-132 (Figure 4b). These results are consistent with those from the HeLa cell extract degradation experiments; both results demonstrate that C/EBPδ protein constructs with intact LZ domains exhibit increased stability compared with C/EBPδ protein constructs lacking the LZ domain. In addition, these results confirm and extend the functional role of the proteasome in C/EBPδ protein degradation.
C/EBPδ degradation by the proteasome is ubiquitin-independent
Ubiquitination has been extensively characterized as a mechanism to facilitate protein degradation by the proteasome [17]. In the present study we found that the LZ domain is required for C/EBPδ ubiquitination and, paradoxically, that the LZ domain stabilizes the C/EBPδ protein. These observations suggested that C/EBPδ ubiquitination is not directly linked to proteasome degradation and that proteasome-mediated degradation of C/EBPδ does not require ubiquitination. Ubiquitin-independent proteasome degradation has been previously reported for several key cell growth regulatory proteins such as ODC, p21, c-Jun and calmodulin [24,28,29]. To investigate the role of ubiquitination in C/EBPδ proteasome-mediated degradation, we first assessed the stability of C/EBPδ full-length (FL) and C/EBPδ ΔLZ (lacking the LZ domain) constructs in the presence or absence of Me-Ub using the HeLa S100 in vitro degradation system. Me-Ub blocks polyubiquitination of protein substrates, and as such, blocks polyubiquitin-mediated proteasome degradation [45]. Consistent with previous results (Figure 4a), in the presence of the polyubiquitination inhibitor Me-Ub the t1/2 of the C/EBPδ FL construct is unchanged, remaining at ∼43 min (Figure 5a). Similarly, the t1/2 of the C/EBPδ ΔLZ construct, a construct lacking the leucine zipper stabilizing domain, is ∼15 min in the presence or absence of Me-Ub (Figure 5a). These results are consistent with the hypothesis that polyubiquitination is not required for C/EBPδ protein degradation.
Figure 5. Proteasome mediated C/EBPδ degradation is unaffected by inhibition of polyubiquitination.
(a) Me-Ub does not inhibit C/EBPδ degradation in vitro. An excess amount of Me-Ub was used to replace ubiquitin. Quantified Western blot results are shown in the right panel. (b) Dominant-negative ubiquitin (HA-UbR7) does not inhibit C/EBPδ degradation in vivo. Degradation of cyclin B1 is shown as a positive control. Protein contents were normalized to β-actin. Quantified Western blot results are presented in the right panel.
To further investigate the role of polyubiquitination in C/EBPδ proteasomal degradation we transfected HC11 cells with HA-UbR7, a dominant-negative ubiquitin construct in which all seven lysine residues in the ubiquitin molecule have been mutated to arginine residues [46]. Alteration of all ubiquitin lysine residues effectively blocks the formation of polyubiquitin chains on target protein substrates and inhibits polyubiquitin-dependent proteasome degradation [46]. The results demonstrated that the stability of C/EBPδ was nearly identical in the presence or absence of the polyubiquitination inhibitor HA-UbR7 (Figure 5b). In contrast with C/EBPδ, cyclin B, a well-characterized cell-cycle-regulatory protein that is polyubiquitinated by the APC (anaphase-promoting complex)/cyclosome E3 ligase system and degraded by the proteasome [47], was stabilized by the HA-UbR7 treatment (Figure 5b). To confirm that C/EBPδ and cyclin B1 are degraded by the proteasome we treated HC11 cells with the proteasome inhibitor MG-132 and demonstrated that the degradation of both C/EBPδ and cyclin B1 is blocked (Figure 5c). These results are consistent with a model in which proteasome degradation of C/EBPδ is polyubiquitin-independent.
