Abstract
ATP-competitive inhibitors of PKC (protein kinase C) such as the bisindolylmaleimide GF 109203X, which interact with the ATP-binding site in the PKC molecule, have also been shown to affect several redistribution events of PKC. However, the reason why these inhibitors affect the redistribution is still controversial. In the present study, using immunoblot analysis and GFP (green fluorescent protein)-tagged PKC, we showed that, at commonly used concentrations, these ATP-competitive inhibitors alone induced redistribution of DAG (diacylglycerol)-sensitive PKCα, PKCβII, PKCδ and PKCϵ, but not atypical PKCζ, to the endomembrane or the plasma membrane. Studies with deletion and point mutants showed that the DAG-sensitive C1 domain of PKC was required for membrane redistribution by these inhibitors. Furthermore, membrane redistribution was prevented by the aminosteroid PLC (phospholipase C) inhibitor U-73122, although an ATP-competitive inhibitor had no significant effect on acute DAG generation. Immunoblot analysis showed that an ATP-competitive inhibitor enhanced cell-permeable DAG analogue- or phorbol-ester-induced translocation of endogenous PKC. Furthermore, these inhibitors also enhanced [3H]phorbol 12,13-dibutyrate binding to the cytosolic fractions from PKCα–GFP-overexpressing cells. These results clearly demonstrate that ATP-competitive inhibitors cause redistribution of DAG-sensitive PKCs to membranes containing endogenous DAG by altering the DAG sensitivity of PKC and support the idea that the inhibitors destabilize the closed conformation of PKC and make the C1 domain accessible to DAG. Most importantly, our findings provide novel insights for the interpretation of studies using ATP-competitive inhibitors, and, especially, suggest caution about the interpretation of the relationship between the redistribution and kinase activity of PKC.
Keywords: ATP-binding site, diacylglycerol, protein kinase C, protein kinase inhibitor, translocation
Abbreviations: aPKC, atypical PKC; cPKC, conventional PKC; DAG, diacylglycerol; DiC8, 1,2-dioctanoyl-sn-glycerol; GFP, green fluorescent protein; HRP, horseradish peroxidase; nPKC, novel PKC; PDBu, phorbol 12,13-dibutyrate; PKC, protein kinase C; PLC, phospholipase C; PLD, phospholipase D
INTRODUCTION
PKC (protein kinase C), of which there are at least ten isoforms, is a lipid-dependent serine/threonine kinase regulating intercellular signalling in numerous cellular processes. All isoforms of PKC have an N-terminal regulatory cofactor-binding domain and a C-terminal catalytic kinase domain. In contrast with their highly conserved catalytic domain, which contains an ATP-binding site and a substrate-binding site, the regulatory domain is more variable. PKC isoforms are divided into three groups on the basis of differences in the regulatory domain: cPKC (conventional PKC; α, βI, βII and γ), nPKC (novel PKC; δ, ϵ, η and ζ) and aPKC (atypical PKC; ζ and ι/λ). The regulatory domains of cPKCs and nPKCs contain two conserved domains, the C1 domain (C1A and C1B) and the C2 domain. The C1 domain binds DAG (diacylglycerol) or tumour-promoting phorbol esters such as PMA. The C2 domain binds acidic phospholipids and also, in cPKCs, Ca2+. The C2 domain of nPKCs lacks amino acids involved in Ca2+ binding. Therefore, under physiological conditions, the activation of cPKCs is regulated by both DAG and Ca2+, whereas the activation of nPKCs is Ca2+-independent. aPKCs have a DAG-insensitive C1 domain and lack the C2 domain in the regulatory domain. Therefore, the activation of aPKCs is both DAG- and Ca2+-independent [1,2].
PKCs are mainly localized in the cytosol in the inactive state, and, upon activation, translocate from the cytosol to the plasma membrane or other intracellular compartments to phosphorylate the substrates [3]. It has been established that cPKCs preferentially translocate to the plasma membrane in response to acute increases in membrane DAG and intracellular Ca2+ resulting from the activation of PLC (phospholipase C) [4]. PLC mediates the hydrolysis of membrane Ptd(4,5)P2 to DAG and Ins(1,4,5)P3, which elevates intracellular Ca2+ by binding to its intracellular receptors [1,2]. Phorbol esters have also been shown to induce translocation of cPKCs and nPKCs to the plasma membrane [5,6]. Another lipid mediator, ceramide, has been reported to induce the translocation of PKCδ and PKCϵ to perinuclear regions such as the Golgi complex [7].
In addition, there is increasing evidence that PKCs also regulate intracellular signalling through protein–protein and/or protein–lipid interactions, without the involvement of phosphorylation, in several cellular processes. For example, Hu and Exton [8] have provided in vivo evidence that PKCα activates PLD1 through a protein–protein interaction. Larsson and co-workers [9,10] have shown, in neuroblastoma cells, that the regulatory domain of PKCθ induces apoptosis and, furthermore, that PKCϵ induces neurite-like processes through its regulatory domain. Induction of apoptosis by PKCδ was shown to be independent of the kinase activity in vascular smooth-muscle cells [11].
The microbial alkaloid staurosporine and its synthetic analogues such as the bisindolylmaleimides GF 109203X and Ro-31-8220 are known as potent PKC inhibitors [12–14]. Staurosporine-related Gö 6976 is also known as cPKC specific inhibitor [15]. These compounds interact with the ATP-binding site of PKCs and inhibit the kinase activity [12–15]. Crystal structures of the staurosporine-complexed PKCθ kinase domain and GF 109203X-complexed atypical PKCι catalytic domain have been reported [16,17]. Therefore, these inhibitors have been widely used to investigate the involvement of the kinase activity of PKC in cellular processes. However, recent evidence indicates that these staurosporine-related compounds (described as ‘ATP-competitive inhibitors’) not only inhibit the kinase activity of PKC, but also affect its redistribution after initial translocation [18–24]. It is well known that ATP-competitive inhibitors prolong the plasma-membrane translocation of cPKC in response to receptor stimulation or to the cell-permeable DAG analogue DiC8 (1,2-dioctanoyl-sn-glycerol) [18, 20]. Our previous study [21] and that of Signorelli et al. [22] showed that pervanadate- or ceramide-induced PKC reverse translocation to the cytosol was prevented by treatment with ATP-competitive inhibitors. In addition, surprisingly, Hu and Exton [23] reported that treatment with the staurosporine analogues Ro-31-8220 and GF 109203X alone apparently induces translocation of PKCα to the membrane fraction. However, it is still controversial whether all those events were simply caused by inhibition of PKC kinase activity by the inhibitors. In addition, as described above, since several PKC functions are not dependent on the kinase activity, but rather mediated by protein–protein interaction, it is important to determine whether ATP-competitive inhibitors also induce translocation of the other PKC isoforms and, if so, the mechanisms involved. However, to date, these issues also remain unclear.
In the present study, in order to understand why ATP-competitive inhibitors affect PKC redistribution, we first examined whether the inhibitors cause translocation of several isoforms of PKC. The result and the subsequent experiments clearly show that ATP-competitive inhibitors induce redistribution of DAG-sensitive PKCs to the membrane containing endogenous DAG by altering DAG sensitivity of PKC.
