Abstract
Leishmania donovani is an intracellular protozoan parasite that causes kala-azar in humans. During infection the extracellular insect forms (promastigotes) undergo rapid differentiation to intracellular amastigotes that proliferates in phagolysosomes of mammalian macrophages. We used microarray-based expression profiling to investigate the time-course of changes in RNA abundance during promastigote-to-amastigote differentiation in a host-free system that mimics this process. These studies revealed that several hundred genes underwent an ordered progression of transient or permanent up- and down-regulation during differentiation. Genes that were permanently up-regulated in amastigotes were enriched for transporters and surface proteins, but under-represented in genes involved in protein and other metabolism. Most of these changes occurred late in the differentiation process, when morphological differentiation was essentially complete. Down-regulated genes were over-represented in those involved in cell motility, growth and/or maintenance, and these changes generally occurred earlier in the process. Genes that were transiently up- or down-regulated during differentiation included those encoding heat shock proteins, ubiquitin hydrolases, RNA binding proteins, protein kinases, a protein phosphatase, and a histone deacetylase. These results suggest that changes in mRNA abundance may be important in signal transduction, as well as protein and mRNA turnover, during differentiation. In addition to these mRNA changes, other transcripts including one or more rRNAs and snoRNAs, and non-coding RNAs from several telomeres, also showed substantial changes in abundance during the differentiation process. This paper provides the first genome-scale quantitative analysis of gene expression during the transition from promastigotes to amastigotes and demonstrates the utility of the host-free differentiation system.
Keywords: L. donovani, differentiation, axenic amastigotes, gene expression, transcript abundance
INTRODUCTION
Intracellular parasitism is a process by which microorganisms cycle between vectors (that transmit the parasites) and hosts (to whom they are pathogenic). As a result, the parasites encounter extreme environmental changes during their lifecycle, to which they respond by differentiating into highly adapted forms that enable them to invade and proliferate inside their hosts. Leishmania donovani, the causative agent of visceral leishmaniasis (known as kala azar in humans), is a parasitic protozoan that cycles between the alimentary tract of sand flies and mammalian macrophages. In the insect vector, the parasites grow as extracellular flagellated promastigotes, which differentiate into intracellular aflagellate amastigotes upon entering the phagolysosome of the host macrophages [1,2]. The amastigotes are adapted to grow and proliferate in the hydrolytic environment inside phagolysosomes [3,4].
Promastigote-to-amastigote differentiation is a complex process that is accompanied by a number of morphological and biochemical changes. Parasites change shape from elongated to spherical and lose most of their flagellum. They undergo a major shift in metabolism, especially in the rate and pH optima for several processes, including DNA synthesis [5] and nutrient uptake [6]. A number of amastigote-specific genes have been identified, including a 3′-nucleotidase [7], the A2 gene family [8,9], HSP100 [10], and a MAP kinase, LMPMK [11]. In addition, certain members of the GP63 and PSA-2 gene families are differentially expressed in amastigotes, and there are differences in the GPI anchor of the latter [12]. In contrast, some processes are down-regulated in amastigotes [13,14], most notably lipophosphglycan (LPG) biosynthesis, resulting in its replacement by glycoinositol phosholipid (GIPL) as the major component of the parasite surface coat [13].
The differentiation process can be mimicked in axenic culture by shifting promastigotes from an insect-like (26°C, pH 7) to an intralysosomal-like (37°C, pH 5.5 and 5% CO2) environment [15-19]. These axenic amastigotes resemble animal-derived amastigotes and have been widely used for investigating parasites activities without the complication of host cell material [20-23]. Time-course analysis of L. donovani differentiation showed that promastigotes expressed the amastigote-specific A2 protein family within an hour of exposure to the intralysosomal environment and at five hours they start to transform to amastigote-shaped cells [19]. This morphological transformation occurs synchronously, while the cells are arrested at the G1 stage of the cell cycle, and is complete within 24 hrs. Differentiation proceeds for an additional two or three days until the parasites compete their shedding of LPG and begin expressing amastigote-specific activities [15,24]. Little is known about the molecular processes that mediate promastigote-to-amastigote differentiation, but it is likely that exposure to the higher temperature and lower pH of the intralysosomal environment initiates a series of changes in gene expression that lead to the morphological changes associated with amastigotes.
Regulation of gene expression in Leishmania is unusual because their protein-coding genes are transcribed as polycistronic RNAs with tens-to-hundreds of adjacent genes on the same DNA strand [25-29]. Mature mRNAs are subsequently obtained from coordinated polyadenylation and trans-splicing, which adds a 39-nt spliced leader (SL) sequence to the 5' end of all mRNAs [30,31]. As a consequence of this unusual gene organization, Leishmania gene expression appears to not be regulated at the level of transcription [32], but stage-specific expression of a number of genes has been shown to be regulated via mRNA stability [8,33-38].
