Abstract
Mutations in the LRRK2 (leucine-rich repeat kinase-2) gene cause late-onset PD (Parkinson's disease). LRRK2 contains leucine-rich repeats, a GTPase domain, a COR [C-terminal of Roc (Ras of complex)] domain, a kinase and a WD40 (Trp-Asp 40) motif. Little is known about how LRRK2 is regulated, what its physiological substrates are or how mutations affect LRRK2 function. Thus far LRRK2 activity has only been assessed by autophosphorylation and phosphorylation of MBP (myelin basic protein), which is catalysed rather slowly. We undertook a KESTREL (kinase substrate tracking and elucidation) screen in rat brain extracts to identify proteins that were phosphorylated by an activated PD mutant of LRRK2 (G2019S). This led to the discovery that moesin, a protein which anchors the actin cytoskeleton to the plasma membrane, is efficiently phosphorylated by LRRK2, at Thr558, a previously identified in-vivo-phosphorylation site that regulates the ability of moesin to bind actin. LRRK2 also phosphorylated ezrin and radixin, which are related to moesin, at the residue equivalent to Thr558, as well as a peptide (LRRKtide: RLGRDKYKTLRQIRQ) encompassing Thr558. We exploited these findings to determine how nine previously reported PD mutations of LRRK2 affected kinase activity. Only one of the mutations analysed, namely G2019S, stimulated kinase activity. Four mutations inhibited LRRK2 kinase activity (R1941H, I2012T, I2020T and G2385R), whereas the remainder (R1441C, R1441G, Y1699C and T2356I) did not influence activity. Therefore the manner in which LRRK2 mutations induce PD is more complex than previously imagined and is not only caused by an increase in LRRK2 kinase activity. Finally, we show that the minimum catalytically active fragment of LRRK2 requires an intact GTPase, COR and kinase domain, as well as a WD40 motif and a C-terminal tail. The results of the present study suggest that moesin, ezrin and radixin may be LRRK2 substrates, findings that have been exploited to develop the first robust quantitative assay to measure LRRK2 kinase activity.
Keywords: ezrin/radixin/moesin family of proteins (ERM proteins), kinase substrate tracking and elucidation screening (KESTREL screening), leucine-rich repeat kinase 2 (LRRK2), mass spectrometry (MS), Parkinson's disease (PD), protein kinase
Abbreviations: BUBR1, Bub (budding uninhibited by benomyl)-related 1; COR, C-terminal of Roc (Ras of complex); CRMP2, collapsin response mediator protein 2; ERM, ezrin/radixin/moesin; FERM, four-point-one/ezrin/radixin/moesin; GbpC, cGMP-binding protein C; GST, glutathione S-transferase; KESTREL, kinase substrate tracking and elucidation; LDS, lithium dodecyl sulfate; LRRK2, leucine-rich repeat kinase 2; LRRKtide, RLGRDKYKTLRQIRQ; MALDI-TOF, matrix-assisted laser-desorption ionization–time-of-flight; MBP, myelin basic protein; NCBI, National Center for Biotechnology Information; PD, Parkinson's disease; RIPK, Rho-interacting protein kinase; ROCK-II, Rho-associated kinase-2; TSSK1, testis-specific serine kinase 1; WD40, Trp-Asp 40
INTRODUCTION
There has been much interest raised by the recent discovery that different autosomal dominant point mutations within the gene encoding LRRK2 (leucine-rich repeat protein kinase-2) predispose humans to develop late-onset PD [Parkinson's disease; OMIM™ (Online Mendelian Inheritance in Man™) accession number 609007], with a clinical appearance indistinguishable from idiopathic PD [1–4]. The genetic analysis undertaken to date indicates that mutations in LRRK2 are relatively frequent, not only accounting for 5–10% of familial PD, but also found in a significant proportion of sporadic PD cases [5,6]. Little is known about how LRRK2 is regulated in cells, what are its physiological substrates and how mutations in LRRK2 cause or increase risk of PD. In mammals there are two isoforms of the LRRK protein kinase, namely LRRK1 (2038 residues) and LRRK2 (2527 residues). They belong to a protein family containing both Roc and COR [C-terminal of Roc (Ras of complex)] domains that has also been termed ROCO [7]. Thus far, mutations in LRRK2, but not LRRK1, have been linked to PD.
The LRRK/ROCO class of protein kinases was initially characterized in the social amoeba Dictyostelium discoideum, as a protein termed GbpC (cGMP-binding protein C) that comprised an unusual member of the Ras/GTPase superfamily distinct from other small GTPase domains, as it possesses other domains, including a protein kinase [7,8]. Subsequent studies suggested that GbpC regulates chemotaxis and cell polarity in Dictyostelium [9], but the physiological substrates for this enzyme have not been elucidated. The defining feature of the LRRK/ROCO proteins is that they possess LRR (leucine-rich repeat) motif, a Ras-like small GTPase, a region of high amino acid conservation that has been termed the COR domain, and a protein kinase catalytic domain [7,10]. The protein kinase domain of LRRK2 belongs to the tyrosine-like serine/threonine protein kinases and is most similar to the RIPKs (receptor-interacting protein kinases) that play key roles in innate immunity signalling pathways [11]. Other domains are also found on specific members of the LRRK kinases. For example, the GbpC possesses an additional DEP [dishevelled, EGL-10 (egg-laying defective protein 10), pleckstrin], cGMP-binding and Ras-GEF (guanine nucleotide exchange factor) domains that are not found in mammalian LRRK1 and LRRK2. Human LRRK1 possesses three ankyrin repeats at its N-terminus, whereas LRRK2 lacks these domains, but possesses a WD40 (Trp-Asp 40) repeat, located towards its C-terminus, not found in LRRK1 [7].
Human LRRK2 consists of leucine-rich repeats (residues 1010–1287), a small GTPase domain (residues 1335–1504), a COR domain (residues 1517–1843), a serine/threonine protein kinase domain (residues 1875–2132) and a motif that has low resemblance to a WD40 repeat (2231–2276) (Figure 1A). To date ∼20 single amino acid substitution mutations have been linked to autosomal-dominant PD, and these have been found within, or in close proximity to, conserved residues of the small GTPase, COR, protein kinase and WD40 domains [3,4].
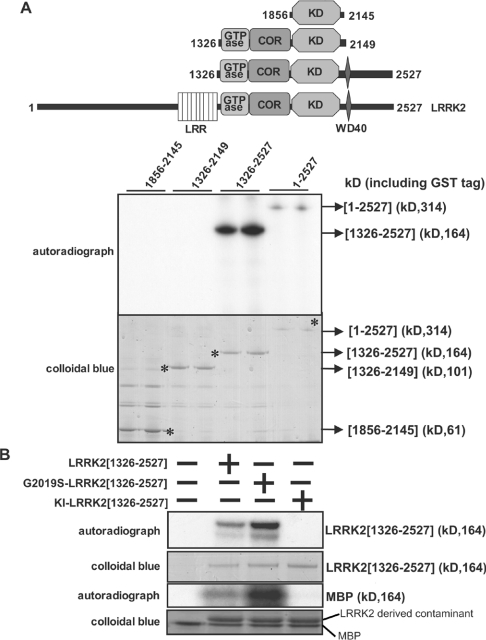
Figure 1. Generation of an active LRRK2 fragment for KESTREL screen.
(A) Upper panel: schematic representation of the domain structure of LRRK2 showing predicted functional domains. Numbering of residues corresponds to human LRRK2 (accession number AAV63975). Abbreviations: LRR, leucine-rich repeat; KD, serine/threonine protein kinase domain. Lower panel: HEK-293 cells were transfected with constructs encoding the indicated forms of GST–LRRK2. At 36 h after transfection, LRRK2 kinases were affinity-purified, subjected to PAGE and stained with Colloidal Blue to quantify relative protein levels. GST–LRRK2 was assayed by measuring autophosphorylation of LRRK2 following PAGE and subsequent autoradiography of the Colloidal Blue-stained bands corresponding MBP or LRRK2. Asterisks on the lower panel correspond to LRRK2 Colloidal Blue-staining bands. kD=kDa. (B) As in (A), except that LRRK2 was also assayed by measuring phosphorylation of MBP. Abbreviation: KI, kinase-inactive (D2017A) LRRK2. Similar results were obtained in three separate experiments.
