Abstract
The ‘de novo methyltransferase’ Dnmt3a (DNA methyltransferase 3a) has been shown to mediate transcriptional repression. Post-translational modification of Dnmt3a by SUMOylation affects its ability to transcriptionally repress. However, very little is known about how the SUMOylation process is regulated. In the present study, we identified a PcG (Polycomb group) protein, Cbx4 (chromobox 4), as a specific interaction partner of Dnmt3a. Co-expression of Cbx4 and SUMO-1 (small ubiquitin-related modifier-1) along with Dnmt3a in transfected cells results in enhanced modification of Dnmt3a with SUMO-1. Purified Cbx4 also promotes SUMOylation of Dnmt3a in vitro. The modification occurs in the N-terminal regulatory region, including the PWWP (Pro-Trp-Trp-Pro) domain. Our results suggest that Cbx4 functions as a SUMO E3 ligase for Dnmt3a and it might be involved in the functional regulation of DNA methyltransferases by promoting their SUMO modification.
Keywords: chromobox 4 (Cbx4), DNA methyltransferase 3a (Dnmt3a), E3 ligase, small ubiquitin-related modifier (SUMO), SUMOylation, Pro-Trp-Trp-Pro domain (PWWP domain)
Abbreviations: Cbx, chromobox; CtBP, C-terminal-binding protein; Dnmt, DNA methyltransferase; EZH2, Enhancer of Zeste homologue 2; GAL4, galactosidase 4 protein; GAL4AD, GAL4 activation domain; GAL4BD, GAL4 DNA-binding domain; GFP, green fluorescent protein; GST, glutathione S-transferase; H3K9, histone H3 Lys9; HA, haemagglutinin; HDAC, histone deacetylase; HEK-293T, human embryonic kidney 293T; HIPK2, homeodomain-interacting protein kinase 2; HP1, heterochromatin protein 1; IPTG, isopropyl β-D-thiogalactopyranoside; NLS, nuclear localization signal; Ni-NTA, Ni2+-nitrilotroacetate; PcG, Polycomb group; PHD, plant homeodomain; PIAS, protein inhibitor of activated STAT (signal transducer and activator of transcription); PWWP, Pro-Trp-Trp-Pro; SD, synthetic defined; SIP1, Smad-interacting protein 1; SUMO, small ubiquitin-related modifier; TDG, thymine DNA glycosylase
INTRODUCTION
Vertebrate genomes are modified by DNA methylation at the C5 position of cytosine residues, mostly within cytosine–guanine dinucleotides. DNA methylation is involved in the regulation of gene expression, genomic imprinting, silencing of transposable elements and endogenous retroviruses, X-chromosome inactivation and the maintenance of heterochromatin [1,2].
The establishment and maintenance of methylation patterns in mammals rely on three Dnmts (DNA methyltransferases). Whereas Dnmt1 maintains the existing methylation patterns during DNA replication, Dnmt3a and Dnmt3b are responsible for the generation of new methylation patterns during gametogenesis and early development and therefore also known as ‘de novo methyltransferases’ [2]. The functions of Dnmt3a and Dnmt3b are crucial, since mice lacking the corresponding genes failed to complete early development [3]. Structurally, Dnmt3a and Dnmt3b contain an N-terminal regulatory domain fused to the C-terminal catalytic domain [2]. Moreover, it has been further demonstrated that the N-terminal region contains a PWWP (Pro-Trp-Trp-Pro) domain which is required for chromatin targeting and methylation of heterochromatin DNA repeats [4,5] and a cysteine-rich PHD (plant homeodomain) zinc-finger domain, which interacts with other proteins, such as HDACs (histone deacetylases), HP1β (heterochromatin protein 1β) and SUV39H1 [suppressor of variegation 3-9 homologue 1 (Drosophila)] [6–9].
A number of proteins have been found to be modified by covalent attachment of SUMO (small ubiquitin-related modifier). SUMOylation regulates protein–protein interaction, enzyme activity, protein stability and subcellular localization [10,11]. An overwhelming majority of the SUMOylated proteins are involved in nuclear processes, including maintenance of nuclear integrity, control of nuclear transport and regulation of chromatin functions, such as transcription and DNA repair [10,11] and heterochromatin assembly [12].
Four SUMO family members, SUMO-1, -2, -3 and -4, have been currently identified in mammals [11]. Mature SUMO is generated firstly by cleavage of a short C-terminal extension from the precursor by a processing protease. Following the cleavage is the conjugation process. SUMO is first activated by an E1 activating enzyme, which consists of an Aos1/Uba2 heterodimer [13], and is then transferred to Ubc9, the E2 conjugating enzyme [14]. Finally, SUMO is covalently linked to the ε-amino group of a given lysine residue in the substrate protein. Although SUMO can be conjugated to its substrate by E1 and Ubc9 in vitro, an additional SUMO E3 ligase is usually required for efficient and specific modifications in vivo. Currently, three classes of E3 ligases have been described. Members of the first class characterized are PIASs [protein inhibitors of activated STAT (signal transducer and activator of transcription)] that promote SUMOylation of numerous proteins, such as p53 and c-Jun [15,16]. The second class is represented by RanBP2 (RAN binding protein 2), which is found to stimulate SUMO modification of proteins such as Sp100 (speckled protein of 100 kDa) and HDAC4 [17,18]. Cbx (chromobox) 4 (Pc2) is the third class of SUMO E3 ligases with no obvious sequence similarity to other known E3s [19]. Cbx4 is a component of PcG (Polycomb group) proteins, which were originally identified in Drosophila as stable repressors of homeotic genes [20]. It interacts with other PcG proteins and functions as a transcriptional repressor [21,22]. Recently, the function of Cbx4 as an E3 ligase to promote SUMOylation of transcriptional repressors CtBP (C-terminal binding protein), SIP1 (Smad-interating protein 1) and HIPK2 (homeodomain interacting kinase 2) was identified both in vivo and in vitro [19,23–25].
Dnmt3a and Dnmt3b interact with Ubc9 and SUMO-1, and SUMOylation of both enzymes has been detected [9,26]. SUMOylation of Dnmt3a disrupts its ability to interact with HDACs and impairs its capacity to repress transcription [9]. However, the regulation of SUMOylation of Dnmt3 proteins remains unknown. In the present study we show that Cbx4 interacts specifically with Dnmt3a and enhances SUMOylation of Dnmt3a both in vivo and in vitro. These data suggest that Cbx4 functions as a SUMO E3 ligase toward Dnmt3a. The modification sites of Dnmt3a are mapped to the N-terminal region encompassing the PWWP domain. Our results suggest that the Polycomb protein Cbx4 might have an impact on the function of Dnmt3a by promoting its SUMO modification.