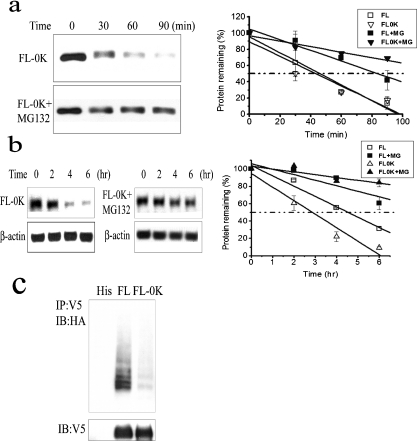
To directly address the role of ubiquitination and C/EBPδ proteasomal degradation, we produced a C/EBPδ construct in which all 12 lysine residues were mutated to arginine residues (FL-0K). To facilitate Western blot detection, C/EBPδ FL-0K was cloned as a fusion construct with a V5 tag in which the single lysine residue present in the V5 tag was also mutated to an arginine residue. We compared the relative stability of C/EBPδ FL-0K construct with C/EBPδ FL (wild-type, 12 lysine residues intact) using both the HeLa S100 extract system and in HC11 cells. The t1/2 values of C/EBPδ FL and C/EBPδ FL-0K were nearly identical (∼43 min) in the HeLa cell extract system (Figure 6a); however, the t1/2 values in HC11 cells were ∼280 min for C/EBPδ FL and ∼180 min for C/EBPδ FL-0K (Figure 6b). These results suggest that the C/EBPδ FL-0K is more unstable than C/EBPδ FL in intact cells, possibly due to the creation of unstructured regions within the mutant C/EBPδ FL-0K protein by the replacement of wild-type lysine residues with arginine residues [48]. The degradation of both C/EBPδ FL and C/EBPδ FL-0K is proteasome-dependent; however, as both exhibit increased stability in MG-132-treated HeLa cell extracts and in HC11 cells (Figures 6a and 6b).
Figure 6. Lysine-less C/EBPδ (FL-0K) undergoes proteasome-mediated degradation.
(a) In vitro degradation of FL-0K with or without MG-132. Quantified Western blot results are shown in the right panel. (b) In vivo degradation of FL-0K in HC11 cells with or without MG-132. Protein contents were normalized to β-actin. Quantified Western blot results are shown in the right panel. (c) Ubiquitination of FL-0K. The bottom panel shows the protein levels of transfected C/EBPδ constructs.
Despite alterations of all available lysine residues in the C/EBPδ protein it is conceivable that C/EBPδ could undergo N-terminal ubiquitination, as has been demonstrated for p21 and other proteins [49,50]. We assessed ubiquitination of C/EBPδ FL and C/EBPδ FL-0K constructs and found that the C/EBPδ FL construct undergoes polyubiquitination, but the C/EBPδ FL-0K is minimally ubiquitinated (Figure 6c). These results demonstrate that C/EBPδ degradation is proteasome-dependent but does not require ubiquitination.
Taken together these results indicate that C/EBPδ protein degradation fits the recently described degradation ‘by default’ model [48]. This model describes a subset of inherently unstable proteins that are targeted for ‘automatic’ degradation by the proteasome [48]. These unique proteins exhibit the following properties: they are inherently unstable, rarely found in ‘free’ (monomeric) form in cells and exhibit ubiquitin-dependent and ubiquitin-independent proteasome degradation [48]. Importantly, inherently unstable proteins that undergo ‘by default’ proteasome degradation are also characterized by protection from degradation by assembly into protein complexes, such as LZ domain-mediated homodimers. It has been hypothesized that the increased stability of proteins in protein complexes provides a mechanism to increase or maintain protein function. In contrast, the increased susceptibility of ‘free’, monomeric proteins to proteasome degradation provides a mechanism to rid the cell of ‘excess’ proteins that are not incorporated into functional complexes and may interfere, or compete, with the protein complex function [48]. To gain further insights into the mechanisms controlling proteasome-mediated degradation of C/EBPδ we are investigating potential interactions between C/EBPδ and proteasome subunits. The initial experiments suggest that C/EBPδ does not directly interact with Rpn10, subunit α5 and Rpt4; however, additional interactions between C/EBPδ and candidate proteasome subunits are under investigation.