The most prevalent mutant form of LRRK2, accounting for ∼6% of familial PD and 3% of sporadic PD cases in Europe, comprises an amino acid substitution of Gly2019 located within the conserved Asp-Tyr-Gly-Mg2+-binding motif, in subdomain VII of the kinase domain, to a serine residue [3]. Recent reports suggest that this mutation moderately enhances (∼2–3-fold) the autophosphorylation of LRRK2, as well as its ability to phosphorylate MBP (myelin basic protein) [12,13]. These findings suggest that over-activation of LRRK2 predisposes humans to develop PD, implying that drugs which inhibited LRRK2, could be utilized to delay the onset of, or even treat, some forms of PD. The study of LRRK2 has been hampered by the difficulty in expressing active recombinant enzyme and by the lack of a robust quantitative assay. In the present study we have developed a method to express active recombinant LRRK2 and utilized this in a KESTREL (kinase substrate tracking and elucidation) screen that has recently been developed to identify physiological substrates of protein kinases (reviewed in [14]). This led to the identification of moesin, which, when denatured, was efficiently phosphorylated by LRRK2 at Thr558, a previously characterized physiologically relevant phosphorylation site. Although further investigation is required to determine whether moesin is a physiological substrate, we have utilized these findings to develop a robust and quantitative assay for LRRK2. Using this methodology we demonstrated that several LRRK2 mutations identified in patients with PD either do not affect, or actually inhibit rather than activate, LRRK2. We also demonstrated a requirement for an intact GTPase, COR and C-terminal region to maintain LRRK2 in a catalytically active conformation.
MATERIALS AND METHODS
Materials
Protease-inhibitor cocktail tablets were obtained from Roche; P81 phosphocellulose paper was from Whatman; [γ-32P]ATP and all protein-chromatography media were purchased from Amersham Biosciences. MBP was from Invitrogen, and precast SDS/polyacrylamide/Bis-Tris gels were from Invitrogen; tissue-culture reagents were from Life Technologies; Millipore Immobilon-P was from Fisher Scientific. Active rat ROCK-II, Rho-associated kinase 2 (residues 2–543) was expressed in baculovirus by the Division of Signal Transduction Therapy Unit, University of Dundee, Dundee, Scotland, U.K. The LRRKtide peptide was synthesized by Dr Graham Bloomberg (Department of Biochemistry, School of Medical Science, University of Bristol, Bristol, U.K.).
Antibodies
The anti-GST (glutathione S-transferase) antibody was raised in sheep against the GST protein. The secondary antibodies coupled to horseradish peroxidase used for immunoblotting were obtained from Pierce.
General methods
Tissue culture, transfection, immunoblotting, restriction-enzyme digests, DNA ligations and other recombinant DNA procedures were performed using standard protocols. All mutagenesis was carried out using the Quik-Change site-directed-mutagenesis kit (Stratagene). DNA constructs used for transfection were purified from Escherichia coli DH5α using Qiagen plasmid Mega or Maxi kit according to the manufacturer's protocol. All DNA constructs were verified by DNA sequencing, which was performed by The Sequencing Service, School of Life Sciences, University of Dundee, Scotland, U.K., using DYEnamic ET terminator chemistry (Amersham Biosciences) on Applied Biosystems automated DNA sequencers.
Buffers
Lysis Buffer contained 50 mM Tris/HCl, pH 7.5, 1 mM EGTA, 1 mM EDTA, 1% (w/v) Triton X-100, 1 mM sodium orthovanadate, 10 mM sodium β-glycerophosphate, 50 mM NaF, 5 mM sodium pyrophosphate, 0.27 M sucrose, 0.1% 2-mercaptoethanol and Complete™ proteinase-inhibitor cocktail (one tablet/50 ml; Boehringer). Buffer A contained 50 mM Tris/HCl, pH 7.5, 0.1 mM EGTA and 0.1% (v/v) 2-mercaptoethanol. Extraction Buffer contained 50 mM Tris/HCl, pH 7.5, 5% (v/v) glycerol, 10 mM 2-mercaptoethanol, 1 mM EDTA, 1 mM EGTA, 0.03% (v/v) Brij-35, Complete™ proteinase inhibitor cocktail (one tablet/50 ml). Sample buffer was 1×NuPAGE® LDS (lithium dodecyl sulfate) sample buffer (Invitrogen) containing 1% (v/v) 2-mercaptoethanol.
Plasmids
A full-length cDNA clone encoding LRRK2 corresponding to NCBI (National Center for Biotechnology Information) accession no. AAV63975 was generously provided by Dr Michel Goedert (MRC Laboratory of Molecular Biology, Hills Road, Cambridge, U.K.). The full-length LRRK2 gene and its fragments utilized in the present study were amplified from the LRRK2 cDNA fragment by standard PCR methods using KOD (Thermococcus kodakaraensis) polymerase (Novagen). The resulting PCR products were subcloned into mammalian pEBG2T and pCMV5 expression vectors as BamH1–Not1 fragments. cDNA encoding full-length as well as C-terminal fragments of human moesin (NCBI accession no. NP_002435), ezrin (NCBI accession no. P15311) and radixin (NCBI accession no. NM_002906) were amplified by PCR from expressed sequence tagged IMAGE clones 4908580, 4819793 and 5284438 respectively that were ordered from Geneservice, Cambridge Science Park, Cambridge, U.K. The PCR products were ligated into different expression vectors as Not1–Not1 (moesin), Spe1–Not1 (ezrin) and BamH1–Not1 (radixin) fragments.
Expression and purification of GST–LRRK2
Typically 10–100 10-cm-diameter dishes of HEK-293 (human embryonic kidney 293) cells were cultured and each dish transfected with 5 μg of the pEBG-2T construct encoding wild-type or different mutant forms of LRRK2 using the polyethylenimine method [15]. The cells were cultured for a further 36 h and lysed in 0.5 ml of ice-cold lysis buffer, the lysates pooled and centrifuged at 4°C for 10 min at 26000 g. The GST-fusion proteins were purified by affinity chromatography on glutathione–Sepharose (10 μl per dish of HEK-293 cells) and were eluted in Buffer A containing 20 mM glutathione and 0.27 M sucrose. The enzyme was snap-frozen in small aliquots and stored at −80°C.