EXPERIMENTAL
Mammalian expression constructs
Dnmt3a cDNA was cloned into pcDNA3-HA as described previously [4]. Different regions of Dnmt3a, namely 1–277, 94–482, 192–482, 270–482 and 474–908, were cloned into pcDNA3-HA by PCR amplification using pcDNA3-HA-Dnmt3a as a template (HA is haemagglutinin). pcDNA3-HA-Dnmt3a (amino acids 1–482) has been described previously [4]. The Dnmt3a point mutant K573R was introduced by PCR amplification using pcDNA3-HA-Dnmt3a as a template and cloned into pcDNA3-HA. Expression constructs for CtBP [27] and Cbx2, Cbx7, Cbx6 and Cbx8 [28] have been described previously. Full-length Cbx4 was PCR amplified from mouse brain cDNA (Clontech) and cloned into pFlag-CMV-2 (Sigma; where CMV is cytomegalovirus). SUMO-1 (amino acids 1–97) was PCR-amplified from mouse testis cDNA (Clontech) and cloned into pEGFP-C1 (Clontech) and pcDNA4-Flag respectively. All PCR-cloned constructs were verified by DNA sequencing.
Cell culture and transfection
HEK-293T (human embryonic kidney-293T) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum. Plasmids were transfected into the cells using Lipofectamine™ (Invitrogen) according to the manufacturer's instructions.
Antibodies
Polyclonal antibody specific for Dnmt3a was raised in rabbits and affinity-purified as described previously [4]. Polyclonal antibody to GST (glutathione S-transferase)-tagged Cbx4 (amino acids 144–362) was raised in rabbit and affinity-purified by binding to recombinant-protein-coupled glutathione–Sepharose 4B columns (Amersham Biosciences). Polyclonal anti-GFP (green fluroescent protein) antibody was purchased from Santa Cruz Biotechnology. Monoclonal anti-histidine antibody, anti-HA antibody, anti-Flag antibody and anti-GST antibody were purchased from Sigma. Monoclonal anti-myc antibody was purchased from Wolwo Biotech Co. Ltd, Shanghai, China.
Yeast two-hybrid constructs and assays
For bait construction, the full-length Dnmt3a coding sequence was cloned into pGBKT7 (Clontech). Two-hybrid screening was performed by mating yeast strain AH109 harbouring GAL4BD [GAL4 (galactosidase 4 protein) DNA-binding domain]-Dnmt3a with strain Y187, which carries a human brain expression library (Clontech). Colonies were selected on the SD (synthetic defined) medium lacking leucine, tryptophan, adenine and histidine according to the manufacturer's (Clontech) instructions. The prey plasmids from positive colonies were then isolated and transformed into Escherichia coli strain Top10. Amplified plasmids were identified by DNA sequencing. For mapping the regions of the proteins involved in the interaction, full-length Cbx4 coding sequence was subcloned into prey vector pGADT7 (Clontech) from pFlag-CMV. Fragments of Cbx4 were cloned into pGADT7 by PCR amplification using pFlag-CMV-Cbx4 as a template. Dnmt3a fragments 1–591, 1–277, 270–482, 270–430, 478–598 and 592–908 were subcloned into pGBKT7 from pcDNA3-HA mammalian expression constructs. Dnmt3a fragment 431–482 was cloned into pGBKT7 by PCR amplification using pcDNA3-HA-Dnmt3a as a template. For examining the interactions between Dnmt3a and other PcG proteins, Cbx2, Cbx6, Cbx7 and Cbx8 coding sequences were cloned in-frame with GAL4AD (GAL4 activation domain) on the prey vector pGADT7, respectively. All PCR-cloned constructs were verified by DNA sequencing.
Protein expression and purification
The Dnmt3a coding sequences for the full-length and an N-terminal region (amino acids 1–482) were cloned into pGEX-4T-3 (Amersham Biosciences) for the expression of the GST fusion proteins in the E. coli strain BL21 (DE3). Purification was performed using glutathione–Sepharose 4B (Amersham Biosciences) as described in the manufacturer's instructions, following the induction of the cells with 0.5 mM IPTG (isopropyl β-D-thiogalactopyranoside) at 27°C for 3 h. For purification of histidine-tagged Cbx4, full-length Cbx4 coding sequence was cloned into pET28a (Novagen), then transformed into the E. coli strain BL21 (DE3) CodonPlus-RP (Stratagene) and cultured at 37°C. When the attenuance (D600) reached 0.6, IPTG was added to a final concentration of 5 μM and the culture was grown for an additional 12 h at 18°C. The cells were harvested by centrifugation (at 4000 g for 10 min) and washed with a lysis buffer [50 mM sodium phosphate, pH 8.0, 300 mM NaCl and 10% (v/v) glycerol] and resuspended in the lysis buffer supplemented with 10 mM 2-mercaptoethanol, 1 μg/ml leupeptin and 1 mM PMSF. After sonication (200 W; 10 s on/10 s off; 40 cycles; JY92-II ultrasonic homogenizer; Ningbo Scientz Biotechnology Co. Ltd, Ningbo, China), the lysate was centrifuged (20000 g for 30 min) and the supernatant was loaded on to an Ni-NTA (Ni2+-nitrilotroacetate) column (Qiagen). The column was washed with the lysis buffer supplemented with 10 mM 2-mercaptoethanol and 20 mM imidazole and eluted with the lysis buffer containing 10 mM 2-mercaptoethanol and 250 mM imidazole. Aos1, Uba2, Ubc9 and SUMO-1 (amino acids 1–97) were expressed as histidine-tagged fusion proteins. pET30-Aos1 and pET30-Uba2 have been described previously [29]. The Ubc9 coding sequence was cloned into pET28a by PCR using pACT2-Ubc9 (H. Jin, unpublished work) as a template. pET30-SUMO-1 was kindly provided by Zhihong Zhang (Institute of Health Sciences, Chinese Academy of Sciences, Shanghai, China). The E. coli strain BL21 (DE3) containing the expression constructs was grown at 37°C. When D600 had reached 0.6, IPTG was added to a final concentration of 0.5 mM and the culture was grown for an additional 3 h at 30°C. The fusion proteins were purified with the use of an Ni-NTA column (Qiagen) according to the manufacturer's instructions. The purified proteins were dialysed and concentrated. All purification steps were carried out at 4°C.
GST pull-down assays
The GST pull-down assay was performed essentially as described [30]. The GST–Dnmt3a fusion protein purified from bacteria was incubated with glutathione–Sepharose 4B beads (Amersham Biosciences) in the binding buffer [20 mM Tris/HCl, pH 7.9, 0.1 M NaCl, 1 mM EDTA, 5 mM MgCl2, 0.1% (v/v) Nonidet P40, 1 mM dithiothreitol, 0.2 mM PMSF and 20% (v/v) glycerol] in a total volume of 200 μl at 4°C for 2 h. The GST–Dnmt3a slurry was washed with the binding buffer containing 1 M NaCl twice and equilibrated with the binding buffer twice. After the purified Cbx4 protein and the GST-Dnmt3a slurry were treated with micrococcal nuclease at a final concentration of 0.1 units/μl at 30°C for 10 min, 500 ng of Cbx4 protein was added to the slurry and incubated at 4°C for another 2 h. The Sepharose beads were vigorously washed five times with the binding buffer, and bound proteins were resolved by SDS/PAGE and visualized by Western blotting using anti-histidine antibody.
