Abstract
A potent anticoagulant protein, IX-bp (Factor IX binding protein), has been isolated from the venom of Trimeresurus flavoviridis (habu snake) and is known to bind specifically to the Gla (γ-carboxyglutamic acid-rich) domain of Factor IX. To evaluate the molecular basis for its anticoagulation activity, we assessed its interactions with various clotting factors. We found that the anticoagulation activity is primarily due to binding to the Gla domains of Factors IX and X, thus preventing these factors from recognizing phosphatidylserine on the plasma membrane. The present study suggests that ligands that bind to the Gla domains of Factors IX and X may have the potential to become novel anticoagulants.
Keywords: anticoagulant, blood coagulation, γ-carboxyglutamic acid (Gla)-rich domain (Gla domain), Factor IX-binding protein (IX-bp), Factor X, phospholipid membrane
Abbreviations: APTT, activated partial thromboplastin time; Gla, γ-carboxyglutamic acid; Gla domain, Gla-rich domain; IX-bp, Factor IX-binding protein; SPR, surface plasmon resonance
INTRODUCTION
In the process of blood coagulation, various factors are known to interact in a sequential fashion to initiate the formation of a haemostatic plug following injury. Although the concept of two clotting pathways (intrinsic and extrinsic) has long served as a useful model for understanding the coagulation process, previous evidence has shown that these two pathways are actually highly connected [1]. For example, Factor VIIa–tissue Factor complex not only activates Factor X but also Factor IX, in consideration of the effect of Mg2+ ions. The result of these two pathways is the same, to produce Factor Xa, which then catalyses the conversion of prothrombin into thrombin. This reaction is generally enhanced in the presence of Ca2+ ions, phospholipids and Factor Va [2]. Thrombin finally cleaves fibrinogen, an inactive circulating plasma protein, to form soluble fibrin monomers. These monomers spontaneously aggregate to form an insoluble fibrin polymer.
Among the blood coagulation factors, Factors IX and X seem to have the ability to interact with more factors (Factors XIa, VIIIa and Va and Factor VIIa–tissue Factor complex) than any other factors in the cascade, and therefore, they are vital in the overall coagulation process. Both of these factors are homologous proteins and contain an N-terminal Gla (γ-carboxyglutamic acid-rich) domain. The Gla domain is responsible for binding to phospholipid membranes. It allows the protein to anchor on the surface of the membrane, which seems to be critical for the function of these proteins. Their binding to the membrane not only assists in localizing these proteins to sites of injury, but also helps in properly positioning these proteins for interactions with various factors. The Gla domain comprises approx. 45 amino acids, and is characterized by nine to twelve Gla residues that are modified post-translationally by a vitamin K-dependent carboxylase [3]. Other Gla domain-containing vitamin K-dependent proteins include prothrombin, Factor VII, protein C, protein S and protein Z. The Gla domains in these coagulation proteins share a high degree of sequence similarity, but their binding affinities to the membranes vary over several orders of magnitude (equilibrium dissociation constant, Kd, ranging from millimolar to micromolar). The Gla domains of these proteins apparently bind to phosphatidylserine and are essential for their maximal binding to the membrane. Furthermore, the Gla domain binds to phosphatidylserine only in the presence of bivalent metal ions [1,4–10].
It appears that in the absence of metal ions, the Gla domain is essentially disordered [11] and unstructured, but in the presence of bivalent metal ions, a stable structure is formed [6–8]. In addition, the folding of the native tertiary structure of the Gla domain required not only Ca2+ ions but also Mg2+ ions [1,4,5]. Although the actual numbers of bivalent metal ions found to be associated with the Gla domains of Factors IX and X and prothrombin vary, according to different structural studies, the presence of five calcium ions aligned at the core of the Gla domain and their co-ordination with the carboxy groups of the Gla residues were observed consistently. Huang et al. [9] provided direct evidence to show that the serine head group has multiple interactions with calcium ions 5 and 6, and Arg10 and Arg16 of the Gla domain in a prothrombin fragment, based on X-ray crystallography and NMR spectroscopy.
Snake venoms obtained from Trimeresurus flavoviridis (habu snake) and Deinagkistrodon acutus (sharp-nosed pit viper) contain proteins that bind to the Gla domains of Factors IX and X [12–14]. Homologous proteins were also discovered in the snake venoms of Echis carinatus leucogaster (saw-scaled viper) and Trimeresurus stejnegeri (Stejneger's bamboo pit viper) [15,16]. These proteins, IX-bp (Factor IX-binding protein) and X-bp (Factor X-binding protein), are known to bind to Factors IX and X, respectively. Both proteins displayed efficient anticoagulant activity and are highly homologous with each other. Although these two proteins belong to the C-type lectin superfamily [17], no lectin activity has been detected so far. The crystal structures of these two proteins complexed with the respective Gla domains of Factors IX and X have been solved [1,18]. They revealed important interactions not only with hydrophilic residues but also between hydrophobic residues, including phenylalanine (Factor X), and leucine and valine (in Factors IX and X) located within the ω-loop of the Gla domain, and the IX-bp and X-bp proteins [1,18]. These results suggest that the anticoagulation activity for the IX-bp and X-bp proteins probably originated by blocking the membrane interaction of the Gla domains of Factors IX and X, especially the hydrophobic residues located within the ω-loop of the Gla domain. However, due to a lack of structural data about the full-length IX–IX-bp and X–X-bp complexes and the limited information about the interactions of IX-bp and X-bp with different proteins involved in blood coagulation, it is not clear whether the anticoagulant activity is solely due to blocking the membrane binding or altering the association rates with other coagulation proteins. Therefore, in the present study, we sought to address the molecular basis for the anticoagulant activity of IX-bp.