The goal of the present study was to use DNA microarray technology to investigate the possible role of changes in RNA abundance during promastigote-to-amastigote differentiation of L. donovani. Microarray expression profiling has been previously used to compare procyclics, metacyclics and amastigotes of L. major [39-41], L. donovani [42], L. infantum [43] and L. mexicana [44], but none of these studies examined changes in gene expression during the process of differentiation. The results of the present study indicate that there is an ordered progression of specific changes in gene expression during L. donovani promastigote-toamastigote differentiation, with some genes changing expression within 5 hr after exposure to the differentiation signal, and others changing only after 24 hours. We also find that a significant number of genes are transiently up- or down-regulated between 5 and 24 hours; an unexpected behavior given the unusual gene organization of the Leishmania genome. Interestingly, we also observed large changes in snoRNA abundance and telomeric transcripts during differentiation. These results suggest that changes in RNA abundance are important during differentiation, and raise the possibility that mechanisms other than changes in mRNA stability play a role in this process.
MATERIALS AND METHODS
Leishmania strain and growth conditions
A cloned line of L. donovani MHOM/SD/00/1SR (LdoS) was used in all experiments [15]. This cell-line was maintained as a clone by inoculating single colonies of promastigotes from medium 199 agar plates. Promastigotes were grown in medium 199 and supplemented with 10% fetal calf serum at 26°C. Promastigote-to-amastigote differentiation in a host-free culture and the maintenance of axenic amastigotes were performed by inoculating late log phase promastigotes in medium 199 at pH 5.5 containing 25% fetal calf serum and incubating them at 37°C in 5% CO2 environment [19]. Axenic amastigotes were routinely sub-cultured for 10 weeks before differentiation back to promastigotes, by changing the growth medium. For microarray analysis, RNA was harvested at 0 (promastigotes), 5, 10, 24 hrs during differentiation and from fully-formed (2 weeks) axenic amastigotes using Trizol reagent (Gibco BRL).
Preparation of labeled cDNA and microarray hybridization
Fluorescently labeled cDNA was prepared and hybridized to microarrays as previously described [39]. Multiple replicates of all hybridizations were performed to account for sample heterogeneity and possible variation due to hybridization and reciprocal labeling experiments were also carried out to identify dye-bias. The amastigote versus promastigote comparison was repeated nine times with Cy5-labeled amastigote RNA and Cy3-labeled promastigote RNA, and six times with reciprocal labeling; the 5 hr and 24 hr versus promastigote comparisons were each carried out six times in the Cy5 vs. Cy3 direction and four in the reciprocal; and the 10 hr versus promastigote comparison was repeated three times in the Cy5 vs. Cy3 direction. At least two different RNA samples were used for all comparisons except for 10 hr versus promastigote. Slides were scanned using a GenePix Pro 4000 scanner (Axon Instruments) and the data was extracted and initially analyzed using the software supplied with the scanner. Local background was subtracted from the intensity value of each spot on the array and the data was exported as Excel files for further statistical analysis.
Microarray data analysis
Statistical analysis was performed using GenePlus (Enodar Biologic Corporation), as described previously [39]. After data normalization and calculation of Cy5/Cy3 ratios [45], the data for replicates of each GSS element were pooled to obtain the mean fold-change for every time-point relative to promastigotes. Two-group panel and pair-wise analyses of the data score were carried out, using a Bonferroni correction in the latter case. The 9078 GSS elements with data from at least three time points after the differentiation signal were analyzed using the TIGR TMeV software package [46]. The variance filter was used to exclude those elements within the bottom 90th percentile of standard deviations of log2-transformed relative signal intensity. The remaining 907 elements were clustered using two-way Hierarchical Clustering (HCL) with complete linkage with the Pearson Correlation distance metric and divided into 12 clusters by using a distance threshold of 0.272.
In order to test for possible co-regulation the proportion of adjacent genes showing similar up- or down-regulation at each time-point was compared to that for non-adjacent genes using a standard chi-square analysis. Similar analyses were performed for genes within the same polycistronic genes cluster (PGC), i.e. all protein-coding sequences (CDSs) on the same DNA strand, without interruption by a CDS on the opposite strand.