LRRK2 KESTREL screen
Brains derived from 50 rats were minced and homogenized with 4 vol. of Extraction Buffer. Insoluble material was sedimented by centrifugation for 20 min at 28000 g at 4°C, and the protein in the supernatant precipitated for 2 h by stirring with 60% (w/v) (NH4)2SO4. The precipitated protein was collected by centrifugation for 20 min at 28000 g, resuspended in Extraction Buffer, desalted by chromatography on Sephadex-G25 (Fine grade) into 30 mM Mops, pH 6.9, 10% (v/v) glycerol, 10 mM 2-mercaptoethanol and 0.03% (v/v) Brij-35 and chromatographed in the latter buffer on heparin–Sepharose. The flow-through of the heparin column was titrated with 1 M NaOH to pH 7.5 and applied to an 8 ml Source™ 15 Q column (GE Healthcare), which was developed in 30 mM Tris/HCl, pH 7.5, 10% (v/v) glycerol, 10 mM 2-mercaptoethanol and 0.03% (v/v) Brij-35 with a 136 ml gradient to 1 M NaCl. Aliquots of all fractions were diluted 10-fold in 50 mM Tris/HCl, pH 7.5, 10 mM 2-mercaptoethanol, 10 μg/ml leupeptin and 1 mM Pefabloc, incubated at 65°C for 15 min prior to incubation for 5 min with 3 mM MnCl2, 1 MBq/ml [γ-32P]ATP in the absence or presence of 2 μg of GST–LRRK2[1326–2527,G2019S] (purity of the LRRK2 enzyme estimated at 2–5% of total protein). The reactions were terminated by addition of LDS sample buffer, subjected to PAGE and electro-transferred to Immobilon P membrane. The membranes were dried and autoradiographed. All fractions and test aliquots were frozen at −80°C. Substrate containing Q-fractions 5 and 6 were diluted 5-fold in 30 mM Tris/HCl, pH 8.2, 10% (v/v) glycerol, 10 mM 2-mercaptoethanol, 0.03% (v/v) Brij-35 and applied to a 1 ml Source 15 Q column. This column was developed in 30 mM Tris/HCl, pH 7.5, 10% (v/v) glycerol, 10 mM 2-mercaptoethanol, 0.03% (v/v) Brij-35 with a 10 ml gradient to 1 M NaCl and 0.5 ml fractions were collected and aliquots were screened with LRRK2 as described above. Substrate-containing Q-column fractions 6 and 7 were applied to a 120 ml Superdex-200 column and 1.2 ml aliquots were collected and screened. Substrate-containing Superdex fractions 12–15 were pooled, concentrated and desalted by filtration in a 2 ml VivaScience (now Sartorius) spin filter. Aliquots (4 μg each) were denatured or left native and tested for the presence of the substrate by phosphorylation in the presence or absence of LRRK2. The samples were electrophoresed on a polyacrylamide gel, stained with Colloidal Blue and analysed by autoradiography. The protein band corresponding to the substrate signal was excised, digested with trypsin and subjected to protein identification by MS fingerprinting.
Expression and purification of human ERM proteins in E. coli
The pGEX expression constructs encoding wild-type and mutant forms of human moesin were transformed into E. coli BL21 cells and 1-litre cultures were grown at 37°C in Luria broth containing 100 μg/ml ampicillin until the attenuance (D600) reached 0.8. Induction of protein expression was carried out by adding 100 μM isopropyl β-D-galactoside and the cells were cultured for a further 16 h at 26°C. Cells were isolated by centrifugation (14000 g, 15 min), resuspended in 15 ml of ice-cold Lysis Buffer and lysed in one round of freeze/thawing, followed by sonication (Branson Digital Sonifier; ten 15 s pulses with a setting of 45% amplitude) to fragment DNA. The lysates were centrifuged at 4°C for 30 min at 26000 g, and the recombinant proteins were affinity-purified on 0.2 ml of glutathione–Sepharose and were eluted in 0.4 ml of Buffer A containing 20 mM glutathione and 0.27 M sucrose.
Mapping the sites on moesin phosphorylated by the G2019S mutant of LRRK2
Moesin (4 μg) was treated at 65°C for 15 min and then incubated at 30°C with 1.5 μg of GST–LRRK2[1326–2527,G2019S] in Buffer A containing 10 mM MgCl2 and 100 μM [γ-32P]ATP (10000 c.p.m./pmol) in a total reaction volume of 50 μl. The reaction was terminated after 40 min by adding Sample Buffer to a final concentration of 1% (w/v) LDS/10 mM dithiothreitol, and the samples heated at 100°C for 1 min and cooled on ice. 4-Vinylpyridine was added to a concentration of 50 mM, and the sample was left on a shaking platform for 30 min at room temperature to alkylate cysteine residues. The samples were subjected to electrophoresis on a Bis-Tris/4–12% (w/v) polyacrylamide gel, which was stained with Colloidal Blue and then autoradiographed. The phosphorylated moesin band was excised, cut into smaller pieces, washed sequentially for 15 min on a vibrating platform with 1 ml of the following: water, a 1:1 (v/v) mixture of water and acetonitrile, 0.1 M ammonium bicarbonate, a 1:1 (v/v) mixture of 0.2 M ammonium bicarbonate and acetonitrile, and finally acetonitrile. The gel pieces were dried with an Edwards Speedivac vacuum pump and incubated in 0.1 ml of 50 mM ammonium bicarbonate, 0.1% (w/v) n-octyl glucoside containing 1 μg of MS-grade trypsin (Promega). After 16 h, 0.1 ml of acetonitrile was added and the mixture incubated on a shaking platform for 10 min. The supernatant was removed and the gel pieces were further washed for 10 min in 0.3 ml of 50 mM ammonium bicarbonate and 0.1% (v/v) trifluoroacetic acid. The combined supernatants, containing >90% of the 32P radioactivity, were chromatographed on a Vydac 218TP5215 C18 column (Separations Group, Hesperia, CA, U.S.A.) equilibrated in 0.1% trifluoroacetic acid in water. The column was developed with a linear acetonitrile gradient (diagonal broken line in Figure 4B) at a flow rate of 0.2 ml/min and fractions of 0.1 ml were collected. Phosphopeptides were further purified by immobilized metal-chelate affinity chromatography on Phospho-Select resin (Sigma).
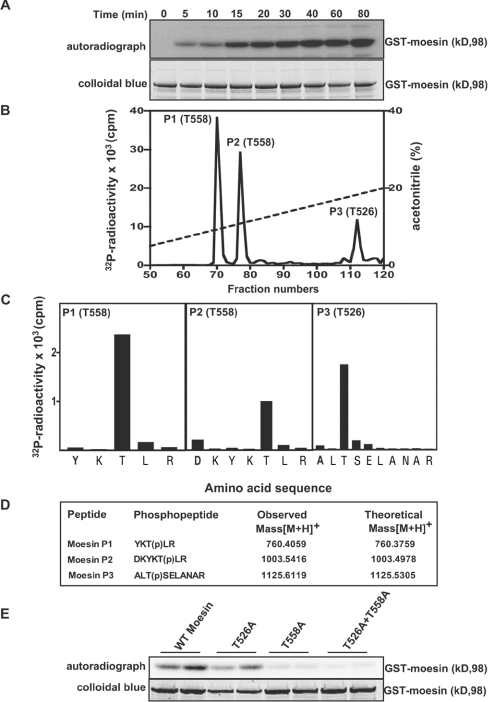
Figure 4. Identification of residues on moesin that are phosphorylated by LRRK2.
(A) E. coli-expressed moesin was incubated at 65°C for 15 min before phosphorylation with GST–LRRK2[1326–2527,G2019S] and [γ-32P]ATP for the indicated times. Phosphorylation of the moesin protein was determined after PAGE and subsequent autoradiography of the Colloidal Blue-stained bands corresponding to moesin. Similar results were obtained in three separate experiments. (B) 32P-labelled moesin after phosphorylation with the GST–LRRK2[1326–2527,G2019S] for 40 min was digested with trypsin and chromatographed on a C18 column. Fractions containing the major 32P-labelled tryptic peptide (P1), peptide P2 and peptide P3 are shown, and no other major 32P-labelled peptides were observed in other fractions of the chromatography. (C) The indicated peptides were subjected to solid-phase sequencing, and the 32P radioactivity released after each cycle of Edman degradation was measured. (D) Peptides were also analysed by MALDI–TOF and MALDI–TOF–TOF-MS (the latter spectra for peptide P2 are shown in Supplementary Figure 1 at http://www.BiochemJ.org/bj/405/bj4050307add.htm) and the inferred amino acid sequence and the site of phosphorylation denoted by ‘(p)’ is indicated, together with the observed and theoretical mass of each peptide. (E) As in (A), except the indicated wild-type and mutant forms of moesin were phosphorylated with GST–LRRK2[1326–2527,G2019S] for 30 min. Similar results were obtained in two separate experiments.