For ethidium bromide treatment of the protein preparations, the GST–Dnmt3a fusion protein was incubated with glutathione–Sepharose 4B beads (Amersham Biosciences) in the binding buffer in a total volume of 200 μl at 4°C for 2 h. Then the GST–Dnmt3a slurry was washed with the binding buffer containing 1 M NaCl twice and equilibrated with the binding buffer twice. The slurry was incubated with 500 ng of Cbx4 protein at 4°C for 2 h in the presence of 50 μg/ml ethidium bromide. The Sepharose beads were washed five times with the binding buffer. The original concentration of ethidium bromide was maintained during the washing steps.
Immunoprecipitation and Western blotting
For co-immunoprecipitation of Dnmt3a and Cbx4, HEK-293T cells were plated in 60-mm-diameter dishes and were transfected with the mammalian expression constructs pcDNA3-HA-Dnmt3a and pFlag-CMV-Cbx4. After 48 h of incubation, 5×106 transfected cells were washed with PBS and lysed in lysis buffer [20 mM Tris/HCl, pH 7.4, 150 mM NaCl, 0.1% (v/v) Nonidet P40, 1 μg/ml leupeptin, 1 μg/ml aprotinin and 1 mM PMSF]. After brief sonication (100 W; 10 s on/10 s off; seven cycles), the lysates were cleared by centrifugation (20000 g, at 4°C for 20 min). Supernatants were incubated with the anti-Flag or anti-HA antibodies at 4°C for 4 h. Protein G–agarose beads (Roche) and the mixture were rotated at 4°C overnight. The beads were washed four times with the lysis buffer and the immunoprecipitated proteins were resolved by SDS/PAGE, transferred to nitrocellulose membranes (Bio-Rad) and probed with antibodies. The ECL (enhanced chemiluminescence) kit (Millipore) was used to visualize immunoreactive proteins. For direct analysis of whole-cell extracts, 1×106 cells were washed with PBS and lysed in SDS-loading buffer and analysed by SDS/PAGE and Western blotting.
For detection of SUMOylation by immunoprecipitation, a procedure described previously [31] was used. Briefly, 5×106 of transfected HEK-293T cells were washed with PBS and lysed in lysis buffer [50 mM Tris/HCl, pH 7.4, containing 150 mM NaCl, 1% (w/v) sodium deoxycholate, 1% (v/v) Triton X-100, 0.1% SDS, 10 mM N-ethylmaleimide, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A and 1 mM PMSF]. After brief sonication, the lysates were cleared by centrifugation at 4°C for 20 min. The supernatants were incubated with mouse monoclonal anti-HA antibody and Protein G–agarose beads (Roche) for 4 h at 4°C with gentle rotation. The beads were washed four times with the lysis buffer. Bound proteins were analysed by SDS/PAGE and Western blotting.
For analysis of expression of GAL4AD and GAL4BD fusion proteins in yeast, cells were grown in 30 ml of SD medium lacking leucine and tryptophan were harvested at a D600 of 0.8–1.0, washed with PBS, and lysed by glass-bead disruption in lysis buffer containing 50 mM Tris/HCl, pH 7.5, 50 mM NaCl, 10% (v/v) glycerol, 1 mM EDTA, 0.5% (v/v) Nonidet P40, 1 mM dithiothreitol, 1 μg/ml aprotinin, 1 mM benzamidine, 1 μg/ml leupeptin, 1 μg/ml pepstatin A and 1 mM PMSF. Cell extracts were then clarified by centrifugation at 4°C and analysed by SDS/PAGE and Western blotting.
In vitro SUMOylation assays
In vitro SUMOylation reactions were performed using purified recombinant proteins according to a previously described procedure [19] with some modifications. The reaction mixtures (20 μl each) contained 50 mM Tris/HCl, pH 7.5, 5 mM MgCl2, 2 mM ATP, 250 ng of Aos1/Uba2, 100 ng of Ubc9, 3 μg of SUMO-1 and 1μg of GST–Dnmt3aN (amino acids 1–482). Reactions were carried out at 30°C for 2 h and terminated by the addition of SDS loading buffer. Products were analysed by SDS/PAGE and Western blotting. For analysis of the effect of Cbx4 on Dnmt3a SUMOylation, reactions were performed as described above, except that the amount of Ubc9 was decreased to 20 ng and 25–100 ng of Cbx4 was added.
RESULTS
Dnmt3a interacts with Cbx4 both in vitro and in vivo
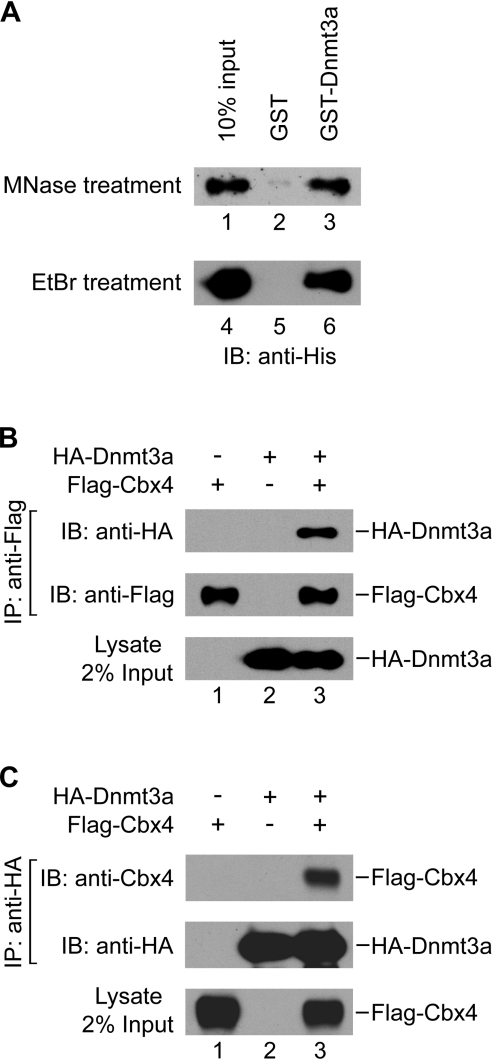
To gain insight into the regulation of the function of the de novo Dnmts, we sought to identify proteins that interact with Dnmt3a. We carried out 2 yeast two-hybrid screen, using the full-length Dnmt3a as the bait. Two positive clones from a human brain library were identified as encoding a Cbx-containing PcG protein termed Cbx4 or Pc2 [22]. To validate the interaction between Dnmt3a and Cbx4, we applied a GST pull-down assay with recombinant mouse Cbx4 and Dnmt3a proteins purified from E. coli. As shown in Figure 1(A), a significant amount of His-tagged Cbx4 (about 10% of the input) was co-precipitated with GST–Dnmt3a (lane 3) but not with GST alone (lane 2), indicating a direct interaction between the two proteins. The protein preparations had been treated with micrococcal nuclease before the pull-down assay to eliminate the possibility of a false positive interaction, which may arise due to contamination of nucleic acid present in the samples [30]. The activity of micrococcal nuclease was verified by complete digestion of the λ-DNA (results not shown).