We found that IX-bp interacts with both Factors IX and X, and does not inhibit the interactions with other coagulation proteins during the activation process. We also found that IX-bp appears to play a vital role in specifically blocking the interaction between the phospholipids, the seat of the overall coagulation process, and the Gla domains of Factors IX and X.
METHODS
Proteins
Human Factors IX, IXa and XIa and lipidated recombinant human tissue Factor were purchased from American Diagnostica (Stamford, CT, U.S.A.). Human Factor VIIa was obtained from Haematologic Technologies (Essex Junction, VT, U.S.A.). Human Factor X was purchased from Innovative Research. All clotting assay reagents were purchased from Roche (Mannheim, Germany). Phospholipids (L-α-phosphatidylcholine, L-α-phosphatidylethanolamine and L-α-phosphatidylserine) were purchased from Avanti Polar Lipids (Alabaster, AL, U.S.A.). The IX-bp was purified from the snake venom of T. flavoviridis, following the published purification protocol [19].
Clotting assay
The APTT (activated partial thromboplastin time) clotting assay was performed using a model Start, from Diagnostica Stago. Initially, a 25 μl aliquot of normal human plasma was mixed with various concentrations of IX-bp (0.1–250 nM, final concentrations) and deficient human plasma to a final volume of 50 μl. To this reaction mixture, a 25 μl aliquot of APTT was added and incubated for 3 min at 37°C. The clotting reaction was initiated by adding 25 μl of 25 mM CaCl2. Results are expressed as the relative change in clotting time. The control reaction was performed with reaction buffer.
Binding analysis of IX-bp with coagulation factors using SPR (surface plasmon resonance)
The binding analyses of IX-bp with human coagulation factors were conducted using the SPR method on a Biacore 2000, with a CM4 sensor chip, from Biacore (Uppsala, Sweden). Initially, the carboxylmethylated surface of the CM4 chip was activated with 70 μl of a mixture of 50 mM NHS (N-hydroxysuccinimide) and 200 mM EDAC [1-ethyl-3-(3-dimethylaminopropyl)carbodi-imide]. The activated chip was coated with 20 μg/ml IX-bp, dissolved in 10 mM sodium acetate buffer (pH 4.0), by injecting 60 μl of the solution at a flow rate of 10 μl/min. The remaining unchanged functional groups on the IX-bp-coated surface were blocked with 1 M ethanolamine hydrochloride, at a similar flow rate for 7 min. To analyse the binding of human coagulation factors, 40 μl aliquots (100 nM final concentration) of factors were injected at a flow rate of 20 μl/min for 2 min. For the kinetic analysis of binding, Factors IX and X, at final concentrations ranging from 50 to 500 nM, were injected.
Intrinsic pathway cleavage assay
The ‘intrinsic pathway’ reactions were performed in 10 μl of reaction buffer (50 mM Hepes buffer, pH 7.4, 1 mM MgCl2 and 5 mM CaCl2). Factor IX (4.4 μM) was incubated in reaction buffer at various concentrations (molar ratio of IX-bp/Factor IX, 0:1, 0.125:1, 0.25:1 and 0.5:1). To these reaction mixtures, Factor XIa (620 nM) was added, and the mixtures incubated for 30 min at 25°C. Afterwards, the products were resolved by SDS/PAGE (gradient of 14–16%) and the gel was stained with Bio-Safe Coomassie solution (Bio-Rad, Hercules, CA, U.S.A.). The intensities of the Factor IX-cleaved products on the gel were quantified using the ImageJ software [NIH (National Institutes of Health), Bethesda, MD, U.S.A.].
Extrinsic pathway cleavage assay
The ‘extrinsic pathway’ reactions were carried out in 10 μl of reaction buffer (50 mM Hepes buffer, pH 7.4, with 1 mM MgCl2 and 5 mM CaCl2). Factor IX (final concentration of 4.4 μM) was mixed with various concentrations of FIX-bp (molar ratio of IX-bp/Factor IX, 0:1, 0.125:1, 0.25:1 and 0.5:1). To this reaction mixture, Factor VIIa (800 nM) and lipidated recombinant human tissue Factor (500 pM) were added. After incubation for 24 h at 25°C, the reaction products were resolved by SDS/PAGE (gradient of 14–16%) and the gel was stained with Bio-Safe Coomassie solution (Bio-Rad). The intensities of the Factor IX-cleaved products on the gel were quantified using the ImageJ software (NIH). Similarly, we have carried out activation of Factor X by tissue factor–VIIa complex in the absence and presence of IX-bp. All conditions used here were similar to that of Factor IX. In short, Factor X (final concentration of 4.4 μM) was mixed with various ratios of IX-bp in the presence of 1 mM MgCl2 and 5 mM CaCl2. To this reaction mixture, Factor VIIa (800 nM) and lipidated recombinant human tissue factor (500 pM) were added. After incubation for 6 h at 25°C, the reaction products were separated, stained and quantified as mentioned above.