Phosphopeptide sequence analysis
Isolated phosphopeptides were analysed by MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight)-MS on an Applied Biosystems 4700 Proteomics Analyser using 5 mg/ml α-cyano-4-hydroxycinnamic acid as the matrix. Spectra were acquired in reflector mode and the phosphopeptides were analysed further by performing MALDI–TOF–TOF on selected masses. The characteristic loss of phosphoric acid (M−98 Da) from the parent phosphopeptide was seen. The site of phosphorylation of all the 32P-labelled peptides was determined by solid-phase Edman degradation on an Applied Biosystems 494C sequencer of the peptide coupled to Sequelon-AA membrane (Milligen) as described previously [16].
Assay of LRRK2 using moesin or MBP as substrates
Assays were set up in a total volume of 25 μl of Buffer A containing 0.5–0.7 μg of either wild-type or mutant forms of LRRK2, 1 μM moesin (full-length or indicated mutants that had been left on ice or incubated at 65°C for 15 min before assay) or 1 μM MBP, 10 mM MgCl2 and 0.1 mM [γ-32P]ATP (300 c.p.m./pmol). After incubation for 30 min at 30°C, the reactions were stopped by the addition of LDS sample buffer. The incorporation of phosphate into moesin or MBP substrates as well as LRRK2 autophosphorylation was determined after electrophoresis of samples on 4–12% polyacrylamide gels and autoradiography of the dried Coomassie Blue-stained gels. The phosphorylated substrates were also excised from the gel and 32P incorporation quantified by Cerenkov counting.
Assay of LRRK2 using LRRKtide as substrate
Assays were set up in a total volume of 50 μl of Buffer A containing 0.5–0.7 μg of either wild-type or mutant forms of LRRK2, 10 mM MgCl2 and 0.1 mM [γ-32P]ATP (300 cpm/pmol) in the presence of 300 μM or the indicated concentration of LRRKtide peptide substrate. After incubation for 30 min at 30°C, reactions were terminated by applying 40 μl of the reaction mixture on to P81 phosphocellulose paper and phosphorylation of LRRKtide was quantified following washing the P81 phosphocellulose in 50 mM phosphoric acid and cerenkov counting. A unit of LRRK2 activity was defined as the amount of enzyme that catalysed the incorporation of 1 nmol of 32P into LRRKtide. Km and Vmax parameters were determined by performing the assay described above using various concentrations of LRRKtide. The Km and Vmax parameters were calculated using the Graph-Pad Prism program.
Immunoblotting
Samples were heated at 70°C for 5 min in Sample Buffer, subjected to PAGE and transferred to a nitrocellulose membrane. Membranes were blocked for 30 min in 50 mM Tris/HCl, pH 7.5, 0.15 M NaCl and 0.2% (v/v) Tween (TBST Buffer) containing 10% (w/v) dried skimmed milk. The membranes were probed with 1 μg/ml of anti-GST antibody for 16 h at 4°C in TBST Buffer containing 5% (w/v) skimmed milk. Detection was performed using horseradish-peroxidase-conjugated secondary antibodies and an enhanced-chemiluminescence reagent.
RESULTS
Expression of an active fragment of LRRK2 for use in KESTREL
As a source of protein kinase for the KESTREL screen, we expressed GST-fusions of LRRK2 in HEK-293 cells. Following affinity purification on glutathione–Sepharose, the expression level of full-length LRRK2 was low, but an LRRK2 fragment encompassing residues 1326–2527, lacking the leucine-rich repeats, but still containing the GTPase, COR, kinase, WD40 and C-terminal tail was significantly higher (Figure 1A). The LRRK2[1326–2527] fragment autophosphorylated when incubated with magnesium and [γ-32P]ATP and phosphorylated MBP, albeit weakly (Figure 1B). A kinase-inactive mutant of LRRK2[1326-2527, D2017A], in which the Mg2+-binding Asp residue was mutated, failed to autophosphorylate or phosphorylate MBP in a parallel reaction (Figure 1B). We also found that the common PD mutant LRRK2[1326–2527,G2019S], mentioned in the Introduction, displayed about a 3-fold higher level of autophosphorylation and MBP phosphorylation compared with non-mutated LRRK2[1326–2527] (Figure 1B), consistent with previous results indicating that this mutation stimulated LRRK2 activity [12,13].
LRRK2 KESTREL screen
To search for proteins in brain that are phosphorylated by LRRK2, an extract derived from 50 rat brains was precipitated with 60% (w/v) (NH4)2SO4, desalted, chromatographed on a heparin–Sepharose column (Figure 2A), followed by a Source-Q column at pH 7.5 (Figure 2B), a Source-Q column at pH 8.2 (Figure 2C) and finally, a Superdex 200 column (Figures 2D and 2E). Aliquots of each column fraction were incubated for 15 min at 65°C in order to inactivate endogenous protein kinases that might phosphorylate proteins and hence decrease background levels of phosphorylation that can otherwise interfere with the KESTREL analysis [17]. Each fraction was then incubated in the presence or absence of GST–LRRK2[1326–2527] or GST–LRRK2[1326–2527,G2019S] and [γ-32P]ATP as described in the Materials and methods section. Utilizing purified non-mutated GST–LRRK2[1326–2527], no significant phosphorylation of any rat brain protein was detected (results not shown). Deploying the more active GST–LRRK2[1326–2527,G2019S] mutant, three proteins were observed to be phosphorylated (Figure 2). These proteins were purified, subjected to PAGE and the identity of the Coomassie Blue-stained band phosphorylated by LRRK2 in each preparation was established by tryptic-peptide MS fingerprinting procedures (Figure 3). This revealed that the proteins phosphorylated by LRRK2 were CRMP2 (collapsin response mediator protein-2), creatine kinase and moesin. CRMP2 and creatine kinase were observed to be 50–100-fold more abundant in brain extracts than moesin (A. Knebel, unpublished work). To examine the relative phosphorylation of these proteins by LRRK2, similar amounts of purified CRMP2, creatine kinase and moesin proteins were phosphorylated with GST–LRRK2[1326–2527,G2019S] and, under these conditions, moesin was phosphorylated to a markedly greater extent than was CRMP2 or creatine kinase (Figure 3). Because CRMP2 and creatine kinase are highly abundant proteins and were phosphorylated by LRRK2 much less efficiently than was moesin, we focused on studying the phosphorylation of moesin by LRRK2.
Figure 2. LRRK2[G2019S] KESTREL screen.
(A–E) Proteins extracted from rat brain that did not bind to heparin–Sepharose were sequentially chromatographed on the indicated columns. The specified fractions were phosphorylated in the presence (+) or absence (−) of GST–LRRK2[1326–2527,G2019S] and [γ-32P]ATP as described in the Materials and methods section. Phosphorylation of substrates was analysed following PAGE of the samples and autoradiography. The identity of the moesin, CRMP2 and creatine kinase as the phosphorylated substrates was established by MS as described in the legend to Figure 3. kD=kDa.
Figure 3. Identification of moesin as an LRRK2 substrate.
Fractions 10–15 from a Superdex 200 column containing the 62 kDa substrate (CRMP2) that interacted with heparin–Sepharose (Figure 2A) and which was further purified in Source-Q prior to Superdex, fractions 12–15 of a Superdex 200 column containing a 68 kDa substrate (moesin) (Figure 2) and fraction 11 of a Q-column containing a 43 kDa substrate (creatine kinase) were concentrated using a VivaScience spin filter. The samples were phosphorylated in the absence (−) or presence (+) of GST–LRRK2[1326-2527,G2019S] and [γ-32P]ATP as described in the Materials and methods section. Phosphorylation of substrates was analysed after PAGE of the samples and autoradiography. All samples were run on the same gel, but the bands shown were cut and pasted together to simplify the data. The black lines indicate where the gel was cut. The Colloidal Blue-stained bands that were phosphorylated by LRRK2 (marked with an asterisk) were excised from the gel, digested in-gel with trypsin, and their identities determined by tryptic-peptide MS fingerprinting. The Mascot score is where a value of >63 is considered significant (P<0.05). kD=kDa.