Figure 1. Cbx4 interacts with Dnmt3a in vitro and in vivo.
(A) Direct interaction of Cbx4 with Dnmt3a: Western-blotting analysis of the His-tagged Cbx4 in fractions obtained from GST pull-down assays using GST (lanes 2 and 5) or GST–Dnmt3a (lanes 3 and 6). About 10% of the input Cbx4 was loaded (lanes 1 and 4). To eliminate the possibility of false-positive interactions caused by contaminating nucleic acid, micrococcal nuclease (MNase) or ethidium bromide (EtBr) was used to treat the protein preparations during pull-down assays. (B) Co-immunoprecipitation (IP) of Dnmt3a with Cbx4 expressed from transfected cells. HEK-293T cells were transiently transfected with pcDNA3-HA-Dnmt3a and pFlag-CMV-Cbx4 alone or in combination as indicated at the top. Whole-cell extracts were immunoprecipitated with anti-Flag antibody. The precipitated proteins were analysed by Western blotting using anti-HA or anti-Flag antibody as indicated. (C) Co-immunoprecipitation of Cbx4 with Dnmt3a from transfected cells. Samples immunoprecipitated with anti-HA antibody were analysed by Western blotting using anti-Cbx4 or anti-HA antibody as indicated.
To further demonstrate that the interaction is not caused by contamination of DNA, we performed GST pull-down assays in the presence of 50 μg/ml ethidium bromide, which is usually used to disrupt protein–DNA interactions [32]. Ethidium bromide had no effect on Dnmt3a and Cbx4 association, excluding the possibility that DNA contributes to the interactions (Figure 1A, lane 6).
To confirm this interaction in vivo, we co-transfected Flag-tagged Cbx4 and HA-tagged Dnmt3a into HEK-293T cells; afterwards, co-immunoprecipitation assays were conducted. We found that the transfected Dnmt3a could be immunoprecipitated by Cbx4 (Figure 1B, lane 3), and vice versa (Figure 1C, lane 3). This interaction is specific, because there was no precipitate detected when either Dnmt3a or Cbx4 was transfected alone (Figures 1B and 1C). Taken together, our data show that Dnmt3a specifically interacts with Cbx4 both in vitro and in vivo.
Mapping of interaction domains of Cbx4 and Dnmt3a
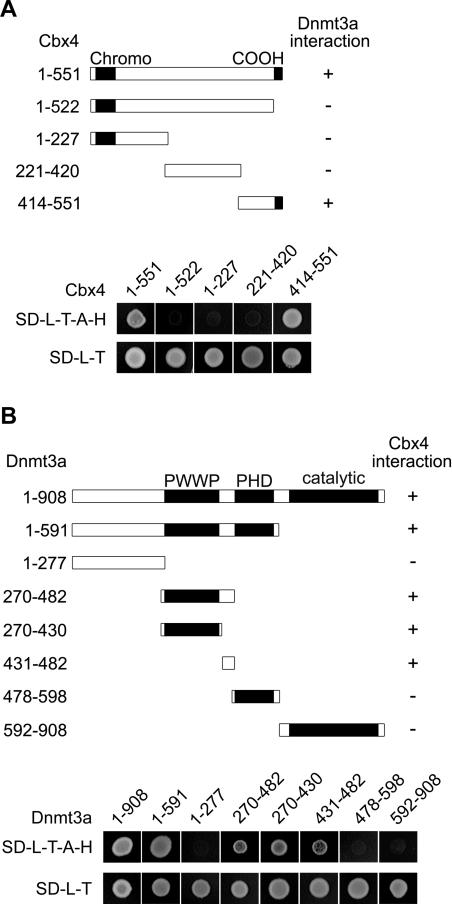
To define domains that are responsible for the interaction, we constructed a series of Cbx4 and Dnmt3a deletion mutants for yeast two-hybrid assays. Fragments of Cbx4 were expressed as fusion proteins with the GAL4 activation domain (GAL4AD) and tested for interaction with full-length Dnmt3a fused with GAL4BD. Cbx4 contains two previously characterized functional domains (Figure 2A). The N-terminal region is a highly conserved chromodomain, which binds to histone H3 when methylated at Lys9 [28]. The C-terminus contains a conserved COOH box that is involved in transcriptional silencing and binding to other PcG proteins [33,34]. We found that yeast expressing the C-terminal part (amino acids 414–551) could grow on the selective medium lacking leucine, tryptophan, adenine and histidine, but neither the N-terminal (amino acids 1–227) nor the middle region (amino acids 221–420) endowed growth potential (Figure 2A). The deletion mutant (amino acids 1–522) lacking the C-terminal region of 29 amino acids lost the capacity to interact. These data indicate that the C-terminal region, including the COOH box, is required for the interaction with Dnmt3a.
Figure 2. Mapping of interaction domains in Cbx4 and Dnmt3a.
(A) Schematic representation of Cbx4 fragments with their Dnmt3a-interacting abilities (upper panel, shown on the right) based on the data from yeast two-hybrid assays (lower panel). The full-length Dnmt3a was fused to GAL4BD. The fusion construct was co-transformed into yeast strain AH109 with the indicated portions of Cbx4, which were fused to the GAL4AD. Growth of colonies on the SD minimal medium (SD-L-T-A-H) lacking leucine, tryptophan, adenine and histidine reflects positive interaction of Cbx4 fragments with Dnmt3a. Regions of conservation (the chromodomain and the COOH box) are depicted with black shading in the bars. (B) Schematic representation of Dnmt3a fragments with their Cbx4-interacting abilities (upper panel, shown on the right) based on the data from yeast two-hybrid assays (lower panel). Full-length Cbx4 was fused to GAL4AD and was co-transformed into yeast strain AH109 with the indicated portions of Dnmt3a, which are fused to the GAL4BD. Growth of colonies on the SD minimal medium (SD-L-T-A-H) reflects positive interactions.
Similarly we mapped the Dnmt3a interaction region for Cbx4 (Figure 2B). Fragments of Dnmt3a were expressed as fusion proteins with GAL4BD and tested for interaction with full-length Cbx4 fused with the GAL4AD. Analysis of seven deletion mutants led to the identification of two continuous regions which are able to mediate the interaction independently. One is the PWWP domain (amino acids 270–430) and the other is the linker region (amino acids 431–482) between the PWWP and PHD domains. Neither the N-terminal region (amino acids 1–277), the PHD domain (amino acids 478–598) nor the C-terminal catalytic domain (amino acids 592–908) could interact with Cbx4. From these results we conclude that the extended PWWP region in Dnmt3a (amino acids 270–482) is responsible for the interaction with Cbx4.