EXPERIMENTAL
Materials
DMSO was purchased from Kanto Chemical Co. (Tokyo, Japan). PMA was purchased from Wako Pure Chemical Co. (Tokyo, Japan). [3H]Phorbol 12,13-dibutyrate ([3H]PDBu) was purchased from American Radiolabelled Chemicals (St Louis, MO, U.S.A.). [γ-32P]ATP was obtained from ICN Biomedicals (Irvine, CA, U.S.A.). GF 109203X, Ro-31-8220 and Gö 6976 were purchased from Calbiochem (San Diego, CA, U.S.A.). PDBu, staurosporine, U-73122, U-73343, DiC8 and histamine were purchased from Sigma Chemical Co. (St Louis, MO, U.S.A.). Anti-PKCδ polyclonal antibody (C-20), anti-PKCζ polyclonal antibody (C-20), anti-PKCα monoclonal antibody (H-7), anti-actin monoclonal antibody (C-2), goat anti-rabbit IgG– HRP (horseradish peroxidase) conjugate and and goat anti-mouse IgG–HRP conjugate were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). Anti-GFP polyclonal antibody was purchased from Molecular Probes (Eugene, OR, U.S.A.).
Plasmid constracts
The expression plasmids bearing cDNA of GFP, GFP-tagged PKCs and GFP-tagged deletion and point mutants of PKCδ and PKCϵ were prepared as described previously [9,25]. PKCα and PKCδ were mutated in the autophosphorylation site using QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene). The following mutagenic primers were used: 5′-C ACA CGA GGG CAG CCT GTC CTC GAG CCA CCA GAT CAG CTG GTC-3′ (for αT638E); 5′-C CAG TCT GAT TTT GAA GGG TTC GAA TAT GTC AAC CCC CAG TTT GTG C-3′ (for αT657E); 5′-C GAT GAG GTT CTT GTC ACT GAA CTC GAG TTG GGG TTT CTC ATT CAG G-3′ (for δS643E); 5′-G CTC ATA TTT GGG GTT CAC GAA TTC GAA GCC CTT GAA GGC TGT CTG G-3′ (for δS662E). GFP-tagged deletion mutants of PKCα and PKCδ lacking the C1A or C1B domain were constructed using QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) [26] with PKCα–GFP or PKCδ–GFP as a template. The primers used for the deletion were as follows: 5′-G AAG AAC GTG CAT GAG GTG AAA GAC CCG GGT GCG GAT AAG GGA CCT GAC-3′ and 5′-GTC AGG TCC CTT ATC CGC ACC CGG GTC TTT CAC CTC ATG CAC GTT CTT C-3′ (for αC1A); 5′-GAC ACT GAT GAC CCC AGA AGC AAG GGA ATG GAT CAC ACA GAG AAG AGG-3′ and 5′-CCT CTT CTC TGT GTG ATC CAT TCC CTT GCT TCT GGG GTC ATC AGT GTC-3′ (for αC1B); 5′-GCC AAG ATT CAC TAC ATC AAG AAC ACT GGC ACT GCT ACC AAT AGC CGG G-3′ and 5′-C CCG GCT ATT GGT AGC AGT GCC AGT GTT CTT GAT GTA GTG AAT CTT GGC-3′ (for δC1A). All PCR products were sequenced to comfirm that they contained the right mutations. In addition, using immunoblot analysis with anti-GFP antibody (described below), we confirmed that the molecular mass of each GFP-tagged mutants was appropriate.
Cell culture and transfection
HL-60 cells were maintained in RPMI 1640 medium supplemented with 20% (v/v) heat-inactivated fetal bovine serum. HeLa cells were maintained in Eagle's minimum essential medium supplemented with 10% fetal bovine serum. COS-7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. All media were supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin. Glass-bottomed culture dishes 3.5 cm in diameter were seeded with 1×105 cells 16–24 h before transfection, and transient transfection was performed with FuGENE™TM 6 (Roche) according to the manufacture's instructions. Transfected cells were cultured at 37°C for 16–48 h prior to imaging and immunoblotting.
Subcellular fractionation and immunoblot analysis
Subcellular fractionation was performed as described previously [27] with slight modification. Briefly, HL-60 cells were washed once with ice-cold PBS and resuspended in ice-cold homogenization buffer (5 mM EGTA, 2 mM EDTA, 1 mM PMSF and protease-inhibitor cocktail tablets in PBS) before homogenization. HeLa cells were usually stimulated with inhibitor 1 h after serum starvation [replaced with PBSG (PBS with 5 mM glucose) containing 1 mM MgCl2]. HeLa cells were washed once with ice-cold homogenization buffer and harvested using ice-cold homogenization buffer. After homogenization using a sonicator, the cell lysate was centrifuged at 500 g for 10 min at 4°C to remove nuclei and unbroken cells. The supernatant was then centrifuged at 100 000 g for 30 min at 4°C to separate the cytosolic and particulate fractions. Immunoblot analysis was performed as described previously [27].
Confocal microscopy
The culture medium was replaced with normal Hepes buffer (135 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM Hepes and 10 mM glucose, pH 7.3) just before stimulation. The fluorescence of GFP was monitored under a Zeiss LSM 510 confocal laser-scanning fluorescence microscope at 488 nm excitation with a 505/550 nm bandpass barrier filter. All experiments were performed at 37°C. DiC8 was well mixed using the sonicator before use.
Measurement of intracellular DAG level
Total lipid extraction and determination of the DAG content were performed using a classical DAG kinase assay as described previously [28], with modification. HeLa cells were harvested and resuspended in 100 μl of PBS and 100 μl of 1 M NaCl. The samples were extracted with 375 μl of chloroform/methanol (1:2, v/v). Then 125 μl of chloroform and 125 μl of 1 M NaCl were added and the chloroform phases were separated by centrifugation at 5000 g and dried under N2. The dried lipid samples were solubilized in 20 μl of a detergent solution (125 mM octyl β-D-glucoside and 200 μg of phosphatidylserine) by sonication. The lipid solution was added to 30 μl of reaction buffer {83 mM Mops (pH 7.2) 33 mM NaF, 1.7 mM dithiothreitol, 17 mM MgCl2, 0.33 mg/ml recombinant DAG kinase (a gift from Dr Naoaki Saito, Biosignal Research Center, Kobe University, Kobe, Japan), 1.7 mM ATP and [γ-32P]ATP}. The samples were incubated at 25°C for 30 min, and then 20 μl of 1% perchloric acid and 450 μl of chloroform/methanol (1:2, v/v) were added. The samples were mixed, and lipids were extracted from the lower chloroform phase following addition of 150 μl of chloroform and 150 μl of 1% perchloric acid. The lower chloroform phase was washed twice with 700 μl of 1% perchloric acid and spotted on to silica-gel-60 thin-layer-chromatography plates. The plates were developed with chloroform/methanol/acetic acid (65:15:5, by vol.) and air-dried. Phosphatidic acid production was quantified using a Fujifilm BAS-2500 Storage Phosphor Imager.
In Vitro [3H]PDBu binding assay
The PDBu binding assay was carried out using the procedure of Wender et al. [29] with modification. The standard mixture (250 μl) in 1.5 ml Eppendorf tubes contained 20 mM Tris/HCl, pH 7.5, 4 mg/ml BSA, 150 μg/ml 1,2-di-(cis-9-octadecenol)-sn-glycero-3-phospho-L-serine (dioleoyl phosphatidylserine), 1–50 nM [3H]PDBu (20 Ci/mmol), and the cytosolic fractions prepared from equal numbers of COS-7 cells transiently expressing PKCα–GFP (2.5×105 cell equivalents). COS-7 cells were transiently transfected by electroporation and cultured for 24 h and the cytosolic fractions were prepared as described above. Samples were incubated for 90 min at 4°C. The [3H]PDBu-bound complex was separated from free ligand by rapid filtration through a 2.1-cm-diameter Whatman GF/B filter, pretreated with freshly prepared 0.3% polyethylenimine for at least 2 h. The sample tube was rinsed with ice-cold 20 mM Tris/HCl, pH 7.5, and the rinse solution was then passed through the filter. The filter was further washed with ice-cold Tris/HCl, pH 7.5. Each filter was then placed in a scintillation vial. Radioactivity (d.p.m.) was determined with a Beckman liquid-scintillation counter. Non-specific binding was measured using an excess of non-radioactive PDBu (30 μM). The curve was fitted to a hyperbola using the computer software Microcal Origin (version 5.0J; Origin LabCorp, Northampton, MA, U.S.A.).