Mapping phosphorylated residues in moesin phosphorylated by LRRK2
We found that recombinant human GST-moesin expressed in E. coli which had been incubated at 65°C for 15 min (as performed in the KESTREL screen) was phosphorylated by LRRK2 in a time-dependent manner to a maximum stoichiometry of ∼0.1 mol of phosphate/mol of moesin (Figure 4A). [32P]Moesin phosphorylated with LRRK2 was digested with trypsin and chromatographed on a C18 column to isolate 32P-labelled phosphopeptides. This revealed two major peaks (P1 and P2) and one minor peak (P3) (Figure 4B). Solid-phase Edman sequencing (Figure 4C) and MS [Figure 4D and Supplementary Figure 1 (http://www.BiochemJ.org/bj/405/bj4050307add.htm) for the MALDI-TOF-TOF spectrum of P2] of P1 and P2 established their identity as peptides phosphorylated at Thr558 and P3 as a peptide phosphorylated at Thr526. We next assessed how mutation of Thr526 and Thr558 in moesin affected phosphorylation by GST–LRRK2[1326–2527,G2019S]. Mutation of Thr526 moderately decreased phosphorylation of moesin by LRRK2, whereas mutation of Thr558 significantly reduced moesin phosphorylation (Figure 4E), indicating that this was the major site of phosphorylation. No phosphorylation of moesin was observed when both Thr526 and Thr558 residues were mutated.
Further analysis of the phosphorylation of moesin by LRRK2
Moesin is a member of the ERM (ezrin/radixin/moesin) family of proteins that functions to anchor the actin cytoskeleton to the plasma membrane and plays an important role in regulating membrane structure and organization [18,19]. Moesin consists of a band FERM (four-point-one/ezrin/radixin/moesin) domain (residues 1–298) that interacts with several plasma-membrane proteins [18,19], as well as PtdIns(4,5)P2. The FERM domain on moesin is followed by an α-helical domain (residues 298–460), a flexible linker region (residues 460–489) and a conserved C-terminal tail [also termed the ‘C-ERMAD domain’ (C-terminal ERM-associated domain); residues 489–575]. The last 30 amino acids of moesin encompassing Thr558 form an F-actin-binding site [20–22]. Moesin and the other ERM proteins exist in at least two conformational states, namely an active ‘open’ form capable of binding to membranes and F-actin and an inactive or dormant ‘closed’ form incapable of linking the actin cytoskeleton to the plasma membrane, as the actin-binding site is masked. The structure of the closed state of moesin reveals that the FERM domain and C-terminal tail of moesin interact with each other, whereas in the open form the FERM and C-terminal domains are dissociated [23]. Phosphorylation of moesin at Thr558, in conjunction with the FERM domain binding membrane proteins and perhaps with PtdIns(4,5)P2, promotes the dissociation of the C-terminal tail from the FERM domain, enabling moesin to bind to F-actin [24,25]. The kinases that phosphorylate moesin at Thr558 have not been firmly established, although some candidates include the ROCK, which phosphorylates the C-terminal tail of ERM proteins in vitro and when overexpressed in cells [26–28].
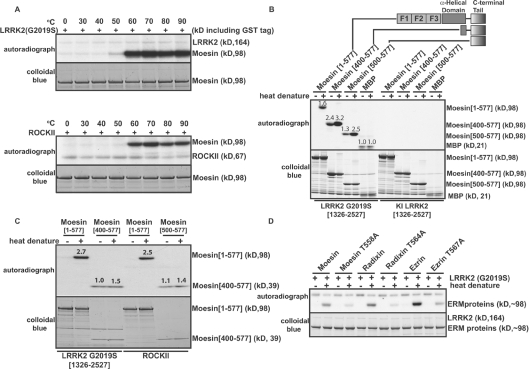
Previous studies indicated that the bacterially expressed C-terminal tail of moesin was phosphorylated by ROCK to a much greater extent than was the full-length moesin protein [26,28]. This is presumably because, when moesin is expressed in E. coli, it will be in the closed conformation in which Thr558 is inaccessible for phosphorylation. We speculated that, in the KESTREL screen, incubating moesin at 65°C before phosphorylation may have induced a conformational change that exposed Thr558. To investigate this, we studied how heating moesin expressed in E. coli affected its phosphorylation by LRRK2 (Figure 5A) as well as ROCK-II (Figure 5B) in parallel reactions. Strikingly, we found that neither LRRK2 nor ROCK-II were capable of phosphorylating moesin that had not been preincubated at a temperature of at least 60°C (Figures 5A and 5B). By contrast, fragments of moesin lacking the FERM domain could be phosphorylated by LRRK2 (Figure 5C) or ROCK-II (Figure 5D) in the absence of heat treatment prior to phosphorylation. Under the conditions employed, the moesin[400–577] C-terminal fragment was phosphorylated to an ∼2-fold greater extent than full-length GST–moesin (Figure 5B).
Figure 5. Analysis of phosphorylation of moesin by LRRK2.
(A) E. coli-expressed GST–moesin (1 μM) was incubated at the indicated temperatures for 15 min before phosphorylation with GST–LRRK2[1326–2527,G2019S] (upper panel) or ROCK-II (lower panel) at 30°C. Phosphorylation of the moesin protein was determined after PAGE and subsequent autoradiography of the Colloidal Blue-stained bands corresponding to moesin. (B and C) As in (A), except the indicated wild-type and truncated forms of moesin (all at a concentration of 1 μM) were heat-denatured by incubating them at 70°C for 15 min before phosphorylation with either active GST–LRRK2[1326–2527,G2019S] or kinase-inactive (KI) GST–LRRK2[1326–2527,D2017A] or ROCKII. Similar results were obtained in two separate experiments. Numbers above the gel bands indicate phosphorylation relative to non-heat-denatured MBP (B) or moesin[500–577] (C). (D) As above, except that phosphorylation of full-length wild-type and indicated mutants of E. coli-expressed GST–ezrin and GST–radixin by GST–LRRK2[1326–2527,G2019S] was analysed.
Ezrin and radixin are phosphorylated by LRRK2 at the residue equivalent to Thr558 of moesin
The amino acid sequence surrounding the Thr558 site of phosphorylation in moesin is identical in ezrin and radixin, suggesting that these proteins will also be phosphorylated by LRRK2. To investigate this, we studied whether LRRK2 would phosphorylate full-length ezrin and radixin that had been expressed in E. coli. Similarly to full-length moesin, ezrin and radixin were only phosphorylated by LRRK2[1326–2527,G2019S] after they were heated at 70°C (Figure 5D). Mutation of the residue equivalent to Thr558 in ezrin (Thr567) and radixin (Thr564) to alanine strongly decreased phosphorylation of these proteins by LRRK2, indicating that these are major phosphorylation sites. Under the conditions used, GST–ezrin was phosphorylated to an about 2-fold greater extent by LRRK2[1326–2527,G2019S] than GST–moesin and GST–radixin, suggesting that this might represent the best in vitro substrate to assess LRRK2 enzymic activity.