H2O2 induces C/EBPδ ubiquitination but not degradation
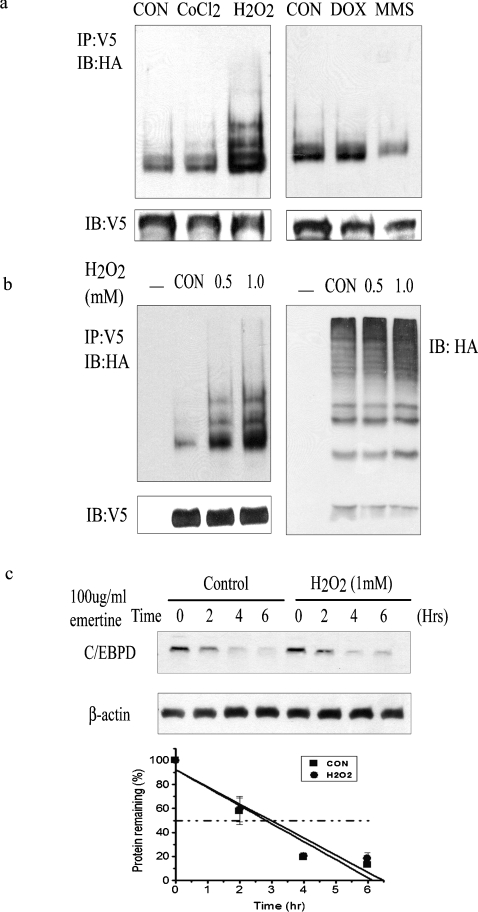
To explore possible functions of C/EBPδ ubiquitination other than protein degradation, we assessed ubiquitination of C/EBPδ in HC11 cells under various stress conditions. We treated HC11 cells with DNA-damaging agents [doxorubicin and MMS (methyl methanesulfonate)], hypoxia (CoCl2) and oxidative stress (H2O2). Only H2O2 treatment induced ubiquitination of C/EBPδ in a dose-dependent manner (Figures 7a and 7b, left). The dose–response data suggest that there is a linear relationship between the level of exposure to H2O2 and C/EBPδ ubiquitination (Figure 7b). H2O2 treatment did not increase total cellular content of ubiquitinated proteins (Figure 7b, right), suggesting that H2O2 induces the ubiquitination of a relatively specific subset of proteins that includes C/EBPδ. Consistent with ubiquitin-independent protein degradation, C/EBPδ protein half-life is not influenced by H2O2 treatment (Figure 7c). These results establish a physiological basis for further investigation into the influence of ubiquitination on C/EBPδ function, cellular quiescence and oxidative stress. Ongoing mechanistic studies are focused on the dose–response relationship between H2O2, C/EBPδ ubiquitination, cellular quiescence and survival.
Figure 7. H2O2 induces C/EBPδ ubiquitination.
(a) Ubiquitination of C/EBPδ in HC11 cells in response to various stress conditions. HC11 cells were co-transfected with HA-Ub and C/EBPδ expression vectors, and then treated with 100 μM CoCl2 (6 h), 1 mM H2O2 (2 h), 2 μM doxorubicin (6 h) or 5 mM MMS (2 h). Control (CON) cells were treated with water. The bottom panel shows the protein levels of transfected C/EBPδ constructs. (b) Ubiquitination of C/EBPδ in response to different H2O2 doses. The left panel shows ubiquitination of C/EBPδ. The bottom panel shows the protein levels of transfected C/EBPδ constructs. The right panel shows total ubiquitinated cellular proteins. (c) H2O2 treatment does not influence C/EBPδ protein stability. HC11 cells were pretreated with water (control) or 1 mM H2O2 for 2 h before addition of emetine, and then harvested at intervals of 2 h after addition of emetine. Protein contents were normalized to β-actin and plotted.
In conclusion, we demonstrated that C/EBPδ is ubiquitinated via a mechanism that requires an intact LZ domain. Despite an essential role in ubiquitination, the LZ domain stabilizes the C/EBPδ protein by facilitating dimerization. Degradation of C/EBPδ is mediated by the proteasome but is independent of ubiquitin. Exposure of quiescent cells to H2O2-induced oxidative stress increases ubiquitination of C/EBPδ. The present study indicates that the function of C/EBPδ in mammary epithelial cells includes a role in the unique protective mechanisms activated to maintain cell viability during cellular quiescence.
Acknowledgments
We thank Dr Xiong Yue and Dr Michele Pagano for the HA-Ub and CS2-UbR7 expression plasmids respectively. This work was supported by NIH grants CA57607 and P30 CA16058.