RESULTS
ATP-competitive inhibitors induce translocation of endogenous PKCα and PKCδ, but not PKCζ, to the particulate fraction
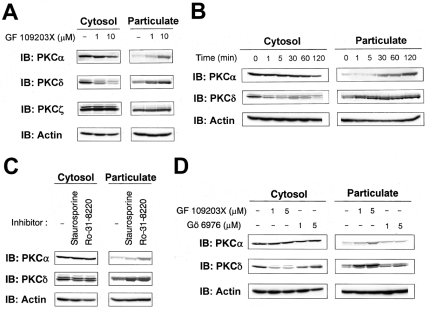
The α, δ, and ζ isoforms of PKC have been shown to be expressed endogenously in HeLa cells [30]. Therefore we first examined whether the ATP-competitive inhibitor GF 109203X induced translocation of endogenous PKCα, PKCδ and PKCζ in HeLa cells. As illustrated in Figure 1(A), treatment of HeLa cells with GF 109203X at 1 μM, a concentration that is commonly used to inhibit the kinase activity of PKC, for 20 min induced translocation of PKCδ from the cytosolic to the particulate fraction. GF 109203X at 10 μM also induced translocation of PKCα, as previously reported by Hu and Exton [23], whereas GF 109203X had no effect on the localization of PKCζ. Surprisingly, translocation of PKCδ began within 1 min of treatment with 5 μM GF 109203X (Figure 1B), whereas translocation of PKCα began within 30 min of treatment. We also examined whether the other ATP-competitive inhibitors staurosporine and Ro-31-8220 induced the translocation of PKCα and PKCδ. Staurosporine and Ro-31-8220 at concentrations of 0.1 μM and 5 μM respectively also induced translocation of PKCα and PKCδ to the particulate fraction (Figure 1C). Furthermore, we examined the effect of Gö 6976, a cPKC-specific inhibitor, on the localization of PKCα and PKCδ. Interestingly, treatment with Gö 6976, at even 1–5 μM, did not induce translocation of PKCδ, whereas GF 109203X, at the same concentrations, induced translocation of PKCδ. Treatment with Gö 6976 caused translocation of PKCα, although the effect was very subtle (Figure 1D). Furthermore, induction of translocation of PKC by the inhibitors was not restricted to HeLa cells, since it was also observed in another cell type, namely HL-60 cells (results not shown). These results indicate that, at commonly used concentrations, ATP-competitive inhibitors induce translocation of endogenous PKCα and PKCδ, but not PKCζ, to the particulate fraction.
Figure 1. Translocation of endogenous PKCs to the particulate fraction by ATP-competitive inhibitors.
(A) HeLa cells were treated with 1 or 10 μM GF 109203X for 20 min. (B) HeLa cells were treated with 5 μM GF 109203X for the indicated times. (C) HeLa cells were treated with 0.1 μM staurosporine or 5 μM Ro-31-8220 for 30 min. (D) HeLa cells were treated with the indicated concentrations of GF 109203X or Gö 6976 for 15 min. Subcellular fractions were obtained as described in the Experimental section and subjected to SDS/PAGE, followed by immunoblotting with anti-PKCα, anti-PKCδ or anti-PKCζ and with anti-actin antibody. Actin was used as a loading control. Results are representative of at least three separate experiments.
ATP-competitive inhibitors selectively induce redistribution of DAG-sensitive PKCs to the endomembrane or the plasma membrane
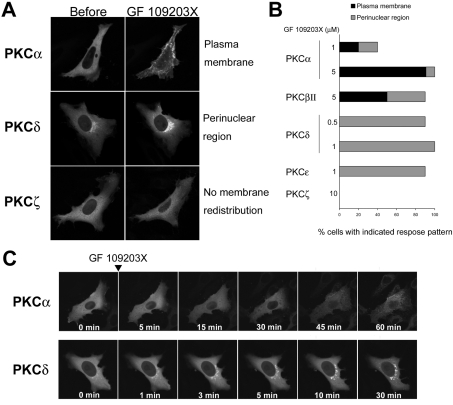
To further characterize the effect of ATP-competitive inhibitors on localization of PKC, we used confocal microscopy to monitor the intracellular movement of several GFP-tagged PKC isoforms (cPKC, α and βII; nPKC, ϵ; aPKC, ζ) transiently transfected into living HeLa cells. Treatment with 0.5–1 μM GF 109203X induced a preferential redistribution of PKCδ- and PKCϵ–GFP to the perinuclear region (Figures 2A–2C). In addition, the response of PKCδ–GFP to the inhibitors was very rapid and prolonged (Figure 2C), consistent with the results for endogenous PKCδ (Figure 1B). PKCα– and PKCβII–GFP redistributed to the plasma membrane as well as to the endomembranes in response to GF 109203X, and the response was slow (Figures 2A–2C), consistent with that of endogenous PKCα (Figure 1B). In addition, higher concentrations (1–5 μM) of the inhibitor were required to induce apparent membrane redistribution (Figure 2B). Conversely, treatment with the inhibitor at 10 μM for 60 min neither induced redistribution of PKCζ–GFP to the plasma membrane nor the endomembrane (see the Discussion) (Figures 2A and 2B), which is also consistent with the response of endogenous PKCζ described above (Figure 1A). Treatment with staurosporine (0.1 μM for δ and ϵ isoforms, 0.5 μM for α and βII isoforms) also induced redistribution of GFP-tagged PKCs in a similar fashion (results not shown). In addition, staurosporine did not induce membrane redistribution of PKCζ–GFP (results not shown). Treatment with 10 μM Gö 6976 did not cause perinuclear redistribution of PKCδ–GFP, although the treatment induced slight plasma membrane redistribution of PKCα–GFP (results not shown). These results indicate that ATP-competitive inhibitors selectively induce redistribution of DAG-sensitive PKCs to the endomembrane or the plasma membrane.
Figure 2. ATP-competitive inhibitors selectively induce membrane redistribution of DAG-sensitive PKCs.
(A) HeLa cells transiently expressing PKCα–, PKCδ– or PKCζ–GFP were treated with GF 109203X at 5 μM for 60 min, 1 μM for 30 min or 10 μM for 60 min respectively. (B) Percentage of cells displaying GF 109203X-induced membrane redistribution of PKC. HeLa cells transiently expressing GFP-tagged cPKCs (PKCα and βII), nPKCs (PKCδ and ϵ), aPKC (PKCζ) were treated with GF 109203X at the indicated concentrations for 60, 15 or 60 min respectively. A total of 10–14 cells were observed. (C) HeLa cells transiently expressing PKCα- or PKCδ–GFP were treated with GF 109203X at 5 or 1 μM respectively for the indicated times.