Development of a peptide-based substrate assay for LRRK2
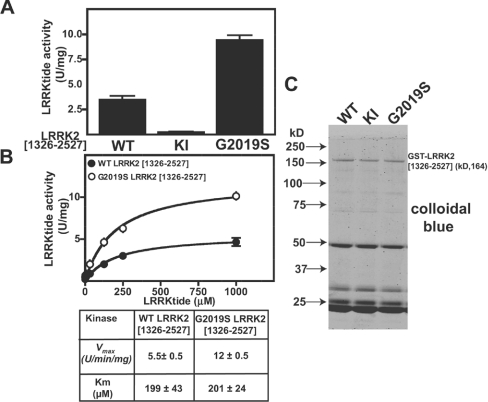
We next investigated whether LRRK2 could phosphorylate a short peptide substrate that encompassed the Thr567/Thr564/Thr558/of ezrin/radixin/moesin (RLGRDKYKTLRQIRQ; sequence identical in all three proteins) in which the underlined and emboldened threonine (T) residue is equivalent to Thr558. We found that GST–LRRK2[1326-2527,G2019S] phosphorylated this peptide at an about 3-fold higher initial rate than the non-mutated GST–LRRK2[1326–2527] under conditions in which a kinase-inactive GST–LRRK2[1326–2527,D2017A] failed to phosphorylate the peptide (Figure 6A). The peptide was termed ‘LRRKtide’ and was phosphorylated by both non-mutated GST–LRRK2[1326–2527] and GST–LRRK2[1326–2527,G2019S] with a similar Km of ∼200 μM (Figure 6B). The Vmax for the phosphorylation of LRRKtide by GST–LRRK2[1326-2527,G2019S] was ∼2.5-fold higher than that by non-mutated GST–LRRK2[1326–2527] (Figure 6B). The GST–LRRK2[1326–2527,G2019S] enzyme showed a Vmax of 10 units/mg, and the purity of the enzyme in this preparation was estimated at ∼5% (Figure 6C), suggesting that a pure preparation of the LRRK2[G2019S] enzyme would phosphorylate LRRKtide with specific activity of 200 units/min per mg – a respectable rate for a relatively active kinase phosphorylating a favourable substrate.
Figure 6. Generation of a peptide substrate for LRRK2.
(A) HEK-293 cells were transfected with constructs encoding the indicated forms of active and kinase-inactive (KI, D2017A) GST–LRRK2. At 36 h after transfection, LRRK2 kinases were affinity-purified, subjected to PAGE and stained with Colloidal Blue to quantify relative protein levels. GST–LRRK2 (1 μg of total protein from each preparation) was assayed by measuring phosphorylation of the LRRKtide peptide (RLGRDKYKTLRQIRQ) at 300 μM as described in the Materials and methods section. Results of the kinase catalytic assays are presented as the mean catalytic activity±S.D. for assays carried out in triplicate. The results presented are representative of two or three independent experiments. (B) As in (A), except that concentrations of LRRKtide were varied in order to enable calculation of the enzymatic parameters Vmax and Km. (C) A 2 μg portion of the indicated forms of GST–LRRK2 assayed in (A) was subjected to PAGE and stained with Colloidal Blue. kD = kDa; U, unit.
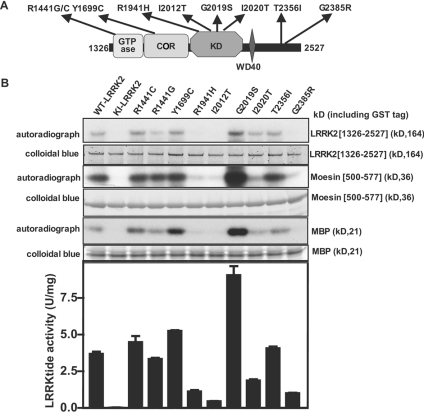
Side-by-side assay of PD mutant forms of LRRK2
Utilizing the assays elaborated on in the present study, we next compared the activity of nine mutant forms of LRRK2 that have been reported in humans suffering from PD (reviewed in [3]). The mutations studied were found in the GTPase domain (R1441C and R1441G), COR region (Y1699C), kinase domain (R1914H, I2012T, G2019S and I2020T) and in a region of the C-terminal tail that lies beyond the WD40 repeat (T2356I and G2385R) (Figure 7A). We found that only the commonly observed G2019S mutation significantly stimulated LRRK2 autophosphorylation as well as phosphorylation of moesin, LRRKtide and MBP (Figure 7B). Four mutants (R1441C, R1441G, Y1699C and T2356I) possessed an activity similar to that of non-mutated LRRK2 in all assays (Figure 7B). Two out of the four mutations in the kinase domain (R1914H and I2012T) were nearly inactive, displaying only marginally greater activity than the kinase-inactive LRRK2[D2017A] used as a control. A third kinase domain mutant, namely I2020T, possessed significantly less activity than non-mutated LRRK2, but had a higher activity than the R1914H and I2012T mutants. Intriguingly, one of the two C-terminal tail LRRK2 mutations, namely G2385R, also possessed very low catalytic activity in all assays (Figure 7B).
Figure 7. Analysis of PD LRRK2 mutants.
(A) Schematic representation showing the location of nine PD-causing mutations in LRRK2 that we analysed (the abbreviations are as given in Figure 1). (B) The non-mutated and indicated mutant forms of GST–LRRK2[1326–2527] were expressed in HEK-293 cells and affinity-purified on glutathione–Sepharose. A 2.0 μg aliquot of each preparation was subjected to PAGE and stained with Colloidal Blue to quantify relative protein levels. Each preparation was assayed by measuring autophosphorylation as well as phosphorylation of MBP, moesin [500-577] and LRRKtide peptide. The data for LRRKtide phosphorylation are presented as the mean specific activity (units per mg of total protein within purified GST–LRRK2 preparation)±S.D. for assays carried out in triplicate. The results presented are representative of two or three independent experiments. Abbreviations: WT, wild-type; KI, kinase-inactive (D2017A) LRRK2.
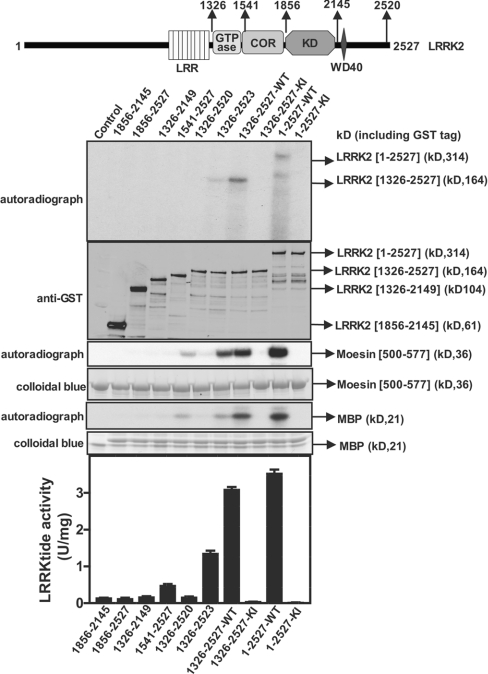
Defining the minimum fragment of LRRK2 that retains protein kinase activity
We compared the activity of full-length and mutant forms of LRRK2 lacking specific domains. Wild-type full-length LRRK2 possessed similar activity towards the moesin, LRRKtide and MBP substrates, as the did the LRRK2[1326–2527] fragment utilized in the rest of the present study. A mutant lacking either the GTPase domain (LRRK2[1541–2527]) or both the GTPase and COR domains (LRRK2[1856–2527]) displayed no autophosphorylation and did not phosphorylate any substrate. Moreover, LRRK2 mutants lacking either the C-terminal WD40 domain (LRRK2[1326–2149]) or just the seven C-terminal amino acids ([LRRK2[1326–2520]) were also inactive. Consistent with the notion that the GTPase, COR, WD40 and C-terminal region of LRRK2 are required for its activity, a fragment of LRRK2 encompassing only the kinase domain (LRRK2[1856–2145]) was devoid of any kinase activity (Figure 8).
Figure 8. Role of non-kinase domains in regulating LRRK2 activity.
Upper panel: schematic representation of the domain structure of LRRK2 showing predicted functional domains and numbering of residues corresponds to human LRRK2 residue (accession number AAV63975). Abbreviations are as given in the legend to Figure 1. Lower panels: the wild-type and indicated fragments of GST–LRRK2 were expressed in HEK-293 cells and affinity-purified on glutathione–Sepharose. A 1.0 μg aliquot of each preparation was subjected to PAGE and immunoblotted with an anti-GST antibody to quantify relative protein levels. Each preparation was also assayed by measuring autophosphorylation as well as phosphorylation of MBP, moesin[500–577] and LRRKtide. The results for the LRRKtide assay are presented as mean specific activities (units per mg of total protein)±S.D. for assays carried out in triplicate. The results presented are representative of two independent experiments. Abbreviations: WT, wild-type; KI, kinase-inactive (D2017A) LRRK2.