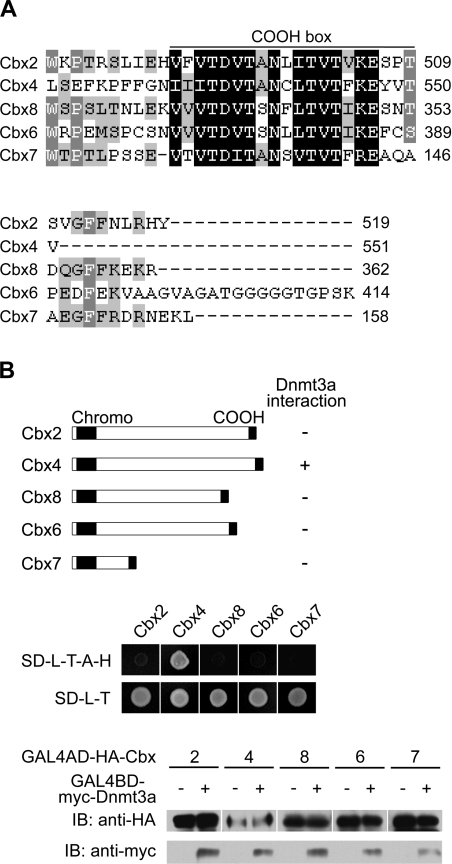
Dnmt3a interacts with Cbx4 specifically
There are five Polycomb proteins identified in the mouse, known as Cbx2 (mPc1 or M33), Cbx4 (mPc2), Cbx8 (mPc3), Cbx6 and Cbx7, all of which contain a chromodomain and a COOH box [35] (Figure 3). Given the high degree of conservation in the COOH box region of Cbx4 (Figure 3A) shown to be required for interaction with Dnmt3a, we then examined whether the other Polycomb proteins are also able to interact with Dnmt3a by 2 yeast two-hybrid assay. As shown in Figure 3(B), all the other four Polycomb proteins failed to interact with Dnmt3a, even though they were well-expressed in yeast, as confirmed by Western blotting (Figure 3B, lower panel). The above data indicate that interaction with Dnmt3a is a unique property of Cbx4. The variable positions in the COOH box and/or adjacent regions of Cbx4 contribute to the specific binding to Dnmt3a.
Figure 3. Dnmt3a interacts with Cbx4 specifically.
(A) Amino acid alignment of the COOH boxes and their flanking regions of the Cbx family members. Identical residues are indicated by black shading and highly conserved residues by grey shading. (B) Schematic representation of Cbx proteins with their Dnmt3a-interacting abilities (upper panel, shown on the right) based on the data from yeast two-hybrid assays (middle panel). Cbx proteins were fused to GAL4AD and co-transformed into yeast strain AH109 with GAL4BD–Dnmt3a. The interactions were accessed by growth of colonies on the SD minimal medium (SD-L-T-A-H). The GAL4AD-Cbx and GAL4BD-Dnmt3a fusion proteins also contain an HA or a myc epitope tag respectively, which were used to confirm the expression of Cbx proteins and Dnmt3a in transformed yeast by Western blotting (bottom panel).
Dnmt3a is SUMOylated in an extended PWWP region
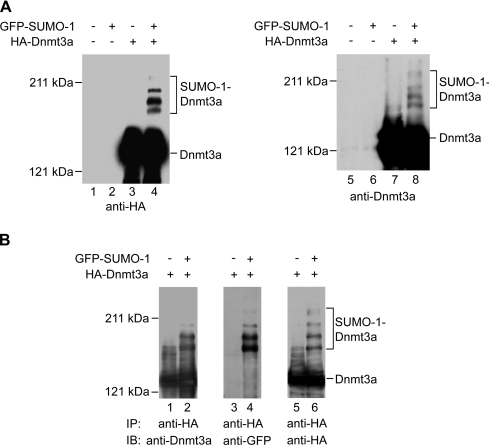
It is known that Dnmt3a can be modified by SUMO-1 [9]. However, the SUMOylated sites remain unknown. Thus we sought to determine the potential regions of SUMOylation using transfections in HEK-293T cells. First, we verified the SUMO-1 modification of Dnmt3a by co-transfection of HEK-293T cells with HA-tagged Dnmt3a and GFP-tagged SUMO-1. Our results showed that at least four high-molecular-mass forms of Dnmt3a could be detected in addition to the major Dnmt3a form of 130 kDa in the protein extract from cells expressing GFP–SUMO-1 with anti-HA antibody (Figure 4A, lane 4). A similar result was obtained when anti-Dnmt3a antibody was used in the Western-blotting analysis (lane 8), suggesting that Dnmt3a might be polySUMOylated in vivo.
Figure 4. Dnmt3a is SUMOylated in HEK-293T cells.
(A) Western-blotting analysis of the SUMOylation of Dnmt3a. HEK-293T cells were transiently transfected with expression plasmids as indicated on the top. Cell lysates were analysed by Western blotting with anti-HA antibody (left panel) or with anti-Dnmt3a antibody (right panel). The positions of Dnmt3a and its SUMOylated forms are indicated. (B) Immunoprecipitation analysis of the SUMOylation of Dnmt3a. HEK-293T cells were transiently transfected with expression plasmids as indicated on the top. The cell extracts were immunoprecipitated with anti-HA antibody. The precipitated proteins were analysed by Western blotting with anti-Dnmt3a antibody (lanes 1 and 2), anti-GFP antibody (lanes 3 and 4) or anti-HA antibody (lanes 5 and 6). The positions of Dnmt3a and its SUMOylated forms are indicated.
To confirm that the high-molecular-mass bands represent SUMOylated forms of Dnmt3a, we then performed an immunoprecipitation for the transfected HA-Dnmt3a, followed by Western blotting against Dnmt3a itself and the GFP–SUMO moiety respectively. The lysates of co-transfeceted cells were immunoprecipitated with anti-HA antibody and the immunoprecipitates were analysed by Western blotting. The high-molecular-mass forms were detected again using the anti-Dnmt3a antibody (Figure 4B, lane 2), suggesting that the extra bands are indeed modified forms of Dnmt3a. More importantly, the same pattern of slowly migrating bands was detected with the anti-GFP antibody (Figure 4B, lane 4), indicating that Dnmt3a is conjugated with GFP–SUMO-1. For further confirmation, we stripped the membrane blotted with anti-GFP antibody and reprobed it with anti-HA antibody. Our data revealed that the high-molecular-mass bands detected by anti-GFP antibody also cross-reacted with anti-HA antibody (Figure 4B, right panel). These results together confirm the polySUMOylation of Dnmt3a in HEK-293T cells.