Formation of a phospholipid bilayer on an L1 chip and analysis of Factors IX and X binding to the membrane
Binding assays of phospholipids against human coagulation factors and IX-bp and their complexes were conducted using the SPR method on a Biacore 2000, with an L1 sensor chip, from Biacore. The liposome preparation was described previously [20]. Phospholipids (L-α-phosphatidylcholine, L-α-phosphatidylethanolamine and L-α-phosphatidylserine, in a 6:3:1 ratio) in chloroform were mixed and evaporated under a stream of nitrogen gas. The phospholipids were suspended (final concentration 1 mM) in a buffer containing 10 mM Hepes (pH 7.4) and 150 mM NaCl in the absence of detergents, and were dispersed by sonication. The lipophilic-group-immobilized L1 chip was coated with this mixture of liposomes at a flow rate of 10 μl/min for 4 min. This process forms a phospholipid bilayer on the L1 chip, as described in [21]. To analyse the binding of human coagulation factors, 40 μl aliquots (100 nM final concentration) of factors were injected at a flow rate of 20 μl/min for 2 min. Similarly, IX-bp was also injected to evaluate the binding. For the kinetic analysis of binding, Factors IX and X, at final concentrations ranging from 50 to 500 nM, were injected. To study the interference of IX-bp in the interactions between Factor IX or X and phospholipids, we made complexes with either Factor IX or X (200 nM) and increasing concentrations of Factor IX-bp (1, 2, 10, 20, 100 and 200 nM), by incubating them in the buffer used for the Biacore analysis in the presence of calcium and magnesium ions for 10 min at room temperature (25°C). After this incubation, the complexed reaction mixture was injected independently on an L1 chip bound with phospholipids, as mentioned above, and analysed. Similar experiments were carried out using IX-bp complexed with Factor X. To evaluate the efficiency of the effector (IX-bp) on the Factor IX or X binding to the phospholipids, EC50 values were determined by fitting the data with a one-site competition equation, as mentioned below, using the GraphPad (San Diego, CA, U.S.A.) Prism 4 software:
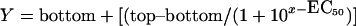 |
where EC50 is the concentration of the effector ligand (IX-bp) required to block half of the specific binding between phospholipids and Factor IX or X [22].
RESULTS
Anticoagulant activity of IX-bp and binding ability to Factors IX and X
To reproduce the previous observations, we purified IX-bp from T. flavoviridis snake venom and analysed its ability to inhibit blood coagulation, using an APTT assay, as described previously [19,23]. The isolated IX-bp protein efficiently prolonged the blood clotting in a concentration-dependent manner, suggesting that the purified protein retained similar biological activity to that observed previously (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/405/bj4050351add.htm). We monitored the coagulation activity from low concentrations to 250 nM of IX-bp. At concentrations above 250 nM, the IX-bp exhibited high anti-coagulation activity with a quite delayed clotting time, which was beyond the assay limit.
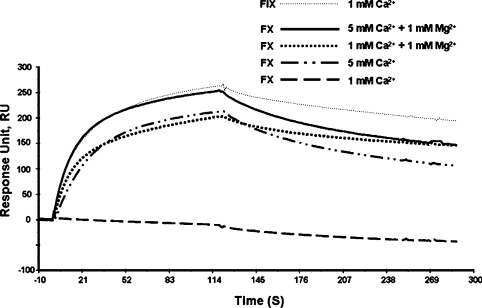
To analyse the binding of IX-bp to the Gla domains of Factor IX and Factor X, we performed an SPR analysis using a CM4 chip, to which IX-bp was covalently attached. The purified Factors IX and X were injected into the flow cell in the presence of 1 mM Ca2+. We carried out the binding analysis in a buffer (10 mM Hepes buffer, pH 7.4, 150 mM NaCl and 1 mM CaCl2) containing Factors IX and X. As observed previously, IX-bp bound specifically to Factor IX, and failed to bind to Factor X [19] (Figure 1). This can be attributed to the insufficient calcium ion concentration needed to attain proper folding of the Gla domain of Factor X, based on the membrane binding studies by Erb et al. [21] in which Factor X required approximately a 3 mM concentration of Ca2+ ions for maximum binding to the membrane. To test whether an insufficient amount of Ca2+ ions during the SPR binding analyses was the sole reason, for the lack of Factor X recognition of IX-bp, we carried out Factor X and IX-bp binding analyses at a 5 mM Ca2+ ion concentration. The SPR analyses clearly showed that a 5 mM concentration of Ca2+ ions facilitated the binding of IX-bp to Factor X (Figure 1). However, such a high concentration of Ca2+ ions is not present in the blood plasma, although Mg2+ ions are present as well [1]. Next, to analyse the interactions of IX-bp with Factor X, using physiologically relevant concentrations of bivalent metal ions (Ca2+ and Mg2+ ions), we repeated these binding analyses in the presence of Mg2+ and Ca2+ ions at 1 mM concentrations. Interestingly, Factor X was recognized efficiently by IX-bp, similar to the level observed for the 5 mM Ca2+ ion concentration (Figure 1). The SPR analyses clearly showed that in the presence of both Ca2+ and Mg2+ ions, Factor IX and Factor X are both recognized by IX-bp, which failed to discriminate between them. To evaluate the efficiency of Factor IX and Factor X recognition by IX-bp, a kinetic analysis was also carried out from the SPR curves. The binding analyses revealed that both the IX-bp–Factor IX and IX-bp–Factor X complexes have equilibrium dissociation constants (Kd) at the low nanomolar level (∼40 nM) (Table 1). These results clearly suggest that IX-bp binds to Factor IX and Factor X with equal efficiency in the presence of Mg2+ and Ca2+ ions.