References
- 1.Johnson P. F. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J. Cell Sci. 2005;118:2545–2555. doi: 10.1242/jcs.02459. [DOI] [PubMed] [Google Scholar]
- 2.Ramji D. P., Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurst H. C. Transcription factors 1: bZIP proteins. Protein Profile. 1995;2:101–168. [PubMed] [Google Scholar]
- 4.Osada S., Yamamoto H., Nishihara T., Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J. Biol. Chem. 1996;271:3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- 5.Hsu W., Kerppola T. K., Chen P. L., Curran T., Chen-Kiang S. Fos and Jun repress transcription activation by NF-IL6 through association at the basic zipper region. Mol. Cell. Biol. 1994;14:268–276. doi: 10.1128/mcb.14.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vallejo M., Ron D., Miller C. P., Habener J. F. C/ATF, a member of the activating transcription factor family of DNA-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to cAMP response elements. Proc. Natl. Acad. Sci. U.S.A. 1993;90:4679–4683. doi: 10.1073/pnas.90.10.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Rourke J., Yuan R., DeWille J. CCAAT/enhancer-binding protein-delta (C/EBP-delta) is induced in growth-arrested mouse mammary epithelial cells. J. Biol. Chem. 1997;272:6291–6296. doi: 10.1074/jbc.272.10.6291. [DOI] [PubMed] [Google Scholar]
- 8.Sivko G. S., DeWille J. W. CCAAT/enhancer binding protein δ (c/EBPδ) regulation and expression in human mammary epithelial cells: I. ‘Loss of function’ alterations in the c/EBPδ growth inhibitory pathway in breast cancer cell lines. J. Cell. Biochem. 2004;93:830–843. doi: 10.1002/jcb.20223. [DOI] [PubMed] [Google Scholar]
- 9.Sivko G. S., Sanford D. C., Dearth L. D., Tang D., DeWille J. W. CCAAT/enhancer binding protein δ (c/EBPδ) regulation and expression in human mammary epithelial cells: II. Analysis of activating signal transduction pathways, transcriptional, post-transcriptional, and post-translational control. J. Cell. Biochem. 2004;93:844–856. doi: 10.1002/jcb.20224. [DOI] [PubMed] [Google Scholar]
- 10.Tang D., Sivko G. S., DeWille J. W. Promoter methylation reduces C/EBPδ (CEBPD) gene expression in the SUM-52PE human breast cancer cell line and in primary breast tumors. Breast Cancer. Res. Treat. 2006;95:161–170. doi: 10.1007/s10549-005-9061-3. [DOI] [PubMed] [Google Scholar]
- 11.O'Rourke J. P., Hutt J. A., DeWille J. Transcriptional regulation of C/EBPδ in G0 growth-arrested mouse mammary epithelial cells. Biochem. Biophys. Res. Commun. 1999;262:696–701. doi: 10.1006/bbrc.1999.1256. [DOI] [PubMed] [Google Scholar]
- 12.Dearth L. R., DeWille J. Posttranscriptional and posttranslational regulation of C/EBPδ in G0 growth-arrested mammary epithelial cells. J. Biol. Chem. 2003;278:11246–11255. doi: 10.1074/jbc.M207930200. [DOI] [PubMed] [Google Scholar]
- 13.Johnson L. F., Williams J. G., Abelson H. T., Green H., Penman S. Changes in RNA in relation to growth of the fibroblast. III. Posttranscriptional regulation of mRNA formation in resting and growing cells. Cell. 1975;4:69–75. doi: 10.1016/0092-8674(75)90135-x. [DOI] [PubMed] [Google Scholar]
- 14.O'Rourke J. P., Newbound G. C., Hutt J. A., DeWille J. CCAAT/enhancer-binding protein δ regulates mammary epithelial cell G0 growth arrest and apoptosis. J. Biol. Chem. 1999;274:16582–16589. doi: 10.1074/jbc.274.23.16582. [DOI] [PubMed] [Google Scholar]
- 15.Jentsch S., Pyrowolakis G. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 2000;10:335–342. doi: 10.1016/s0962-8924(00)01785-2. [DOI] [PubMed] [Google Scholar]
- 16.Welchman R. L., Gordon C., Mayer R. J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 17.Ciechanover A. The ubiquitin–proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickart C. M., Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin–proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 20.Pickart C. M. Ubiquitin in chains. Trends Biochem. Sci. 2000;25:544–548. doi: 10.1016/s0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- 21.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoyt M. A., Coffino P. Ubiquitin-free routes into the proteasome. Cell. Mol. Life Sci. 2004;61:1596–1600. doi: 10.1007/s00018-004-4133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orlowski M., Wilk S. Ubiquitin-independent proteolytic functions of the proteasome. Arch. Biochem. Biophys. 2003;415:1–5. doi: 10.1016/s0003-9861(03)00197-8. [DOI] [PubMed] [Google Scholar]
- 24.Hoyt M. A., Zhang M., Coffino P. Ubiquitin-independent mechanisms of mouse ornithine decarboxylase degradation are conserved between mammalian and fungal cells. J. Biol. Chem. 2003;278:12135–12143. doi: 10.1074/jbc.M211802200. [DOI] [PubMed] [Google Scholar]