PKCs with autophosphorylation sites mutated to glutamate also redistribute in response to ATP-competitive inhibitors
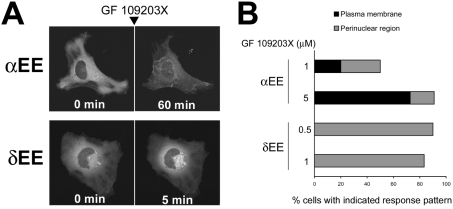
Next we investigated the mechanism by which ATP-competitive inhibitors induce the membrane redistribution of DAG-sensitive PKCs. PKCs are well known to contain autophosphorylation sites in their C-termini [1,18,20] Therefore, to confirm that ATP-competitive inhibitors cause membrane redistribution of PKC independently of the phosphorylation state of autophosphorylation sites, we examined whether ATP-competitive inhibitors also induce membrane redistribution of PKCα and PKCδ with autophosphorylation sites (T638 and S657 for PKCα, S643 and S662 for PKCδ) mutated to glutamate (αEE and δEE respectively), an amino acid commonly used to mimic phosphate. As shown in Figure 3, treatment with GF 109203X also induced membrane redistribution of αEE and δEE mutants. In addition, the time course and distribution pattern of the glutamate mutants were similar to those of wild-types (Figures 3A and 3B). These results indicate that ATP-competitive inhibitors cause PKC membrane redistribution independently of the phosphorylation state of the C-terminus autophosphorylation sites.
Figure 3. ATP-competitive inhibitors induce membrane redistribution of GFP-tagged αEE and δEE mutants.
(A) HeLa cells transiently expressing GFP-tagged αEE or δEE were treated with 10 or 1 μM GF 109203X respectively for the indicated times. (B) Percentage of cells displaying GF 109203X-induced membrane redistribution of PKC. HeLa cells transiently expressing GFP-tagged αEE or δEE were treated with GF 109203X at the indicated concentrations for 60 or 15 min respectively. A total of 10–12 cells were observed.
PKC membrane redistribution by ATP-competitive inhibitors requires its DAG-sensitive C1 domain
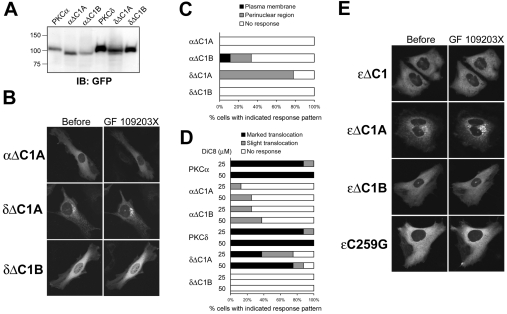
As shown in Figures 1(A) and 2(B), treatment with GF 109203X at 10 μM, a concentration that inhibits the kinase activity of PKCζ [15], did not induce membrane redistribution of endogenous and GFP-tagged PKCζ, whereas such treatment did induce apparent translocation of DAG-sensitive PKCα and PKCδ. Therefore we speculated that interaction of the ATP-competitive inhibitor with the ATP-binding site is not sufficient for the induction of membrane redistribution of PKC and that the DAG-sensitive C1 domain is also required. To assess this possibility, we examined the effect of the deletion of C1A or C1B domain on GF 109203X- or DiC8-induced membrane redistribution of PKCα and PKCδ. Using immunoblot analysis with anti-GFP antibody, we confirmed that the molecular masses of the GFP-tagged mutants were appropriate and no significant degradation products were detected (Figure 4A). Treatment with 5 μM GF 109203X had no effect on the localization of PKCα deletion mutant lacking C1A domain (αΔC1A), although the treatment several times induced membrane redistribution of the C1B domain deletion mutants (αΔC1B) (Figures 4B and 4C). In addition, whereas treatment with 25 or 50 μM DiC8 induced marked transient translocation of wild-type PKCα, the treatment induced slight translocation of αΔC1A and αΔC1B in a few cases (Figure 4D). Unexpectedly, the PKCδ deletion mutant lacking the C1A domain (δΔC1A) was mainly localized in the perinuclear region in unstimulated cells. However, treatment with 1 μM GF 109203X enhanced the perinuclear accumulation of δΔC1A (Figures 4B and 4C). In addition, treatment with 25 or 50 μM DiC8 induced slight translocation of δΔC1A to the plasma membrane and a considerable decrease in the amount of δΔC1A in the perinuclear region (Figure 4D). The C1B domain deletion mutant of PKCδ (δΔC1B) was localized in the cytosol and never translocated in response to GF 109203X or DiC8 (Figures 4B, 4C and 4D). Recently it has been shown that deletion of ϵC1 and ϵC1B domains, but not ϵC1A domain, or the ϵC1B point mutant ϵC259G, decreased membrane translocation of PKCϵ in response to DiC8 [31]. Therefore we further examined the effects of GF 109203X on the localization of these deletion and point mutants of PKCϵ. Whereas treatment with 1 μM GF 109203X caused redistribution of DAG-sensitive ϵΔC1A to the perinuclear region, the treatments had no effect on the localization of ϵΔC1, ϵΔC1B and ϵC259G (Figure 4E). These results indicate that PKC membrane redistribution by ATP-competitive inhibitors requires its DAG-sensitive C1 domain.
Figure 4. PKC membrane redistribution by an ATP-competitive inhibitor requires its DAG-sensitive C1 domain.
(A) Whole-cell lysates were obtained from COS-7 cells transfected with the indicated plasmids using ice-cold homogenization buffer containing 1% Triton X-100 and subjected to SDS/PAGE, followed by immunoblotting with anti-GFP antibody. (B) HeLa cells transiently expressing GFP-tagged αΔC1A, δΔC1A and δΔC1B were treated with GF 109203X at 5 μM for 60 min (αΔC1A) or 1 μM for 20 min (δΔC1A and δΔC1B). A total of seven to nine cells were observed. (C) HeLa cells transiently expressing GFP-tagged αΔC1A, αΔC1B, δΔC1A and δΔC1B were treated with GF 109203X at 5 μM for 60 min (αΔC1A and αAC1B) or 1 μM for 20 min (δΔC1A and δΔC1B). A total of eight cells were observed. (D) HeLa cells transiently expressing GFP-tagged PKCα, αΔC1A, αΔC1B, PKCδ, δΔC1A and δΔC1B were treated with 25 or 50 μM DiC8 for 10 min. The translocation of δΔC1A indicates the decrease from the perinuclear region. (E) HeLa cells transiently expressing GFP-tagged ϵΔC1, ϵΔC1A, ϵΔC1B and ϵC259G were treated with 1 μM GF 109203X for 20 min. Results are representative of at least seven separate experiments.
An inhibitor of PLC, U-73122, prevents PKC membrane redistribution by ATP-competitive inhibitors
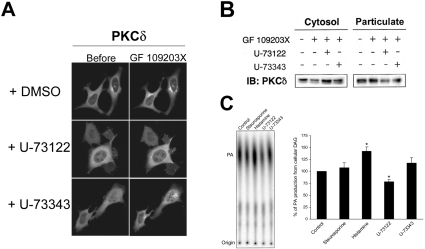
To investigate whether endogenous DAG generated by PLC is involved in membrane redistribution of PKC by ATP-competitive inhibitors, we examined the effects of U-73122, a PLC inhibitor, on the membrane redistribution of PKC by ATP-competitive inhibitors. Treatment with 2 μM U-73122 for 10 min prevented the membrane redistribution of PKCδ–GFP after treatment with ATP-competitive inhibitors, whereas treatment with 2 μM U-73343, the inactive analogue of U-73122, had no effect on the membrane redistribution (Figure 5A). U-73122 also prevented membrane redistribution of PKCα–GFP by ATP-competitive inhibitors (results not shown). These results were further confirmed by an immunoblot analysis showing that treatment with 4 μM U-73122 for 30 min also prevented GF 109203X-induced translocation of endogenous PKCδ (Figure 5B).