Figure 1. SPR-based analyses of the binding of IX-bp to Factors IX and X.
The activated CM4 chip was coated with 20 μg/ml IX-bp, by injecting 60 μl of the solution at a flow rate of 10 μl/min, and was blocked with 1 M ethanolamine hydrochloride at a similar flow rate for 7 min. Then 40 μl aliquots (100 nM final concentration) of factors were injected at a flow rate of 20 μl/min for 2 min. Appropriate concentrations of calcium or magnesium ions were added to the buffer (10 mM Hepes, pH 7.4, and 150 mM NaCl). FIX, Factor IX.
Table 1. Kinetic analysis by SPR.
| Clotting factor | Kd (nM) | ka (M−1·s−1) | kd (s−1) |
|---|---|---|---|
| IX-bp with coagulation factors on a CM4 chip | |||
| Factor IX | 34±5 | 1.6×105 | 2.5×10−3 |
| Factor X | 40±2 | 1.8×105 | 6.7×10−3 |
| Phospholipids with coagulation factors on an L1 chip | |||
| Factor IX | 50±4 | 5.8×104 | 2.3×10−3 |
| Factor X | 140±5 | 9.0×105 | 1.0×10−1 |
Intrinsic and extrinsic pathway cleavage assays
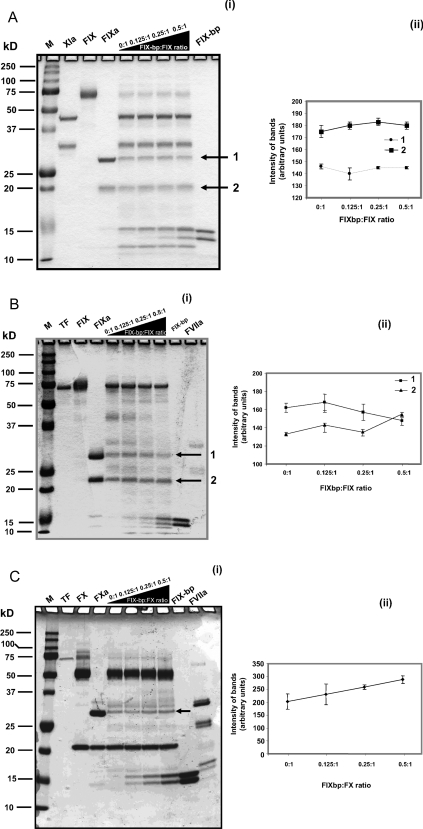
To address whether IX-bp interferes with other protein factor interaction sites that are involved in Factor IX-dependent reactions, we used recently developed gel-based activation assays in the absence of phospholipids [23]. The activation of Factor IX was carried out by incubating Factor IX and Factor XIa in activation buffer (50 mM Hepes buffer, pH 7.4, 1 mM MgCl2 and 5 mM CaCl2) with increasing concentrations of IX-bp for 30 min at 25°C. The resulting cleaved products of Factor IX were separated by PAGE and quantified. As seen in Figure 2(a) (i and ii), it is evident that the intensities of the Factor IX cleavage products (shown by arrows) were not affected significantly, suggesting that the activation of Factor IX by Factor XIa, through an intrinsic pathway, was not inhibited. Next, the ability of IX-bp to block the activation of Factor IX by the Factor VIIa–tissue factor complex was evaluated in vitro, using the assay conditions described previously [23] with some modification as described in the Methods section. The resulting cleavage products of Factor IX [indicated by arrows in Figure 2b (i)] were quantified [Figure 2b (ii)], with increasing IX-bp concentration. The cleavage product levels were not significantly affected, suggesting that the activation of Factor IX by the VIIa–tissue factor, through an extrinsic pathway, was also not inhibited. Alternative to this pathway, we have also analysed the activation of Factor X by the Factor VIIa–tissue factor complex with IX-bp [Figure 2c (i and ii)]. As observed in the case of Factor IX activation, the activation of Factor X was also unaffected in the presence of IX-bp. Our studies further suggest that IX-bp is not involved in binding to proteins involved in these pathways. We also carried out direct binding assays, using an SPR analysis, between IX-bp and other major factors involved in these two pathways, but no binding signal was observed (results not shown). Taken together, these results suggest that IX-bp binding to Factor IX or Factor X and their activation products (Factor IXa and Factor Xa) do not affect the activation mediated by either Factor XIa or Factor VIIa–tissue factor complex in the absence of phospholipids.