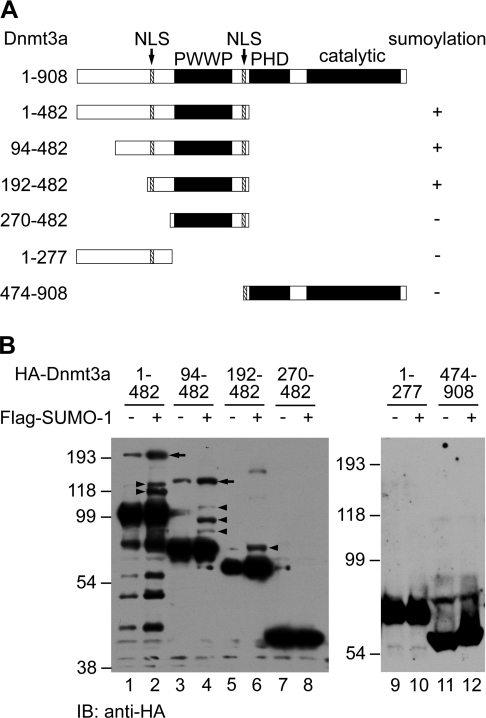
Considering that Dnmt3a is polySUMOylated in HEK-293T cells (Figure 4), we speculated that Dnmt3a might contain multiple accepting sites for SUMO. Furthermore, we searched for potential sites in Dnmt3a similar to the consensus SUMO modification target sequence ΨKXE/D (where Ψ is a hydrophobic residue, preferably L, I, F or V, and X is any residue) [36] and identified only one candidate site (IKED) at Lys573. However, mutation of Lys573 to arginine did not impair the SUMOylation of Dnmt3a (results not shown). In order to determine the location of SUMOylation site(s), we co-expressed a series of N- and C-terminal deletion mutants of Dnmt3a with SUMO-1 in HEK-293T cells and analysed their SUMOylation status by Western blotting. As shown in Figure 5(B), SUMOylation products were detected from mutant proteins harbouring amino acids 1–482, 94–482 and 192–482 (lanes 2, 4 and 6, arrowheads). Notably, mutant protein of amino acids 94–482 has three SUMOylated forms, but the 192–482 protein has only one, indicating that two accepting sites reside between amino acids 94 and 191. Further deletion from the N-terminus abolished SUMOylation of the truncated protein (amino acids 270–482, lane 8). Although the region 1–277 presumably harbours two accepting sites and an NLS (nuclear localization signal) (Figure 5A), which is necessary for SUMO-1 conjugation in vivo [37], it cannot be modified on its own. This suggests that the intact PWWP domain is necessary for a substrate fragment to be SUMOylated. The C-terminal half of Dnmt3a harbouring a PHD and a catalytic domain did not appear to be involved, since no SUMOylation of the 474–908 mutant occurred (Figure 5B, lane 12). Thus we conclude that the conjugation domain in Dnmt3a is located between amino acids 192 and 482 (Figure 5A). Notably, we also detected a large band with an additional molecular mass of ∼80 kDa which cross-reacted with anti-HA antibody (Figure 5B, lanes 1–4, arrows). It migrated more slowly than the largest form of SUMO-1-conjugated Dnmt3a and appeared in the absence of Flag-SUMO-1, suggesting that the band was not derived from SUMOylation. We surmise that an unknown post-translational modification might occur in the N-terminal region of Dnmt3a in HEK-293T cells.
Figure 5. Mapping of the Dnmt3a region necessary for SUMOylation.
(A) Schematic overview of the Dnmt3a deletion mutants used. The NLS sequences are indicated by arrows. The ability to be SUMOylated was assessed on the basis of the data in (B) and is indicated on the right. (B) Western-blotting analysis of the SUMOylation of Dnmt3a deletion mutants. HEK-293T cells were transiently transfected with expression plasmids encoding different regions of Dnmt3a, as indicated on the top. Cell lysates were analysed by Western blotting with anti-HA antibody. The positions of SUMOylated forms of Dnmt3a fragments are marked by arrowheads. Arrows depict the additional bands which might represent an unknown form of post-translational modification of Dnmt3a.
Cbx4 enhances SUMOylation of Dnmt3a in vivo and in vitro
Although we and Ling et al. [9] demonstrated that Dnmt3a can be SUMOylated in mammalian cells, its specific SUMO E3 ligase remains unknown. Considering that human Cbx4 has been shown to act as a SUMO E3 ligase for the SUMOylation of CtBP, SIP1 and HIPK2 [19,23–25] and our findings that Dmnt3a interacts with Cbx4, we further explored the possibility that Cbx4 functions as an E3 ligase toward Dnmt3a.
First we tested our hypothesis by examining whether the SUMOylation of Dnmt3a could be promoted by Cbx4 in intact cells. We confirmed that the mouse homologue of Cbx4 under investigation can also function as a SUMO E3 ligase for CtBP (Figure 6A, lane 4). Then we tested whether Cbx4 could enhance SUMOylation of Dnm3a. A plasmid expressing Flag-tagged Cbx4 was co-transfected along with the expression vectors for HA-tagged Dnmt3a and GFP-tagged SUMO-1 into HEK-293T cells, and the SUMOylation of Dnmt3a was analysed by Western blotting. As shown in Figure 6(B), the intensity of SUMOylated forms of Dnmt3a increased significantly when Cbx4 was co-expressed in the presence of GFP–SUMO-1 (compare lanes 5 and 4). Repeated experiments confirmed that the increase in SUMOylation was not caused by variations in the expression levels of GFP–SUMO-1 and HA–Dnmt3a, both of which were believed to exist in excess in the transfected cells.
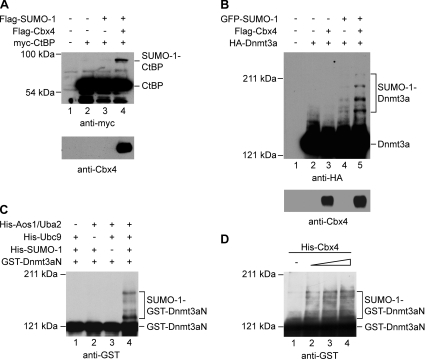
Figure 6. Cbx4 enhances SUMOylation of Dnmt3a in vivo and in vitro.
(A) Cbx4 promotes SUMO-1 conjugation to CtBP in transfected cells. HEK-293T cells were transiently transfected with expression plasmids encoding Flag–SUMO-1, Flag–Cbx4 and myc-CtBP as indicated. The cell lysates were analysed by Western blotting using anti-myc (upper panel) or anti-Cbx4 (lower panel) antibody. (B) Cbx4 promotes SUMO-1 conjugation to Dnmt3a in transfected cells. HEK-293T cells were transiently transfected with expression plasmids encoding GFP–SUMO-1, Flag–Cbx4 and HA–Dnmt3a as indicated. The cell lysates were analysed by Western blotting using anti-HA (upper panel) or anti-Cbx4 antibody (lower panel). (C) SUMOylation of Dnmt3a in vitro. Purified GST-tagged Dnmt3a (amino acids 1–482, GST–Dnmt3aN) was incubated in the absence or presence of 250 ng of recombinant E1 (Aos1/Uba2 heterodimer), 100 ng of recombinant E2 (Ubc9) and 3 μg of recombinant SUMO-1 as indicated. Reactions were terminated by the addition of SDS loading buffer. Proteins were analysed by Western blotting using anti-GST antibody. (D) Cbx4 enhances SUMOylation of Dnmt3a in vitro. Purified GST–Dnmt3aN was incubated with 250 ng of E1 (Aos1/Uba2 heterodimer), 20 ng of E2 (Ubc9) and 3 μg of SUMO-1. Lanes 1–4 contained 0, 25, 50 or 100 ng of purified His-Cbx4 respectively. Note that the amount of Ubc9 used here was 5-fold lower than that used in (C).