Figure 5. PKC membrane redistribution by ATP-competitive inhibitors requires endogenous DAG generated by the spontaneous action of PLC.
(A) HeLa cells transiently expressing PKCδ–GFP were pretreated with 2 μM U-73122 or U-73343 for 10 min and treated with 1 μM GF 109203X for 20 min. (B) HeLa cells were pretreated with 4 μM U-73122 or U-73343 for 30 min and treated with 1 μM GF 109203X for 30 min. Subcellular fractions were obtained as described in the Experimental section and subjected to SDS/PAGE, followed by immunoblotting (IB) with anti-PKCδ antibody. Results are representative of at least three separate experiments. (C) The DAG level in staurosporine (1 μM, 15 min)-, histamine (100 μM, 5 min)-, U-73122 and U-73343 (4 μM, 60 min)-treated cells was analysed by using the DAG kinase assay. The DAG content was determined by quantifying the amount of 32P-labelled phosphatidic acid (PA) using the BAS-2500 Phosphor Imager. Results are means±S.E.M. for four separate experiments. *P<0.01 versus control cells (unpaired Student's t test). We used U-73122 at a lower concentration (2 μM) in the confocal analysis because higher concentrations of U-73122 seemed to affect cell adhesion to some extent, perhaps as a result of the lower cell density.
ATP-competitive inhibitors do not cause acute PLC activation and DAG generation
To further investigate whether the ATP-competitive inhibitors induce acute DAG generation through activation of PLC, we measured intracellular DAG levels by a classical DAG kinase assay. As shown in Figure 5(C), treatment with staurosporine for 15 min had no significant effect on the intracellular DAG level, whereas treatment with histamine, which is known to induce activation of PLC in HeLa cells [32], for 5 min increased intracellular DAG by approx. 40% compared with control cells. These results indicate that treatment with staurosporine does not cause acute activation of PLC and generation of DAG. Furthermore, we examined the effect of U-73122 or U-73343 on the intracellular level of DAG. Interestingly, the DAG level in U-73122-treated cells was decreased by at least 20% compared with control cells, whereas that in U-73343-treated cells was not decreased (Figure 5C). These results suggest that endogenous DAG generated by the basal activity of PLC was required for PKC membrane redistribution by ATP-competitive inhibitors.
ATP-competitive inhibitors enhance DiC8- and PMA-induced translocation of PKC
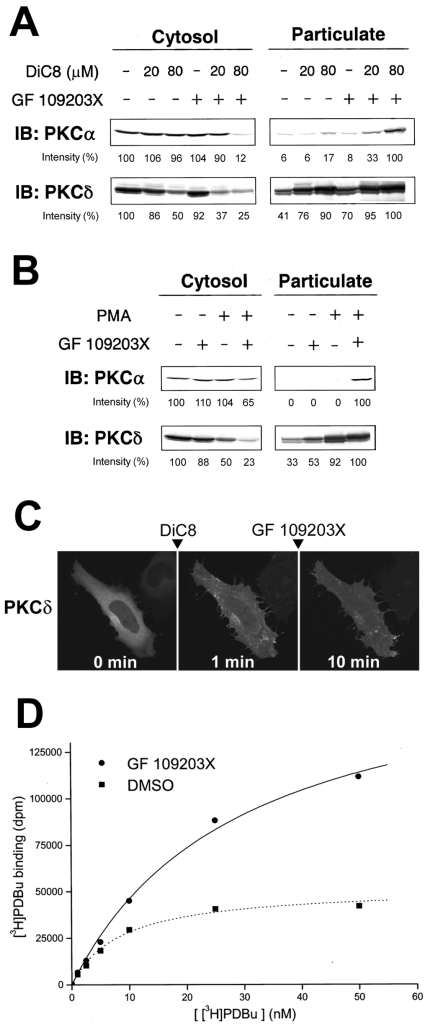
The observations described in Figure 5 prompted us to hypothesize that PKC molecules become more sensitive to DAG when they interact with an ATP-competitive inhibitor and that, as a result, PKC redistributes to the plasma membrane and endomembrane, which contain endogenous DAG generated by the basal activity of PLC. To explore this possibility in vivo, we used immunoblot analysis to examine whether pretreatment with an ATP-competitive inhibitor enhances DiC8-induced translocation of endogenous PKCα and δ in HL-60 cells. As illustrated in Figure 6(A), treatment with 0.5 μM GF 109203X alone had no effect on the localization of PKCα, whereas pretreatment with 0.5 μM GF 109203X caused a marked increase in the amount of PKCα in the particulate fraction after DiC8 treatment (Figure 6A, top). Similarly, induction of translocation of PKCδ by DiC8 in combination with GF 109203X required a lower concentration of DiC8 compared with induction by DiC8 alone (Figure 6A, bottom). We further examined whether an ATP-competitive inhibitor enhances translocation of PKC induced by the phorbol ester PMA. As shown in Figure 6(B), treatment with 10 nM PMA alone for 20 min had no effect on the localization of PKCα, whereas a combination of 1 μM GF 109203X and 10 nM PMA increased the amount of PKCα in the particulate fraction (Figure 6B, top). For PKCδ, although treatment with 10 nM PMA alone induced some translocation, pretreatment with GF 109203X followed by addition of 10 nM PMA caused complete translocation of PKCδ from the cytosol to the particulate fraction (Figure 6B, bottom). Similar results were obtained in HeLa cells (results not shown). We further examined the effect of exogenously added DiC8 on the GF 109203X-induced endomembrane redistribution of PKCδ–GFP. Pretreatment of HeLa cells with 10 μM DiC8 caused transient translocation of PKCδ–GFP to the plasma membrane, whereas subsequent treatment with 1 μM GF 109203X greatly prolonged translocation to the plasma membrane (Figure 6C), but did not induce redistribution to the perinuclear region, as observed with GF 109203X treatment alone (Figure 2C). This effect was also observed in PKCϵ–GFP-overexpressing HeLa cells (results not shown). These results indicate that ATP-competitive inhibitors enhance DiC8- and PMA-induced translocation of PKC.
Figure 6. GF 109203X enhances DiC8- and PMA-induced translocation of endogenous PKC and [3H]PDBu binding to the cytosolic fraction prepared from PKCα–GFP-overexpressing cells.
(A) HL-60 cells were pretreated with 0.5 μM GF 109203X for 20 min and then stimulated with 20 or 80 μM DiC8 for 5 min. (B) HL-60 cells were pretreated with 1 μM GF109203X for 20 min and then stimulated with 10 nM PMA for 15 min. Subcellular fractions were obtained as described in the Experimental section and subjected to SDS/PAGE, followed by immunoblotting (IB) with anti-PKCα and anti-PKCδ antibodies. Densitometric analysis was performed using Scion Image software. In the cytosolic fraction the intensity of the protein bands is shown as a percentage of that of the control cells. In the particulate fraction the intensity of the protein bands is shown as a percentage of that of the 80 μM DiC8-plus-GF 109203X (A)- or PMA-plus-GF 109203X (B)-treated cells. Results are representative of at least three separate experiments. (C) HeLa cells transiently expressing PKCδ–GFP were pretreated with 10 μM DiC8 for 1 min, and then treated with 1 μM GF 109203X for 9 min. Results are representative of at least three separate experiments. (D) The PDBu binding was evaluated by the method described in the Experimental section in the presence of DMSO or 10 μM GF 109203X. The expression of PKCα–GFP was confirmed by immunoblotting with anti-GFP and anti-PKCα antibodies. Results are representative of at least three separate experiments, with duplicate determinations in each experiment.