DISCUSSION
In the present study we employed the most active mutant G2019S form of LRRK2 encompassing residues 1326–2527 for a kinase substrate screen in brain and identified moesin and the related ezrin and radixin proteins as substrates for this enzyme. A peptide encompassing the phosphorylation site Thr558, the sequence of which is identical in moesin, ezrin and radixin, was efficiently phosphorylated by LRRK2. On the basis of these findings we developed the first robust quantitative assay for LRRK2 and analysed the requirements for catalytic kinase activity of LRRK2, by comparing a set of point mutants and deletions and to establish how mutations of LRRK2 found in PD patients have an impact on enzyme activity. The present study is another example of the usefulness of the KESTREL approach in finding substrates for poorly characterized protein kinases. Further work is required to evaluate whether moesin, ezrin or radixin are physiological substrates for LRRK2. To do this rigorously, it would be vital to assess the phosphorylation of moesin at Thr558 in LRRK2 knock-out/down mice or cells or in knock-in mice expressing the G2019S mutation. It may also be necessary to knock out or knock down the expression of the LRRK1 that most likely would also phosphorylate ERM proteins efficiently.
Were the ERM proteins found to be physiological substrates for LRRK2, the role that these might play in neurodegeneration and development of PD would require further investigation. In this regard, moesin and radixin have been implicated as playing a key role in regulating neurite outgrowth, as neurons that are deficient in these proteins display a marked reduction of growth cone size, disappearance of radial striations, retraction of the growth cone and a marked disorganization of actin filaments that invade the central region of growth cones [29]. Recent studies also demonstrate that overexpression of the activated G2019S LRRK2 mutant induces a progressive reduction in neurite length and branching both in primary neuronal-cell cultures and in the rat nigrostriatal pathway, whereas LRRK2 deficiency leads to increased neurite length and branching [30]. Taken together these data suggest that deregulation of moesin phosphorylation by mutant LRRK2 might contribute to the early loss of dopaminergic axon terminals in PD. In humans, inactivating mutations in a gene encoding a protein related to moesin, termed merlin (moesin/ezrin/radixin-like protein), cause neurofibromatosis type 2, a form of cancer affecting predominantly the nervous system [31]. Little is known regarding the identity of the kinases that phosphorylate the residue equivalent to Thr558 in merlin's C-terminal tail. It would be of interest to investigate whether LRRK2 could phosphorylate merlin at this residue.
Our results confirm that the most common G2019S LRRK2 PD mutation enhances kinase activity, a conclusion that is consistent with two recent studies in which LRRK2 activity was assessed by autophosphorylation and phosphorylation of MBP [12,13]. Our kinetic analysis with LRRKtide also indicates that the G2019S mutation stimulates LRRK2 activity by increasing the catalytic Vmax constant rather than enhancing substrate-binding Km affinity. It would be interesting to crystallize the LRRK2 catalytic domain and determine how substitution of the conserved glycine residue near to the Mg2+-aspartic acid residue in subdomain VII of the kinase domain could stimulate the catalytic efficiency of phosphorylation. Analysis of the 518 human kinases indicates that two protein kinases {TSSK1 (testis-specific serine kinase 1), BUBR1 [Bub (budding uninhibited by benomyl)-related 1]}, as well as seven predicted inactive pseudokinases [KSR2 (kinase suppressor of Ras 2), STLK6, RSKL1, SgK071, Domain2_GCN2, SgK269 and SgK196] have a glycine-to-serine substitution motif at subdomain VII of their catalytic domain [11,32]. It would be interesting to establish how mutation of the TSSK1 and BUBR1 subdomain VII serine residue to glycine affected the activity of these kinases. It is possible that such amino acid substitutions were used as an evolutionary mechanism to increase the basal activity of these enzymes. It might also be interesting to investigate the effect of glycine-to-serine mutation in other protein kinases.
Our data indicate that not all PD mutations stimulate the activity of LRRK2. We found that four PD mutations R1441C and R1441G (located in the GTPase domain), Y1699C (located in the COR domain) and T2356I (located in the C-terminal tail) did not significantly influence LRRK2 kinase activity. Moreover, three mutations R1941H and I2012T (located in the kinase domain) and G2385R (located in the C-terminal tail) markedly inhibited LRRK2 kinase activity. Another PD mutation, namely I2020T (in the residue located next to Gly2019) decreased LRRK2 autophosphorylation and phosphorylation of MBP and moesin, but to a lower extent than the R1941H, I2012T and G2385R mutations (Figure 7). A recent report has indicated that the Y1699C mutation achieved ∼50% increased autophosphorylation compared with the wild-type protein [33]. Although our results may indicate that this mutant possesses marginally greater activity than the wild-type protein (Figure 7), it is significantly less active than the G2019S mutant. Another report judged the ability of the I2020T LRRK2 mutant to autophosphorylate to a ∼40% higher level than did the wild-type LRRK2 [34], a finding that contrasts with ours, namely that this mutant possesses lower activity. The reasons for this discrepancy are not clear, but measurement of autophosphorylation is potentially unreliable, not least as it is difficult to ensure that these assays are linear. Moreover, autophosphorylation of a protein kinase is not always proportional to the intrinsic activity of the enzyme. Our results indicate that not all mutations exert their effects in the same manner as the G2019S mutation, which increases protein kinase activity. It is possible that some of the mutations exert their effects by interfering with the cellular interaction of LRRK2 with regulatory binding partners and/or alter LRRK2 cellular stability or localization. The finding that some mutations decrease kinase activity indicates that inhibition of LRRK2 might also have the potential to lead to degeneration of dopaminergic neurons and development of PD. If this were the case, it would suggest that the therapeutic efficacy of LRRK2 inhibitors might be limited to treatment of patients with activating G2019S LRRK2 mutations and that doses of such drugs would need to be utilized that do not decrease the activity of the disease-causing LRRK2[G2019S] enzyme below basal levels.
Our work also demonstrates that an intact C-terminal tail of LRRK2 is required for activity, as truncation of only the seven C-terminal residues of this region ablated LRRK2 activity (Figure 8). We also found that the G2385R PD mutation located C-terminal to the WD40 motif inactivated LRRK2 kinase activity (Figure 7). Recent work suggest that this mutation, which is especially prevalent in ethnic Chinese Taiwanese populations, may represent a polymorphism that increases the risk of developing PD rather than a PD causative mutation [35]. The C-terminal region of LRRK2, apart from the WD40 motif, shows no homology with any other known protein or other functional domain, and further analysis is required to investigate the mechanism by which this domain can regulate LRRK2.
In summary the present results define the minimum fragment of LRRK2 that retains protein kinase activity and also demonstrate that, in vitro, LRRK2 efficiently phosphorylates moesin at Thr558. Although further work is required to establish whether moesin is a physiological substrate of LRRK2, our findings will aid the functional analysis of LRRK2 by providing a more quantitative and robust methodology to assess LRRK2 protein kinase activity. They will also enable a better characterization of how different PD mutations affect LRRK2 activity and assist drug-discovery programmes in screening for LRRK2 inhibitors for the treatment of PD.