The stimulatory effect of Cbx4 on SUMOylation of Dnmt3a raises the question as to whether Cbx4 is directly involved in this process. To address this question, an in vitro SUMOylation assay was performed using a GST fusion of Dnmt3a (amino acids 1–482; Dnmt3aN) that contains the SUMO-1 accepting sites as well as the regions that mediate the interactions with Cbx4 and Ubc9 [9]. First we confirmed that SUMO modification occurred in the presence of the purified enzyme components and Dnmt3a substrate (Figure 6C). At least four high-molecular-mass bands were detected with anti-GST antibody from the reaction containing recombinant E1, Ubc9 and SUMO-1 (lane 4). These bands were not observed when E1, Ubc9 or SUMO-1 was absent from the reaction mixture (lanes 1–3). This result indicated that GST–Dnmt3aN worked as a substrate for SUMOylation in vitro. We then examined whether Cbx4 can increase the SUMOylation efficiency in the reactions containing a diminished amount of Ubc9. Under this condition, only a small amount of SUMOylated Dnmt3aN was formed (Figure 6D, lane 1). However, when purified histidine-tagged Cbx4 was added to the reaction mixtures, the modified forms increased significantly and the increase occurred in a dose-dependent manner (Figure 6D, lanes 2–4). Taken together, these data indicate that SUMOylation of Dnmt3a can be enhanced by Cbx4.
DISCUSSION
Although as many as 3×107 cytosine residues in the mammalian genome are methylated, they are restricted to specific regions. The patterns of methylated and unmethylated DNA are well regulated during mammalian development. In adult tissue cells, repeat sequences, such as intracisternal A particles and satellite DNA, are highly methylated, while most CpG islands are unmethylated. In proliferating embryonic cells, including embryonic stem cells, the methylation level of the genome remains constant, despite the high-level expression of Dnmt3a and Dnmt3b. In developing germ cells and gastrulating embryos, de novo methylation of specific sequences occurs [1,38]. These facts suggest that the activities of Dnmts are tightly regulated. In this regard, SUMO conjugation of the de novo methyltransferase is of particular relevance, as the modification may alter the structure and function of the enzymes [9,26]. In the present paper we report that SUMOylation of Dnmt3a is stimulated by a PcG protein, Cbx4, thus providing an important clue about the mechanism controlling regulation of the post-translational modification.
Cbx4 as E3 ligase for Dnmt3a
Similarly to ubiquitination conjugation, the efficient conjugation of SUMO to its target proteins usually needs three enzymes: an E1 activating enzyme, an E2 conjugating enzyme (Ubc9) and an E3 ligase. While E1 and E2 are sufficient for SUMO modification of target proteins, SUMO E3 ligases are believed to determine substrate specificity and increase SUMOylation efficiency. The PcG protein Cbx4 interacts with Ubc9 and functions as a SUMO E3 ligase for CtBP, SIP1 and HIPK2 [19,23–25]. In the present study we showed that Cbx4 interacts with Dnmt3a via its C-terminus. Co-expression of Cbx4 in cultured cells significantly stimulates the SUMOylation of Dnmt3a, probably as a SUMO E3 ligase for Dnmt3a. Since Dnmt3a also interacts with PIAS proteins, another class of E3 for multiple proteins, Cbx4 may recruit Dnmt3a to heterochromatin domains (see the discussion below), where PIASs catalyse SUMOylation of Dnmt3a. Thus the stimulatory effect of Cbx4 on SUMOylation of Dnmt3a might be indirect. However, this does not seem to be the case, because PIASs lack E3 activity toward Dnmt3a both in vitro and in vivo [9]. Furthermore, our data show that Cbx4 enhances the SUMOylation of purified Dnmt3a in vitro, suggesting that it does possess a catalytic activity for Dnmt3a. All together, these data suggest that Cbx4 functions as a SUMO E3 ligase for Dnmt3a.
Heterochromatin localization of Dnmt3 enzymes may be required for their SUMOylation
We found that the extended PWWP region of Dnmt3a is required for both interaction with Cbx4 E3 ligase and SUMOylation. It is known that this region is also required for the localization of Dnmt3a to pericentromeric heterochromatin [4,5]. Its deletion resulted in diffused nuclear distribution without accumulation in centromeric heterochromatin domains. Since both endogenous and transfected Dnmt3a lacking SUMOylation show heterochromatin distribution [4,5], localization to heterochromatin does not depend on SUMOylation. It is nevertheless an open question as to whether heterochromatin localization is a prerequisite for SUMOylation. Supporting this possibility, recent work showed a high abundance of SUMOs in heterochromatin domains in a cultured human cell line [12]. Heterochromatin localization of Dnmt3a could be facilitated by Cbx4, which is known to specifically recognize H3K9 (histone H3 Lys9) methylation [28], a hallmark for heterochromatin. Therefore Cbx4 can also play a role in the recruitment of Dnmt3a to the subcellular location for SUMOylation, in addition to its function as a SUMO E3 ligase.
Potential functional consequences of SUMOylation in the PWWP domain
The PWWP domain of Dnmt3b is also proposed to bind DNA in vitro [39]. Dnmt3b defective in this domain failed to methylate the minor satellite repeats in the heterochromatin region in ES (embryonic stem) cells [5]. In this regard, SUMOylation of Dnmt3a or Dnmt3b might affect the interaction with their target DNA sequences. One similar case is the SUMOylation of human TDG (thymine DNA glycosylase), which alters the conformation of the enzyme and facilitates its disassociation from the abasic DNA product after the completion of the base excision process [40]. Alternatively, SUMOylation of the PWWP domain can interfere with functions of other domains of the enzyme. It has been shown that SUMOylation of Dnmt3a disrupts the PHD-mediated interaction with HDACs 1/2 and thus abolishes its transcriptional repression activity [9]. It is also possible that SUMO modification might affect the catalytic activity and stability of Dnmt3 enzymes. Elucidation of the biological significance of the modification would rest on the further identification of modified forms for the endogenous Dnmt3 proteins in specific tissues at certain developmental stages.
Implication of Cbx4-Dnmt3a interaction in de novo DNA methylation
DNA methylation and histone methylation are two major epigenetic markers for transcriptional repression. Recent findings reveal H3K9 methylation can serve as a signal for the establishment of DNA methylation patterns in plants and animals. The function of an H3K9 methyltransferase gene, Suv39h, is required for Dnmt3b-dependent CpG methylation at major centromeric satellites in mouse embryonic stem cells [41]. Methylation at H3K9 creates a binding site for HP1, which recognizes methylated H3K9 by its chromodomain and may in turn recruit Dnmts [42,43]. In addition, a component of H3K27 methyltransferase in the PcG complex EZH2 (Enhancer of Zeste homologue 2) interacts with Dnmts and is supposed to serve as a platform for recruiting the Dnmts to EZH2-target promoters [44]. Another PcG protein, EED (embryonic ectoderm development), also interacts with Dnmts [44]. These findings suggest cross-talk between the two epigenetic repression systems. Our finding of the direct interaction between Cbx4 and Dnmt3a could suggest that Cbx4 may function as a linker between histone and DNA methylation. A genomic locus rich in trimethylated H3K9 can be recognized by Cbx4 through its N-terminal chromodomain [28] and Dnmt3a can be recruited to the sites through its C-terminal interaction region. In this process, SUMOylation of Dnmt3a may occur as a regulatory step.