ATP-competitive inhibitors enhance [3H]PDBu binding to the cytosolic fractions prepared from PKCα–GFP-overexpressing cells
Finally, to gain further insight into the mechanism by which ATP-competitive inhibitors make PKC sensitive to DAG, we examined the effect of ATP-competitive inhibitors on [3H]PDBu binding to cytosolic fractions prepared from PKCα–GFP-overexpressing COS-7 cells. As shown in Figure 6(D), [3H]PDBu binding to the cytosolic fractions was increased in the presence of GF 109203X at each concentration of [3H]PDBu (even at 50 nM, in which [3H]PDBu binding in the absence of GF 109203X was almost saturated.). The cytosolic fraction prepared from non-transfected COS-7 cells exhibited much lower [3H]PDBu binding, even in the presence of GF 109203X, than the cytosolic fraction prepared from PKCα–GFP-overexpressing COS-7 cells (results not shown). We also confirmed that overexpression of GFP alone had no significant effect on the [3H]PDBu binding to the cytosolic fraction (results not shown). These results suggest that ATP-competitive inhibitors increase at least the PDBu-binding ability of PKCα.
DISCUSSION
In the present study we showed that ATP-competitive inhibitors, which interact with the ATP-binding site of the PKC molecule, induce membrane redistribution of DAG-sensitive PKCs. The results from subsequent experiments suggest that these inhibitors alter the DAG sensitivity of the PKC molecule, thereby causing redistribution of PKC to the membrane containing DAG generated by basal PLC activity.
Our immunoblot analysis, as well as the in vitro binding assay, suggests that ATP-competitive inhibitors alter the DAG- or phorbol-ester-binding ability of the PKC molecule, although GF 109203X, one of the inhibitors, did not appear to increase the affinity of at least PKCα–GFP for PDBu (Figure 6). In contrast with our results, a previous study has shown that GF 109203X had a slight effect on [3H]PDBu binding to PKC [13]. This difference may have resulted from the conformational state of the enzyme used (as described below) or the experimental system, although the precise reason remains unclear. In fact, in our system, ATP-competitive inhibitors were unsuccessful in greatly increasing the binding of [3H]PDBu to whole-cell lysate that was prepared with homogenization buffer containing 1% Triton X-100 (results not shown). The mechanism by which interaction with ATP-competitive inhibitors alters the DAG sensitivity of PKC molecules is unknown. Our present study showed that, as in the case of wild-type PKCα and PKCδ, αEE and δEE autophosphorylation mutants were also membrane-redistributed in response to GF 109203X (Figure 3). It has been reported that GF 109203X does not affect the phosphorylation of PKCα [18]. In addition, several studies have showed that PKC with autophosphorylation sites mutated to alanine was mainly localized in the cytosol [18,22]. Therefore it is unlikely that dephosphorylation of autophosphorylation sites of PKC is responsible for the membrane redistribution by ATP-competitive inhibitors. A previous report has speculated that ATP-competitive inhibitors destabilize the closed conformation of PKC, thereby exposing the C1a domain and making it available for interaction with DAG [18]. Several studies have shown that, similar to the results obtained using ATP-competitive inhibitors, mutation of an ATP-binding site also prolongs the plasma-membrane localization of cPKC in response to receptor stimulation or DiC8 [18,20]. Interestingly, Perander et al. [33] have shown that an ATP-binding site mutant of λPKC, an isoform of DAG-insensitive aPKC, accumulates in the cell nucleus, whereas the wild-type kinase is mainly cytosolic. In addition, this nuclear accumulation was shown to be not correlated with the activity status of the kinase. Instead, their results indicated that disruption of the intramolecular interactions by mutation of the ATP-binding site of λPKC led to nuclear localization. The reported crystal structure of PKCθ–staurosporine indicates that the binding of staurosporine affects the conformation of the PKCθ kinase domain [16]. Furthermore, the crystal structure of the catalytic domain of atypical PKCι complexed with GF 109203X has been reported. That study also revealed that the bound GF 109203X blocks the ATP-binding site and converts the kinase domain into an intermediate open conformation [17]. Interestingly, we observed that prolonged treatment with higher concentrations of ATP-competitive inhibitors caused nuclear accumulation of PKCζ–GFP (results not shown). Overall, on the basis of the ideas and evidence described above, we also speculate that interaction of ATP-competitive inhibitors with the ATP-binding site of PKCs causes disruption of the intramolecular interactions (closed conformation) and makes the C1 domain accessible to DAG, and that, as a result, DAG-sensitive PKCs become more sensitive to DAG. This speculation also explains why nPKC responses to the inhibitor more rapidly than cPKC, as shown in Figure 1 and 2 (cPKC is regulated by not only its C1 domain, but also its Ca2+-sensitive C2 domain). We observed that some other types of PKC inhibitor also cause PKC redistribution (results not shown). Therefore it is possible that this concept also applies to other type PKC inhibitors. To date, a number of studies, including a previous one of our own, have shown that ATP-competitive inhibitors prevent membrane dissociation or re-localization of PKC after initial translocation in response to various stimuli [18–24]. From results using the inhibitors, some of those studies have concluded that the kinase activity of PKC (to phosphorylate the putative substrate) is required for those events. However, our present results clearly raise the question as to whether those events, including the membrane dissociation, indeed required the kinase activity. In fact our results suggest another possibility, namely that, due to a more open conformation of the PKC–inhibitor complex, PKC merely retains its localization at the membrane containing DAG or phorbol ester.
Recent studies indicate that the C1A and C1B domains of PKCs are functionally distinct. Previous studies by Cho and co-workers indicated that the C1A domain plays a critical role in the DAG-induced membrane binding and activation of PKCα [34,35]. In addition, in-vitro-activity and membrane-binding analyses by Cho and co-workers also showed that the C1A domain is critical for the DAG-induced membrane binding and activation of PKCδ [36]. Consistent with the results obtained by Cho and colleagues, we showed that treatment with DiC8 had no significant effect on the localization of αΔC1A, although the treatment had only minor effects on the localization of αΔC1B (Figure 4D). In addition, ATP-competitive inhibitors had no effect on the localization of αΔC1A (Figures 4B and 4C). In the case of PKCδ, however, the deletion of the C1B domain caused a loss of responsiveness to DiC8, whereas δΔC1A was still sensitive to DiC8 (although δΔC1A mainly localized in the perinuclear region) (Figure 4D). ATP-competitive inhibitors also had no effect on the localization of δΔC1B, whereas the treatment enhanced the perinuclear accumulation of δΔC1A (Figure 4B and C). The reason why αΔC1B also lacks DAG sensitivity is unknown. In addition, it is also unknown what causes the different results between Cho et al.'s and our studies regarding the responsible domain of PKCδ for DAG, although the difference may also have resulted from the experimental system. Importantly, our study showed that the responsiveness of the deletion mutants of PKCα and PKCδ to the inhibitors was highly correlated with that of the mutants to the DiC8. Furthermore, our results showed that GF 109203X had no effect on the localization of ϵΔC1, ϵΔC1B and ϵC259G, whereas the treatment induces redistribution of ϵΔC1A to the perinuclear region. The responsiveness of the mutants to the inhibitor is also correlated with that of the mutants to DiC8 recently reported [31]. Therefore we concluded that the DAG-sensitive C1 domain of individual PKC isoforms is required for the membrane redistribution by the inhibitors.