Online data
Acknowledgments
M.J. undertook most of the experimentation described in the present paper, planned experiments, contributed to writing the paper and generated the Figures; R.J.N. undertook experimentation showing that LRRK2 phosphorylated ezrin and radixin; M.D. generated the expression constructs and participated in helpful discussions; D.G.C. undertook the phosphopeptide mapping and participated in helpful discussions; F.G. supported the KESTREL screen and participated in helpful discussions; A.K. performed the KESTREL screen, participated in discussions and helped to write the manuscript; and D.R.A. conceived the project, helped plan and interpret the experiments and wrote the manuscript. We thank Dr Gerard Manning (Institute for Biological Studies, San Diego, CA, U.S.A.) for discussion on the presence of glycine-to-serine substitutions of subdomain VII motifs within kinases, Dr Douglas J. Lamont (University of Dundee) for undertaking the MS associated with the KESTREL screen, Michel Goedert for providing the LRRK2 cDNA, the Sequencing Service (School of Life Sciences, University of Dundee) for DNA sequencing, the Post Genomics and Molecular Interactions Centre for MS facilities (School of Life Sciences, University of Dundee) and the protein production and antibody purification teams [Division of Signal Transduction Therapy (DSTT), University of Dundee], co-ordinated by Dr Hilary McLauchlan and Dr James Hastie, for generation and purification of antibodies and ROCK-II. We thank the Association for International Cancer Research, Diabetes UK, the Medical Research Council, the Moffat Charitable Trust and the pharmaceutical companies supporting the Division of Signal Transduction Therapy Unit (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck & Co. Inc, Merck KgaA and Pfizer) for financial support.
References
- 1.Paisan-Ruiz C., Jain S., Evans E. W., Gilks W. P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A. M., Khan N., et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 2.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R. J., Calne D. B., et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Mata I. F., Wedemeyer W. J., Farrer M. J., Taylor J. P., Gallo K. A. LRRK2 in Parkinson's disease: protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Taylor J. P., Mata I. F., Farrer M. J. LRRK2: a common pathway for parkinsonism, pathogenesis and prevention? Trends Mol. Med. 2006;12:76–82. doi: 10.1016/j.molmed.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Farrer M., Stone J., Mata I. F., Lincoln S., Kachergus J., Hulihan M., Strain K. J., Maraganore D. M. LRRK2 mutations in Parkinson disease. Neurology. 2005;65:738–740. doi: 10.1212/01.wnl.0000169023.51764.b0. [DOI] [PubMed] [Google Scholar]
- 6.Zabetian C. P., Samii A., Mosley A. D., Roberts J. W., Leis B. C., Yearout D., Raskind W. H., Griffith A. A clinic-based study of the LRRK2 gene in Parkinson disease yields new mutations. Neurology. 2005;65:741–744. doi: 10.1212/01.wnl.0000172630.22804.73. [DOI] [PubMed] [Google Scholar]
- 7.Marin I. The Parkinson disease gene LRRK2: evolutionary and structural insights. Mol. Biol. Evol. 2006;23:2423–2433. doi: 10.1093/molbev/msl114. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg J. M., Bosgraaf L., Van Haastert P. J., Smith J. L. Identification of four candidate cGMP targets in Dictyostelium. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6749–6754. doi: 10.1073/pnas.102167299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosgraaf L., Russcher H., Smith J. L., Wessels D., Soll D. R., Van Haastert P. J. A novel cGMP signalling pathway mediating myosin phosphorylation and chemotaxis in Dictyostelium. EMBO J. 2002;21:4560–4570. doi: 10.1093/emboj/cdf438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosgraaf L., Van Haastert P. J. Roc, a Ras/GTPase domain in complex proteins. Biochim. Biophys. Acta. 2003;1643:5–10. doi: 10.1016/j.bbamcr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 12.West A. B., Moore D. J., Biskup S., Bugayenko A., Smith W. W., Ross C. A., Dawson V. L., Dawson T. M. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greggio E., Jain S., Kingsbury A., Bandopadhyay R., Lewis P., Kaganovich A., van der Brug M. P., Beilina A., Blackinton J., Thomas K. J., et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Cohen P., Knebel A. KESTREL: a powerful method for identifying the physiological substrates of protein kinases. Biochem. J. 2006;393:1–6. doi: 10.1042/BJ20051545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durocher Y., Perret S., Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell D. G., Morrice N. A. Identification of protein phosphorylation sites by a combination of mass spectrometry and solid phase Edman sequencing. J. Biomol. Techn. 2002;13:121–132. [PMC free article] [PubMed] [Google Scholar]
- 17.Troiani S., Uggeri M., Moll J., Isacchi A., Kalisz H. M., Rusconi L., Valsasina B. Searching for biomarkers of Aurora-A kinase activity: identification of in vitro substrates through a modified KESTREL approach. J. Proteome Res. 2005;4:1296–1303. doi: 10.1021/pr050018e. [DOI] [PubMed] [Google Scholar]
- 18.Bretscher A., Edwards K., Fehon R. G. ERM proteins and merlin: integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 19.Polesello C., Payre F. Small is beautiful: what flies tell us about ERM protein function in development. Trends Cell Biol. 2004;14:294–302. doi: 10.1016/j.tcb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Gary R., Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol. Biol. Cell. 1995;6:1061–1075. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pestonjamasp K., Amieva M. R., Strassel C. P., Nauseef W. M., Furthmayr H., Luna E. J. Moesin, ezrin, and p205 are actin-binding proteins associated with neutrophil plasma membranes. Mol. Biol. Cell. 1995;6:247–259. doi: 10.1091/mbc.6.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turunen O., Wahlstrom T., Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J. Cell Biol. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson M. A., Reczek D., Bretscher A., Karplus P. A. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 24.Huang L., Wong T. Y., Lin R. C., Furthmayr H. Replacement of threonine 558, a critical site of phosphorylation of moesin in vivo, with aspartate activates F-actin binding of moesin. Regulation by conformational change. J. Biol. Chem. 1999;274:12803–12810. doi: 10.1074/jbc.274.18.12803. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura F., Huang L., Pestonjamasp K., Luna E. J., Furthmayr H. Regulation of F-actin binding to platelet moesin in vitro by both phosphorylation of threonine 558 and polyphosphatidylinositides. Mol. Biol. Cell. 1999;10:2669–2685. doi: 10.1091/mbc.10.8.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsui T., Maeda M., Doi Y., Yonemura S., Amano M., Kaibuchi K., Tsukita S., Tsukita S. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J. Cell Biol. 1998;140:647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshiro N., Fukata Y., Kaibuchi K. Phosphorylation of moesin by rho-associated kinase (Rho-kinase) plays a crucial role in the formation of microvilli-like structures. J. Biol. Chem. 1998;273:34663–34666. doi: 10.1074/jbc.273.52.34663. [DOI] [PubMed] [Google Scholar]
- 28.Tran Quang C., Gautreau A., Arpin M., Treisman R. Ezrin function is required for ROCK-mediated fibroblast transformation by the Net and Dbl oncogenes. EMBO J. 2000;19:4565–4576. doi: 10.1093/emboj/19.17.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paglini G., Kunda P., Quiroga S., Kosik K., Caceres A. Suppression of radixin and moesin alters growth cone morphology, motility, and process formation in primary cultured neurons. J. Cell Biol. 1998;143:443–455. doi: 10.1083/jcb.143.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 31.McClatchey A. I., Giovannini M. Membrane organization and tumorigenesis – the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- 32.Boudeau J., Miranda-Saavedra D., Barton G. J., Alessi D. R. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 33.West A. B., Moore D. J., Choi C., Andrabi S. A., Li X., Dikeman D., Biskup S., Zhang Z., Lim K. L., Dawson V. L., Dawson T. M. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum. Mol. Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 34.Gloeckner C. J., Kinkl N., Schumacher A., Braun R. J., O'Neill E., Meitinger T., Kolch W., Prokisch H., Ueffing M. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum. Mol. Genet. 2006;15:223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 35.Farrer M. J., Stone J. T., Lin C. H., Dachsel J. C., Hulihan M. M., Haugarvoll K., Ross O. A., Wu R. M. Lrrk2 G2385R is an ancestral risk factor for Parkinson's disease in Asia. Parkinsonism Relat. Disord. 2007;13:89–92. doi: 10.1016/j.parkreldis.2006.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.