Acknowledgments
We thank Dr Eric N. Olson (Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX, U.S.A.) for the CtBP plasmid, Dr C. David Allis and Dr Emily Bernstein (The Rockefeller University, New York, NY, U.S.A.) for Cbx2, Cbx7, Cbx6 and Cbx8 plasmids, Dr Zhihong Zhang for SUMO-1 plasmids and discussions, Dr Mary Dasso (National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, U.S.A.) for Aos1 and Uba2 plasmids. We also thank Dr Songkai Liu (China Novartis Institutes for Biomedical Research, Shanghai, China), Dr Zhaoxia Chen (Division of Biology, Beckman Research Institute of the City of Hope, Duarte, CA, U.S.A.), Dr Jianping Ding (Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China) and Dr Xiaolan Zhao (Molecular Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY, U.S.A.) for critical reading of the manuscript before its submission. Work in G.X.'s laboratory is supported by grants from the China National Science Foundation, the Ministry of Science and Technology of China (National Basic Research Program 2005CB522400), the Shanghai Municipal Commission for Science and Technology, and the Max-Planck-Gesellschaft zur Förderung der Wissenschaften (the Max Planck Society for the Advancement of Science).
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Bestor T. H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 3.Okano M., Bell D. W., Haber D. A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 4.Ge Y. Z., Pu M. T., Gowher H., Wu H. P., Ding J. P., Jeltsch A., Xu G. L. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 2004;279:25447–25454. doi: 10.1074/jbc.M312296200. [DOI] [PubMed] [Google Scholar]
- 5.Chen T., Tsujimoto N., Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachman K. E., Rountree M. R., Baylin S. B. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 7.Fuks F., Burgers W. A., Godin N., Kasai M., Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuks F., Hurd P. J., Deplus R., Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ling Y., Sankpal U. T., Robertson A. K., McNally J. G., Karpova T., Robertson K. D. Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription. Nucleic Acids Res. 2004;32:598–610. doi: 10.1093/nar/gkh195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson E. S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 11.Dohmen R. J. SUMO protein modification. Biochim. Biophys. Acta. 2004;1695:113–131. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 12.Uchimura Y., Ichimura T., Uwada J., Tachibana T., Sugahara S., Nakao M., Saitoh H. Involvement of SUMO modification in MBD1- and MCAF1-mediated heterochromatin formation. J. Biol. Chem. 2006;281:23180–23190. doi: 10.1074/jbc.M602280200. [DOI] [PubMed] [Google Scholar]
- 13.Johnson E. S., Schwienhorst I., Dohmen R. J., Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson E. S., Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 15.Kahyo T., Nishida T., Yasuda H. Involvement of PIAS1 in the SUMOylation of tumor suppressor p53. Mol. Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt D., Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 18.Kirsh O., Seeler J. S., Pichler A., Gast A., Muller S., Miska E., Mathieu M., Harel-Bellan A., Kouzarides T., Melchior F., Dejean A. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 2002;21:2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kagey M. H., Melhuish T. A., Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 20.Pirrotta V. PcG complexes and chromatin silencing. Curr. Opin. Genet. Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- 21.Satijn D. P., Otte A. P. RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol. Cell Biol. 1999;19:57–68. doi: 10.1128/mcb.19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satijn D. P., Olson D. J., van der Vlag J., Hamer K. M., Lambrechts C., Masselink H., Gunster M. J., Sewalt R. G., van Driel R., Otte A. P. Interference with the expression of a novel human polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol. Cell Biol. 1997;17:6076–6086. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kagey M. H., Melhuish T. A., Powers S. E., Wotton D. Multiple activities contribute to Pc2 E3 function. EMBO J. 2005;24:108–119. doi: 10.1038/sj.emboj.7600506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long J., Zuo D., Park M. Pc2-mediated SUMOylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J. Biol. Chem. 2005;280:35477–35489. doi: 10.1074/jbc.M504477200. [DOI] [PubMed] [Google Scholar]
- 25.Roscic A., Moller A., Calzado M. A., Renner F., Wimmer V. C., Gresko E., Ludi K. S., Schmitz M. L. Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol. Cell. 2006;24:77–89. doi: 10.1016/j.molcel.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Kang E. S., Park C. W., Chung J. H. Dnmt3b, de novo DNA methyltransferase, interacts with SUMO-1 and Ubc9 through its N-terminal region and is subject to modification by SUMO-1. Biochem. Biophys. Res. Commun. 2001;289:862–868. doi: 10.1006/bbrc.2001.6057. [DOI] [PubMed] [Google Scholar]
- 27.Zhang C. L., McKinsey T. A., Lu J. R., Olson E. N. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem. 2001;276:35–39. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein E., Duncan E. M., Masui O., Gil J., Heard E., Allis C. D. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azuma Y., Tan S. H., Cavenagh M. M., Ainsztein A. M., Saitoh H., Dasso M. Expression and regulation of the mammalian SUMO-1 E1 enzyme. FASEB J. 2001;15:1825–1827. doi: 10.1096/fj.00-0818fje. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen T. N., Goodrich J. A. Protein–protein interaction assays: eliminating false positive interactions. Nat. Methods. 2006;3:135–139. doi: 10.1038/nmeth0206-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishida T., Yasuda H. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 2002;277:41311–41317. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- 32.Lai J. S., Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller J., Gaunt S., Lawrence P. A. Function of the Polycomb protein is conserved in mice and flies. Development. 1995;121:2847–2852. doi: 10.1242/dev.121.9.2847. [DOI] [PubMed] [Google Scholar]
- 34.Bunker C. A., Kingston R. E. Transcriptional repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol. Cell Biol. 1994;14:1721–1732. doi: 10.1128/mcb.14.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tajul-Arifin K., Teasdale R., Ravasi T., Hume D. A., Mattick J. S. Identification and analysis of chromodomain-containing proteins encoded in the mouse transcriptome. Genome Res. 2003;13:1416–1429. doi: 10.1101/gr.1015703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melchior F. SUMO – nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez M. S., Dargemont C., Hay R. T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z. X., Riggs A. D. Maintenance and regulation of DNA methylation patterns in mammals. Biochem. Cell Biol. 2005;83:438–448. doi: 10.1139/o05-138. [DOI] [PubMed] [Google Scholar]
- 39.Qiu C., Sawada K., Zhang X., Cheng X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Biol. 2002;9:217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinacher R., Schar P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 2005;15:616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 41.Lehnertz B., Ueda Y., Derijck A. A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A. H. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 42.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 44.Vire E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J. M., et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]