- 25.Murakami Y., Matsufuji S., Hayashi S., Tanahashi N., Tanaka K. Degradation of ornithine decarboxylase by the 26S proteasome. Biochem. Biophys. Res. Commun. 2000;267:1–6. doi: 10.1006/bbrc.1999.1706. [DOI] [PubMed] [Google Scholar]
- 26.Coffino P. Antizyme, a mediator of ubiquitin-independent proteasomal degradation. Biochimie. 2001;83:319–323. doi: 10.1016/s0300-9084(01)01252-4. [DOI] [PubMed] [Google Scholar]
- 27.Zhang M., MacDonald A. I., Hoyt M. A., Coffino P. Proteasomes begin ornithine decarboxylase digestion at the C terminus. J. Biol. Chem. 2004;279:20959–20965. doi: 10.1074/jbc.M314043200. [DOI] [PubMed] [Google Scholar]
- 28.Chen X., Chi Y., Bloecher A., Aebersold R., Clurman B. E., Roberts J. M. N-acetylation and ubiquitin-independent proteasomal degradation of p21Cip1. Mol. Cell. 2004;16:839–847. doi: 10.1016/j.molcel.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Touitou R., Richardson J., Bose S., Nakanishi M., Rivett J., Allday M. J. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 2001;20:2367–2375. doi: 10.1093/emboj/20.10.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jariel-Encontre I., Pariat M., Martin F., Carillo S., Salvat C., Piechaczyk M. Ubiquitinylation is not an absolute requirement for degradation of c-Jun protein by the 26 S proteasome. J. Biol. Chem. 1995;270:11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- 31.Tarcsa E., Szymanska G., Lecker S., O'Connor C. M., Goldberg A. L. Ca2+-free calmodulin and calmodulin damaged by in vitro aging are selectively degraded by 26 S proteasomes without ubiquitination. J. Biol. Chem. 2000;275:20295–20301. doi: 10.1074/jbc.M001555200. [DOI] [PubMed] [Google Scholar]
- 32.Haglund K., Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hicke L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 34.Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 35.Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z. J., Parent L., Maniatis T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 37.Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 38.Hershko A. The ubiquitin pathway for protein degradation. Trends Biochem. Sci. 1991;16:265–268. doi: 10.1016/0968-0004(91)90101-z. [DOI] [PubMed] [Google Scholar]
- 39.Ciechanover A., Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 40.Yang K., Zhu J., Sun S., Tang Y., Zhang B., Diao L., Wang C. The coiled-coil domain of TRAF6 is essential for its auto-ubiquitination. Biochem. Biophys. Res. Commun. 2004;324:432–439. doi: 10.1016/j.bbrc.2004.09.070. [DOI] [PubMed] [Google Scholar]
- 41.Kim Y. E., Kim D. Y., Lee J. M., Kim S. T., Han T. H., Ahn J. H. Requirement of the coiled-coil domain of PML-RARα oncoprotein for localization, sumoylation, and inhibition of monocyte differentiation. Biochem. Biophys. Res. Commun. 2005;330:746–754. doi: 10.1016/j.bbrc.2005.03.052. [DOI] [PubMed] [Google Scholar]
- 42.Hattori T., Ohoka N., Inoue Y., Hayashi H., Onozaki K. C/EBP family transcription factors are degraded by the proteasome but stabilized by forming dimer. Oncogene. 2003;22:1273–1280. doi: 10.1038/sj.onc.1206204. [DOI] [PubMed] [Google Scholar]
- 43.Parkin S. E., Baer M., Copeland T. D., Schwartz R. C., Johnson P. F. Regulation of CCAAT/enhancer-binding protein (C/EBP) activator proteins by heterodimerization with C/EBPγ (Ig/EBP) J. Biol. Chem. 2002;277:23563–23572. doi: 10.1074/jbc.M202184200. [DOI] [PubMed] [Google Scholar]
- 44.Wei W., Yang H., Cao P., Menconi M., Chamberlain C., Petkova V., Hasselgren P. O. Degradation of C/EBPβ in cultured myotubes is calpain-dependent. J. Cell Physiol. 2006;208:386–398. doi: 10.1002/jcp.20684. [DOI] [PubMed] [Google Scholar]
- 45.Hershko A., Ganoth D., Pehrson J., Palazzo R. E., Cohen L. H. Methylated ubiquitin inhibits cyclin degradation in clam embryo extracts. J. Biol. Chem. 1991;266:16376–16379. [PubMed] [Google Scholar]
- 46.Sheaff R. J., Singer J. D., Swanger J., Smitherman M., Roberts J. M., Clurman B. E. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 47.Thornton B. R., Toczyski D. P. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell Biol. 2003;5:1090–1094. doi: 10.1038/ncb1066. [DOI] [PubMed] [Google Scholar]
- 48.Asher G., Reuven N., Shaul Y. 20S proteasomes and protein degradation ‘by default’. BioEssays. 2006;28:844–849. doi: 10.1002/bies.20447. [DOI] [PubMed] [Google Scholar]
- 49.Coulombe P., Rodier G., Bonneil E., Thibault P., Meloche S. N-terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome. Mol. Cell. Biol. 2004;24:6140–6150. doi: 10.1128/MCB.24.14.6140-6150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Breitschopf K., Bengal E., Ziv T., Admon A., Ciechanover A. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998;17:5964–5973. doi: 10.1093/emboj/17.20.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]