Figure 2. In vitro intrinsic and extrinsic cleavage assays.
(a) In vitro intrinsic cleavage assay. (i) The intrinsic cleavage (conversion of Factor IX into IXa by Factor XIa) reaction was carried out in binding buffer (50 mM Hepes, 1 mM MgCl2 and 5 mM CaCl2) with 4.4 μM of Factor IX mixed with 620 nM of Factor XIa, at ambient temperature for 30 min. Similar reactions were also carried out in the presence of the IX-bp (550, 1100 and 2200 nM). For identification, each protein was loaded as a marker. Cleavage products are indicated by arrows. (ii) The intensities of cleavage products 1 and 2 were quantified using the ImageJ software. The average from two experiments is indicated with error bars. (b) In vitro extrinsic cleavage assay. (i) The extrinsic cleavage assay (conversion of Factor IX into IXa by lipidated tissue vactor and Factor VIIa) was carried out in binding buffer, 4.4 μM of Factor IX mixed with 500 pM of tissue factor and 800 nM of Factor VIIa, at ambient temperature for 24 h. Similar reactions were also carried out in the presence of IX-bp (550, 1100 and 2200 nM). For identification, each protein was loaded as a marker. Cleavage products are indicated by arrows. (ii) The intensities of cleavage products 1 and 2 were quantified using the ImageJ software. The average from two experiments is indicated with error bars. (c) In vitro extrinsic cleavage assay. (i) Similar to Figure 2(b), but conversion of Factor X into Xa is assayed. In this case, one cleavage product (indicated by an arrow) was quantified. (ii) The intensity of the cleaved product was quantified using the ImageJ software. The average from two experiments is indicated with error bars. FIX-bp, IX-bp.
Analysis of Factor IX and Factor X Gla-domain interactions with the phospholipid bilayer
We next evaluated whether IX-bp binding to the Gla domains of Factor IX and Factor X prevents these factors from binding to the membrane, by an SPR-based analysis. In this assay, we first prepared the phospholipid vesicles and reconstituted the phospholipid bilayer on the L1 chip. This type of phospholipid bilayer is specifically recognized by Factor IX and Factor X in a buffer (50 mM Hepes buffer, pH 7.4, 1 mM MgCl2 and 5 mM CaCl2) (see Supplementary Figures 2a and 2b at http://www.BiochemJ.org/bj/405/bj4050351add.htm). However, IX-bp failed to bind to either the phospholipid bilayer membrane or Cephalin, under similar conditions (results not shown). Binding kinetics studies revealed that under these conditions, both Factors IX and X bind to these bilayers at nanomolar concentrations (Table 1 and Supplementary Figures 2a and 2b). We also evaluated the effects of Mg2+ ions on the binding abilities of Factors IX and X to these bilayer membranes, as Mg2+ is necessary under physiological conditions. In the absence of Mg2+ ions and with a lower concentration (1 mM) of Ca2+ ions, Factor X was unable to bind to these bilayer membranes (results not shown). Thus, in contrast with the Factor IX Gla domain, Mg2+ ions are an indispensable component for the interaction between Factor X and the phospholipid membrane, as well as IX-bp and Ca2+ ions, suggesting that the Gla domain of Factor X may achieve an active form for its functions. The system reported here is probably analogous to the interactions observed with plasma membranes and Factor IX or Factor X in vivo.
Inhibition of Factor IX and X binding to the membrane by IX-bp
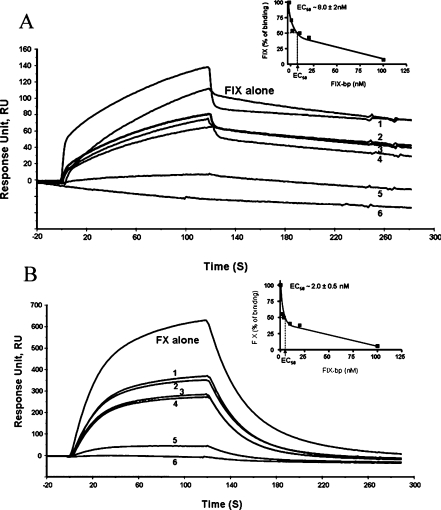
The above results clearly suggest that the described assay could be suitable to determine whether the binding of IX-bp to the Gla domains of Factor IX and Factor X affects the interactions with the membrane. For this, we prepared the Factor IX–IX-bp complex and the Factor X–IX-bp complex, and injected them into the flow cell of the L1 chip, constructed with the phospholipid bilayer membrane. The binding of these complexes to the bilayer membrane was affected in a concentration-dependent manner (Figures 3a and 3b). When this assay was carried out at higher concentrations (100 nM), IX-bp blocked the binding of Factors IX and X to the membrane by over 94%. To evaluate the efficiency of IX-bp on the Factor IX and X binding to the phospholipids, the EC50 values were determined. The EC50 is the concentration of the effector ligand (IX-bp) required to block half of the specific binding between the phospholipids and Factor IX or X. The EC50 values of IX-bp against Factor IX and Factor X were 8±2 and 2±0.5 nM respectively (Figures 3a and 3b).