Our cell-imaging studies showed that the perinuclear region is the preferential targeting site of nPKC in response to ATP-competitive inhibitors (Figure 2). Cell-fractionation studies have shown that DAG can be produced locally in the plasma membrane, internal membranes or the nucleus [2,37,38]. In addition, it has been shown that protein kinase D is recruited to the trans-Golgi network by direct binding of DAG [39] and that Ras guanyl nucleotide-releasing protein 1 translocates to the Golgi apparatus in response to Src-dependent activation of PLCγ1 [40]. Our present results showed that treatment with U-73122, a PLC inhibitor, reduced the intracellular DAG level (Figure 6C). All of the above evidence suggests that DAG can be produced locally in endomembranes such as the Golgi complex, as well as in the plasma membrane, by basal PLC activity under normal growth conditions, which may explain why PKCδ redistributes to the perinuclear region in response to ATP-competitive inhibitors. Using confocal microscopy, we also observed the attenuated perinuclear accumulation of PKCδ–GFP induced by ATP-competitive inhibitors after serum starvation for 1 h or more (replaced with Hepes buffer) (results not shown), although immunoblot analysis showed, even after serum starvation for 1 h, that the inhibitors caused sufficient translocation of endogenous PKCs (Figure 1). On the other hand, however, it is still unclear why nPKC redistributes preferentially to the perinuclear region and not to the plasma membrane. A recent excellent study by Dries et al. [41] identified a single residue in the C1 domain that sensitizes nPKC to cellular DAG production. The residue was also shown to play an important role in the targeting of nPKC to the Golgi. Therefore, one possible explanation is that the affinity of an individual C1 domain for DAG is responsible for the targeting site of ATP-competitive inhibitors-treated PKCs.
In summary, our experiments clearly demonstrate that, at concentrations commonly used to inhibit kinase activity in a large number of previous studies, ATP-competitive inhibitors also affect the localization of PKC isoforms, perhaps by disrupting the closed conformation. That is to say, these inhibitors also induce membrane accumulation of inactive PKCs. Since PKC has also been shown to mediate intracellular signalling by protein–protein interactions, as described in the Introduction, our findings may provide novel insights for the interpretation of the conflicting data with regard to ATP-competitive inhibitors. Furthermore, although it is currently believed that the kinase activity of PKC is required for several redistribution events, including membrane dissociation, our results clearly question the validity of this notion.
Acknowledgments
We thank Dr Yasuhito Shirai for helpful discussions of this work, and we thank Dr Naoaki Saito (Biosignal Research Center, Kobe University, Kobe, Japan) for providing plasmids and recombinant DAG kinase and also for helpful discussions.
References
- 1.Newton A. C. Regulation of protein kinase C. Curr. Opin. Cell Biol. 1997;9:161–167. doi: 10.1016/s0955-0674(97)80058-0. [DOI] [PubMed] [Google Scholar]
- 2.Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 3.Farrar W. L., Anderson W. B. Interleukin-2 stimulates association of protein kinase C with plasma membrane. Nature. 1985;315:233–235. doi: 10.1038/315233a0. [DOI] [PubMed] [Google Scholar]
- 4.Oancea E., Meyer T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell. 1998;95:307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- 5.Sakai N., Sasaki K., Ikegaki N., Shirai Y., Ono Y., Saito N. Direct visualization of the translocation of the γ-subspecies of protein kinase C in living cells using fusion proteins with green fluorescent protein. J. Cell Biol. 1997;139:1465–1476. doi: 10.1083/jcb.139.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohmori S., Shirai Y., Sakai N., Fujii M., Konishi H., Kikkawa U., Saito N. Three distinct mechanisms for translocation and activation of the δ subspecies of protein kinase C. Mol. Cell. Biol. 1998;18:5263–5271. doi: 10.1128/mcb.18.9.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kashiwagi K., Shirai Y., Kuriyama M., Sakai N., Saito N. Importance of C1B domain for lipid messenger-induced targeting of protein kinase C. J. Biol. Chem. 2002;277:18037–18045. doi: 10.1074/jbc.M111761200. [DOI] [PubMed] [Google Scholar]
- 8.Hu T., Exton J. H. Mechanisms of regulation of phospholipase D1 by protein kinase Cα. J. Biol. Chem. 2003;278:2348–2355. doi: 10.1074/jbc.M210093200. [DOI] [PubMed] [Google Scholar]
- 9.Schultz A., Jönsson J.-I., Larsson C. The regulatory domain of protein kinase Cθ localises to the Golgi complex and induces apoptosis in neuroblastoma and Jurkat cells. Cell Death Differ. 2003;10:662–675. doi: 10.1038/sj.cdd.4401235. [DOI] [PubMed] [Google Scholar]
- 10.Zeidman R., Löfgren B., Påhlman S., Larsson C. PKCϵ, via its regulatory domain and independently of its catalytic domain, induces neurite-like processes in neuroblastoma cells. J. Cell Biol. 1999;145:713–726. doi: 10.1083/jcb.145.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goerke A., Sakai N., Gutjahr E., Schlapkohl W. A., Mushinski J. F., Haller H., Kolch W., Saito N., Mischak H. Induction of apoptosis by protein kinase Cδ is independent of its kinase activity. J. Biol. Chem. 2002;277:32054–32062. doi: 10.1074/jbc.M203734200. [DOI] [PubMed] [Google Scholar]
- 12.Ward N. E., O'Brian C. A. Kinetic analysis of protein kinase C inhibition by staurosporine: evidence that inhibition entails inhibitor binding at a conserved region of the catalytic domain but not competition with substrates. Mol. Pharmacol. 1992;41:387–392. [PubMed] [Google Scholar]
- 13.Toullec D., Pianetti P., Coste H., Bellevergue P., Grand-Perret T., Ajakane M., Baudet V., Boissin P., Boursier E., Loriolle F., Duhamel L., et al. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 1991;266:15771–15781. [PubMed] [Google Scholar]
- 14.Davis P. D., Elliott L. H., Harris W., Hill C. H., Hurst S. A., Keech E., Kumar M. K., Lawton G., Nixon J. S., Wilkinson S. E. Inhibitors of protein kinase C. 2. Substituted bisindolylmaleimides with improved potency and selectivity. J. Med. Chem. 1992;35:994–1001.. doi: 10.1021/jm00084a004. [DOI] [PubMed] [Google Scholar]
- 15.Martiny-Baron G., Kazanietz M. G., Mischak H., Blumberg P. M., Kochs G., Hug H., Marmé D., Schächtele C. Selective inhibition of protein kinase C isozymes by the indolocarbazole Gö 6976. J. Biol. Chem. 1993;268:9194–9197. [PubMed] [Google Scholar]
- 16.Xu Z.-B., Chaudhary D., Olland S., Wolfrom S., Czerwinski R., Malakian K., Lin L., Stahl M. L., Joseph-McCarthy D., Benander C., Fitz L., Greco R., et al. Catalytic domain crystal structure of protein kinase C-θ (PKCθ) J. Biol. Chem. 2004;279:50401–50409. doi: 10.1074/jbc.M409216200. [DOI] [PubMed] [Google Scholar]