Figure 3. SPR-based binding analyses of the interference of the binding of Factor IX–IX-bp and Factor X–IX-bp complexes to the phospholipid membrane.
(a) Factor IX and different molar ratios of the IX-bp and Factor IX complex. (b) Factor X and different molar ratios of the IX-bp and Factor X complex. The lipophilic-group-immobilized L1 chip was coated with this mixture of liposomes at a flow rate of 10 μl/min for 4 min. The complexes of IX-bp and Factor IX or Factor X were made by incubating them in the presence of calcium and magnesium ions for 10 min at room temperature, at different ratios. The complexed reaction mixture was injected independently on an L1 chip bound with phospholipids. Injections into the flow cell of various IX-bp concentrations, 1, 2, 10, 20, 100 and 200 nM, complexed with 200 nM of Factors IX or X are indicated as 1, 2, 3, 4, 5 and 6 respectively. The inset shows the efficiency of IX-bp blocking the interactions between the phospholipids and Factor IX or X, calculated as EC50 values. FIX, Factor IX.
DISCUSSION
Previous studies showed that the Gla domain-containing proteins, prothrombin, Factor VII, protein C, protein S, protein Z, and the Factor IX and Factor X proteins, specifically bind to membranes with various affinities [10,24]. Subsequent structural studies revealed that phosphatidylserine is one of the molecules needed for the Gla domain-specific binding on the phospholipid membrane [9]. The importance of Gla domain residues for blood coagulation was reaffirmed by identifying natural substitutions within the Gla domain of the dysfunctional Factor IX protein in an X-linked haemophilia disorder. In these studies, Factor IX mutant replacements of Gla domain residues with other amino acids were observed, especially at the ω-loop region, which showed poor membrane binding ability [25,26]. Previous studies clearly revealed the importance of Mg2+ ions, not only for the stabilization of the Gla domain of Factor IX but also for mediating interactions with the membrane [1]. More specifically, Mg2+ ions mediate the interaction between the Gla residues of Factor IX and the negative head groups of the phosphatidylserine, via Mg2+ bridging.
Proteins that bind to Gla domains have been identified and isolated from various snake venoms [12–14]. One such protein is IX-bp, which reportedly has high anticoagulation activity and is known to bind to the Gla domain of Factor IX and to discriminate the Gla domain of Factor X [19], and thus is classified as IX-bp. Interestingly, the IX-bp and X-bp proteins share high similarity in their primary, secondary and tertiary structures. Moreover, the residues that interact with the Gla-domain residues of either Factor IX or Factor X are highly conserved among the IX-bp and X-bp proteins [16]. In spite of the higher similarities, these proteins that bind to the Gla domains of Factor IX and Factor X have been classified solely on the basis of their binding ability in vitro. In order to understand these discrepancies, we re-evaluated the binding ability of IX-bp to the Gla domains of Factors IX and X, using SPR methodology.
Using an SPR-based analysis, we confirmed the observation that, in the presence of 1 mM CaCl2, IX-bp can discriminate between the coagulation Factors IX and X. Under similar conditions, based on a solid-phase binding analysis, it was previously reported that IX-bp specifically bound to the Gla of Factor IX and failed to bind to Factor X, even at a 100-fold excess concentration [19]. Considering the previously determined importance of the role of Mg2+ for the effective function of the Gla domain [1], we believed that in the earlier studies, the inability of IX-bp to bind to the Gla domain of the Factor X was due to the absence of Mg2+ ions in their binding buffer. When we included 1 mM Mg2+ ions along with 1 mM Ca2+ ions in the binding buffer, the IX-bp failed to discriminate between Factors IX and X. In addition, the crystal structure of IX-bp revealed that it also has Ca2+ binding sites, but these binding sites are not suitable for the Mg2+ ions [27]. Therefore this indicates the requirement of both bivalent metal ions for the reconstitution of a functional Gla domain in Factors IX and X [1,18]. The present study also suggests that Mg2+ ions play a role in the blood coagulation process, by the activation of the Gla domain of Factor X. The concentrations of bivalent metal ions used in the present study are physiologically relevant, as the blood plasma levels of these ions are 0.8–1.2 and 2.2–2.6 mM for Mg2+ and Ca2+ respectively [1]. Taken together, our binding analysis suggests that IX-bp can bind specifically not only to the Gla domain of Factor IX but also to the Gla of Factor X, under physiological cation conditions.
The IX-bp showed efficient anticoagulant activity in the APTT assay. The sole reason for the anticoagulation property of IX-bp was thought to be based on its binding to the ω-loop region of the Factor IX Gla domain, thereby inhibiting the function of Factor IX/IXa. However, previous studies failed to address IX-bp binding to other coagulation factors classified in the intrinsic and extrinsic clotting pathways. In order to determine unambiguously whether IX-bp is capable of binding to other coagulation factors, as well as to other sites of Factor IX/IXa during the coagulation process, we used an in vitro analysis method with purified proteins involved in two coagulation pathways, which we recently developed in our laboratory [23]. The gel-based cleavage assay and the binding assay of the factors by an SPR analysis indicated that IX-bp does not interfere with the interactions between these proteins.