- 17.Messerschmidt A., Macieira S., Velarde M., Badeker M., Benda C., Jestel A., Brandstetter H., Neuefeind T., Blaesse M. Crystal structure of the catalytic domain of human atypical protein kinase C-ι reveals interaction mode of phosphorylation site in turn motif. J. Mol. Biol. 2005;352:918–931. doi: 10.1016/j.jmb.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 18.Stensman H., Rraghunath A., Larsson C. Autophosphorylation suppresses whereas kinase inhibition augments the translocation of protein kinase Cα in response to diacylglycerol. J. Biol. Chem. 2004;279:40576–40583. doi: 10.1074/jbc.M405560200. [DOI] [PubMed] [Google Scholar]
- 19.Tanimura A., Nezu A., Morita T., Hashimoto N., Tojyo Y. Interplay between calcium, diacylglycerol and phosphorylation in the spatial and temporal regulation of PKCα–GFP. J. Biol. Chem. 2002;277:29054–29062. doi: 10.1074/jbc.M201130200. [DOI] [PubMed] [Google Scholar]
- 20.Feng X., Hannun Y. A. An essential role for autophosphorylation in the dissociation of activated protein kinase C from the plasma membrane. J. Biol. Chem. 1998;273:26870–26874. doi: 10.1074/jbc.273.41.26870. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi H., Suzuki K., Namiki H. Phenylarsine oxide and H2O2 plus vanadate induce reverse translocation of phrbol-ester-activated PKCβII. Cell Struct. Funct. 2003;28:123–130. doi: 10.1247/csf.28.123. [DOI] [PubMed] [Google Scholar]
- 22.Signorelli P., Luberto C., Hannun Y. A. Ceramide inhibition of NF-κB activation involves reverse translocation of classical protein kinase C (PKC) isoenzymes: reqirement for kinase activity and carboxyl-terminal phosphorylation of PKC for ceramide response. FASEB J. 2001;15:2401–2414. doi: 10.1096/fj.01-0244com. [DOI] [PubMed] [Google Scholar]
- 23.Hu T., Exton J. H. Protein kinase Cα translocates to the perinuclear region to activate phospholipase D1. J. Biol. Chem. 2004;279:35702–35708. doi: 10.1074/jbc.M402372200. [DOI] [PubMed] [Google Scholar]
- 24.Becker P. B., Hannun Y. A. Isoenzyme-specific translocation of protein kinase C (PKCβII) and not PKCβI to a juxtanuclear subset of recycling endosomes. J. Biol. Chem. 2004;279:28251–28256. doi: 10.1074/jbc.M400770200. [DOI] [PubMed] [Google Scholar]
- 25.Kajimoto T., Shirai Y., Sakai N., Yamamoto T., Matsuzaki H., Kikkawa U., Saito N. Ceramide-induced apoptosis by translocation, phosphorylation and activation of protein kinase Cδ in the Golgi complex. J. Biol. Chem. 2004;279:12668–12676. doi: 10.1074/jbc.M312350200. [DOI] [PubMed] [Google Scholar]
- 26.Burke T. F., Cocke K. S., Lemke S. J., Angleton E., Becker G. W., Beckmann R. P. Identification of a BRCA1-associated kinase with potential biological relevance. Oncogene. 1998;16:1031–1040. doi: 10.1038/sj.onc.1201623. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi H., Suzuki K., Namiki H. Pervanadate-induced reverse translocation and tyrosine phosphorylation of phorbol ester-stimulated protein kinase C βII are mediated by Src-family tyrosine kinases in porcine neutrophils. Biochem. Biophys. Res. Commun. 2004;314:830–837. doi: 10.1016/j.bbrc.2003.12.163. [DOI] [PubMed] [Google Scholar]
- 28.Preiss J., Loomis C. R., Bishop W. R., Stein R., Niedel J. E., Bell R. M. Quantitative measurement of sn-1,2-diacylglycerols present in platelets, hepatocytes and ras- and sis-transformed normal rat kidney cells. J. Biol. Chem. 1986;261:8597–8600. [PubMed] [Google Scholar]
- 29.Wender P. A., Irie K., Miller B. L. Identification, activity and structural studies of peptides incorporating the phorbol ester-binding domain of protein kinase C. Proc. Natl. Acad. Sci. U.S.A. 1995;92:239–243. doi: 10.1073/pnas.92.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kajimoto T., Ohmori S., Shirai Y., Sakai N., Saito N. Subtype-specific translocation of the δ subtype of protein kinase C and its activation by tyrosine phosphorylation induced by ceramide in HeLa cells. Mol. Cell. Biol. 2001;21:1769–1783. doi: 10.1128/MCB.21.5.1769-1783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheeseman K. L., Ueyama T., Michaud T. M., Kashiwagi K., Wang D., Flax L. A., Shirai Y., Loegering D. J., Saito N., Lennartz M. R. Targeting of PKC-ϵ during FcγR-dependent phagocytosis requires the ϵC1B domain and phospholipase C-γ1. Mol. Bol. Cell. 2006;17:799–813. doi: 10.1091/mbc.E04-12-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Violin J. D., Zhang J., Tsien R. Y., Newton A. C. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J. Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perander M., Bjørkøy G., Johansen T. Nuclear import and export signals enable rapid nucleocytoplasmic shuttling of the atypical protein kinase Cλ. J. Biol. Chem. 2001;276:13015–13024. doi: 10.1074/jbc.M010356200. [DOI] [PubMed] [Google Scholar]
- 34.Medkova M., Cho W. Interplay of C1 and C2 domain of protein kinase C-α in its membrane binding and activation. J. Boil. Chem. 1999;274:19852–19861. doi: 10.1074/jbc.274.28.19852. [DOI] [PubMed] [Google Scholar]
- 35.Bittova L., Stahelin B. V., Cho W. Roles of ionic residues of the C1 domain in protein kinase C-α activation and the origin of phosphatidylserine specificity. J. Biol. Chem. 2001;276:4218–4226. doi: 10.1074/jbc.M008491200. [DOI] [PubMed] [Google Scholar]
- 36.Stahelin R. V., Digman M. A., Medkova M., Ananthanarayanan B., Rafter J. D., Melowic H. R., Cho W. Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase Cδ. J. Biol. Chem. 2004;279:29501–29512. doi: 10.1074/jbc.M403191200. [DOI] [PubMed] [Google Scholar]
- 37.Divecha N., Banfic H., Irvine R. F. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. EMBO J. 1991;10:3207–3214. doi: 10.1002/j.1460-2075.1991.tb04883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin T. F. J., Hsieh K.-P., Porter B. W. The sustained second phase of hormone-stimulated diacylglycerol accumulation does not activate protein kinase C in GH3 cells. J. Biol. Chem. 1990;265:7623–7631. [PubMed] [Google Scholar]
- 39.Baron C. L., Malhotra V. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science. 2002;295:325–328. doi: 10.1126/science.1066759. [DOI] [PubMed] [Google Scholar]
- 40.Bivona T. G., Perez, De Castro I., Ahearn I. M., Grana T. M., Chiu V. K., Lockyer P. J., Cullen P. J., Pellicer A., Cox A. D., Philips M. R. Phospholipase Cγ activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424:694–698. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- 41.Dries D. R., Gallegos L. L., Newton A. C. A single residue in the C1 domain sensitizes novel protein kinase C isoforms to cellular diacylglycerol production. J. Biol. Chem. 2006;282:826–830. doi: 10.1074/jbc.C600268200. [DOI] [PubMed] [Google Scholar]