Previous studies provided clear evidence that the Gla domains of Factors IX and X interact with phosphatidylserine in the plasma membrane. This step is essential to allow these factors to anchor on the surface of the membrane, which seems to be critical for their functions. By this process, the Gla domains help Factors IX and X not only in localizing near the sites of injury but also properly positioning these proteins for various interactions with different coagulation factors. To evaluate whether IX-bp blocks these interactions specifically, we constructed phospholipid bilayers, including phosphatidylserine. Both Factors IX and X efficiently bound to the membrane at low concentrations in the presence of Mg2+ and Ca2+ ions. However, when the IX-bp was complexed with either Factor IX or Factor X, the binding was suppressed in a concentration-dependent manner. Thus, the present study clearly indicates that IX-bp does not discriminate between Factors IX and X during the inhibition of the coagulation process, and that the main role of IX-bp is to block the interactions between the phospholipid head group and the Gla domains of coagulation Factors IX and X. As a consequence of these dual target properties, IX-bp shows efficient anticoagulant activity.
Based on the present investigations, we conclude that the IX-bp anticoagulant activity was achieved by the binding of this protein to the Gla domains of both Factors IX and X, and by these interactions, it prevents their binding to the membrane target, phosphatidylserine. Considering the efficiency of inhibiting blood coagulation by IX-bp, it appears that screening for small ligands that bind to the Gla domains of Factors IX and X may yield efficient anticoagulants.
Online data
Acknowledgments
This work was supported by funds from the AIST to P. K. R. K.; S. C.B. G. was partly supported by the JSPS (Japan Society for the Promotion of Science).
References
- 1.Shikamoto Y., Morita T., Fujimoto Z., Mizuno H. Crystal structure of Mg2+ and Ca2+-bound Gla domain of Factor IX complexed with binding protein. J. Biol. Chem. 2003;278:24090–24094. doi: 10.1074/jbc.M300650200. [DOI] [PubMed] [Google Scholar]
- 2.Furie B., Furie B. C. The molecular basis of blood coagulation. Cell. 1988;53:505–518. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 3.Mann K. G., Nesheim M. E., Church W. R., Haley P., Krishnaswamy S. Surface-dependent reactions of the vitamin K-dependent enzyme complexes. Blood. 1990;76:1–16. [PubMed] [Google Scholar]
- 4.Sekiya F., Yamashita T., Atoda H., Komiyama Y., Morita T. Regulation of the tertiary structure and function of coagulation Factor IX by magnesium (II) ions. J. Biol. Chem. 1995;270:14325–14331. doi: 10.1074/jbc.270.24.14325. [DOI] [PubMed] [Google Scholar]
- 5.Sekiya F., Yoshida M., Yamashita T., Morita T. Localization of the specific binding site for magnesium (II) ions in Factor IX. FEBS Lett. 1996;392:205–208. doi: 10.1016/0014-5793(96)00813-7. [DOI] [PubMed] [Google Scholar]
- 6.Freedman S. J., Furie B. C., Furie B., Baleja J. D. Structure of the calcium ion-bound gamma-carboxyglutamic acid-rich domain of Factor IX. Biochemistry. 1995;34:12126–12137. doi: 10.1021/bi00038a005. [DOI] [PubMed] [Google Scholar]
- 7.Freedman S. J., Furie B. C., Furie B., Baleja J. D. Structure of the metal-free gamma-carboxyglutamic acid-rich membrane binding region of Factor IX by twodimensional NMR spectroscopy. J. Biol. Chem. 1995;270:7980–7987. doi: 10.1074/jbc.270.14.7980. [DOI] [PubMed] [Google Scholar]
- 8.Freedman S. J., Blostein M. D., Baleja J. D., Jacobs M., Furie B. C., Furie B. Identification of the phospholipid binding site in the vitamin K-dependent blood coagulation protein Factor IX. J. Biol. Chem. 1996;271:16227–16236. doi: 10.1074/jbc.271.27.16227. [DOI] [PubMed] [Google Scholar]
- 9.Huang M., Rigby A. C., Morelli X., Grant M. A., Huang G., Furie B., Seaton B., Furie B. C. Structural basis of membrane binding by Gla domain of vitamin K-dependent proteins. Nat. Struct. Biol. 2003;10:751–756. doi: 10.1038/nsb971. [DOI] [PubMed] [Google Scholar]
- 10.Huang M., Furie B. C., Furie B. Crystal structure of calcium-stabilized human Factor IX Gla domain bound to a conformation-specific anti-Factor IX antibody. J. Biol. Chem. 2004;279:14338–14346. doi: 10.1074/jbc.M314011200. [DOI] [PubMed] [Google Scholar]
- 11.Sunnerhagen M., Forsen S., Hoffren A. M., Drakenberg T., Teleman O., Stenflo J. Structure of the Ca(2+)-free Gla domain sheds light on membrane binding of blood coagulation proteins. Nat. Struct. Biol. 1995;2:504–509. doi: 10.1038/nsb0695-504. [DOI] [PubMed] [Google Scholar]
- 12.Atoda H., Morita T. A novel blood coagulation Factor IX/Factor X-binding protein with anticoagulant activity from the venom of Trimeresurus flavoviridis (habu snake): isolation and characterization. J. Biochem. (Tokyo) 1989;106:808–813. doi: 10.1093/oxfordjournals.jbchem.a122935. [DOI] [PubMed] [Google Scholar]
- 13.Atoda H., Hyuga M., Morita T. The primary structure of coagulation Factor IX/Factor X-binding protein isolated from the venom of Trimeresurus flavoviridis. Homology with asialoglycoprotein receptors, proteoglycan core protein, tetranectin, and lymphocyte Fc epsilon receptor for immunoglobulin. E. J. Biol. Chem. 1991;266:14903–14911. [PubMed] [Google Scholar]
- 14.Atoda H., Ishikawa M., Mizuno H., Morita T. Coagulation Factor X-binding protein from Deinagkistrodon acutus venom is a Gla domain-binding protein. Biochemistry. 1998;37:17361–17370. doi: 10.1021/bi981177x. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y. L., Tsai I. H. Functional and sequence characterization of coagulation Factor IX/Factor X-binding protein from the venom of Echis carinatus leucogaster. Biochemistry. 1996;35:5264–5271. doi: 10.1021/bi952520q. [DOI] [PubMed] [Google Scholar]
- 16.Lee W.-H., Zhuang Q. Y., Zhang Y. Cloning and characterization of a blood coagulation Factor IX-binding protein from the venom of Trimeresurus stejnegeri. Toxicon. 2003;41:765–772. doi: 10.1016/s0041-0101(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 17.Drickamer K. Evolution of Ca(2+)-dependent animal lectins. Prog. Nucleic Acid Res. Mol. Biol. 1993;45:207–232. [PubMed] [Google Scholar]
- 18.Mizuno H., Fujimoto Z., Atoda H., Morita T. Crystal structure of an anticoagulant protein in complex with the Gla domain of Factor X. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7230–7234. doi: 10.1073/pnas.131179698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atoda H., Ishikawa M., Yoshihara E., Sekiya F., Morita T. Blood coagulation Factor IX-binding protein from snake venom of Trimeresurus flavoviridis: purification and characterization. J. Biochem. 1995;118:965–973. doi: 10.1093/jb/118.5.965. [DOI] [PubMed] [Google Scholar]
- 20.Shikamoto Y., Shibusawa S., Okuyama I., Morita T. Characterization of membrane-associated prothrombin activator in normal and injured murine tissues. FEBS Lett. 1997;412:526–530. doi: 10.1016/s0014-5793(97)00852-1. [DOI] [PubMed] [Google Scholar]
- 21.Erb E. M., Stenflo J., Drakenberg T. Interaction of bovine coagulation Factor X and its glutamic-acid-containing fragments with phospholipid membranes. A surface plasmon resonance study. Eur. J. Biochem. 2002;269:3041–3046. doi: 10.1046/j.1432-1033.2002.02981.x. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto R., Katahira M., Nishikawa S., Baba T., Taira K., Kumar P. K. R. A novel RNA motif that binds efficiently and specifically to the Tat protein of HIV and inhibits the trans-activation by Tat of transcription in vitro and in vivo. Genes Cells. 2000;5:371–388. doi: 10.1046/j.1365-2443.2000.00330.x. [DOI] [PubMed] [Google Scholar]
- 23.Gopinath S. C. B., Shikamoto Y., Mizuno H., Kumar P. K. R. A potent anti-coagulant RNA aptamer inhibits blood coagulation by specifically blocking the extrinsic clotting pathway. Thromb. Haemostasis. 2006;95:767–771. [PubMed] [Google Scholar]
- 24.McDonald J. F., Shah A. M., Schwalbe R. A., Kisiel W., Dahlback B., Nelsestuen G. L. Comparison of naturally occurring vitamin K-dependent proteins: correlation of amino acid sequences and membrane binding properties suggests a membrane contact site. Biochemistry. 1997;36:5120–5127. doi: 10.1021/bi9626160. [DOI] [PubMed] [Google Scholar]
- 25.Giannelli F., Green P. M., High K. A., Sommer S., Poon M. C, Ludwig M. C., Schwaab R., Reitsma P. H., Goossens M., Yoshioka A., Brownlee G. G. Haemophilia B: database of point mutations and short additions and deletions – fourth edition. Nucleic Acids Res. 1993;21:3075–3087. doi: 10.1093/nar/21.13.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang N. S., Zhang M., Thompson A. R., Chen S. H. Factor IX chongqing: a new mutation in the calcium-binding domain of Factor IX resulting in severe hemophilia B. Thromb. Haemostasis. 1990;63:24–26. [PubMed] [Google Scholar]
- 27.Mizuno H., Fujimoto Z., Koizumi M., Kano H., Atoda H., Morita T. Crystal structure of coagulation Factor IX-binding protein from habu snake venom at 2.6 Å: implication of central loop swapping based on deletion in the linker region. J. Mol. Biol. 1999;289:103–112. doi: 10.1006/jmbi.1999.2756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.