Mapping GSSs to genes
End-sequences of the GSS clones [47] were aligned with the Leishmania major genome sequence v5.0 using cross_match [48] to map the GSS elements to the annotated protein-coding genes [29]. In cases where reads were available from only one end of the clone, the other end was assumed to be 2 kb from the mapped end. The mapped GSS clones were classified into four different categories (Table I, and Supplementary Fig. S1), based on their position relative to the CDS of each gene: those representing a single mRNA; those representing two or more mRNAs; those likely containing only intergenic sequence (including RNA genes); and those which could not be mapped to the LmjF genome. A GSS was assumed to represent a single mRNA if one end was located within the region from 200 bp 5′ of the CDS and 500 bp 3′ (the de facto location of the mRNA) and the other end was located within the same region or within the 5′ or 3′ intergenic region (in cases where inter-CDS distance was less than 700 bp, this space was divided proportionately into 3′ and 5′ UTRs of the adjacent genes). A GSS was assumed to represent two or more mRNA if the ends were located in different CDSs. In these cases, the GSS was arbitrarily assigned to the more 5′ gene, but was given a lower confidence score (see Supplementary Methods). A GSS clone was deemed intergenic if the genomic region spanned by the two ends contained no CDS. Each category was further divided into sub-categories based on the location of the GSS. Using this approach, we identified 4031 (39%) GSS elements that represent a single mRNA, 1257 (12%) likely representing two or more adjacent mRNAs, and 1176 (11%) that likely lie in intergenic regions (see Table I). The remaining 3999 (38%) GSS elements could not be mapped to the genome, mostly due to sequence failure. Since the selection of GSS elements for inclusion on the microarray was random, 2360 (28%) genes were be represented by a single GSS, 1508 (18%) were represented by more than one GSS, and 4437 (53%) were not be represented by any GSS elements on these microarrays (see Table II). Of the first two categories, 1271 and 1404 genes, respectively, are represented by at least one GSS that lies within a single specific CDS. Thus, our microarrays represent 33% (2675) of the LmjF genes with a high degree of certainty, while another 9% (123+660) are represented with a lower degree of certainty (i.e. GSS elements spanning two or more genes) and 5% (114+296) are represented only by GSS elements within adjacent intergenic sequences. GSS elements containing telomeric or sub-telomeric sequences were identified by using cross_match [48] to align the end sequences against the telomeric hexamer repeat (THR), telomere associated sequence (TAS) and Leishmania sub-telomeric repeats (LST-R) sequence [49,50].
Table I.
Assignment of GSS elements to LmjF genes.
| Category | Sub-category | Elements | Total |
|---|---|---|---|
| Single mRNA | Internal | 2074 | 4031 |
| + 5' flank | 1040 | ||
| + 3' flank | 832 | ||
| + 5'and 3' flanks | 85 | ||
| Two or more mRNAs | + UTR of adjacent gene(s) | 601 | 1257 |
| + CDS of adjacent gene(s) | 656 | ||
| Intergenic | Intergenic only | 829 | 1176 |
| + 5'and/or 3' UTR | 347 | ||
| Unknown | Sequence failed | 3952 | 3999 |
| No sequence match | 21 | ||
| kDNA | 26 |
Table II.
Coverage of LmjF genes by GSS elements
| Category | Number | Total |
|---|---|---|
| ≥2 GSSs | ||
| single mRNA only | 662 | |
| single mRNA + others | 609 | 1508 |
| ≥ 2 mRNAs ± intergenic | 123 | |
| intergenic only | 114 | |
| 1 GSS | ||
| single mRNA | 1404 | |
| ≥ 2 mRNAs | 660 | 2360 |
| intergenic | 296 | |
| No GSSs | 4437 | 4437 |
| Total | 8305 | 8305 |
Northern blot analyses
Leishmania donovani total RNA was prepared and subjected to northern blotting as previously described [19]. Probes were labeled with [P32] by random priming of the plasmid pDA-PET 33/7 [51] for LmjF23.1080 (which encodes SHERP1); or DNA fragments that were PCR-amplified from genomic DNA using GCATCGCTGTTGGCACCTTCG and CGTCACTGGGAAGCATCATCA primers for LmjF34.0070 (encoding a ascorbate-dependent peroxidase); ATGGTCGGCGTATGCTTGAGGC and CTGAAGTCACGCATGAAGGGAT primers for LmjF17.1090 (encoding a putative ubiquitin hydrolase); and GCATCGCTGTTGGCACCTTCG and CGTCACTGGGAAGCATCATCA primers for LmjF35.1200 (encoding an RNA helicase).
RESULTS
Identification of stage-regulated genes
In order to determine changes in mRNA abundance during axenic differentiation of L. donovani 1SR (LdoS) promastigotes into amastigotes, RNA was isolated at 5, 10 and 24 hrs after exposure to the differentiation signal (i.e. increasing temperature from 26°C to 37°C and reducing pH from 7 to 5.5), as well as late-log stage promastigotes and fully differentiated axenic amastigotes, and used to probe microarrays containing PCR-amplified DNA from genome survey sequence (GSS) clones of L. major Friedlin (LmjF)[39]. Even though these arrays contained LmjF DNA, more than 90% of the GSS elements showed similar signals to that obtained with LmjF DNA when probed with DNA from LdoS (unpublished data). Plots of the log10 normalized intensity values obtained for each DNA element probed with Cy3- vs. Cy5-labeled promastigote RNA showed a tight clustering (r2=0.99) around the line of best fit (Fig. 1A), indicating excellent reproducibility and little, or no, dye bias. The clustering remained tight for promastigotes vs. 5, 10, 24 hr, and amastigotes, but progressively widened with more outliers, representing RNAs that are differentially regulated during differentiation (Fig. 1B-E). All hybridizations, except the promastigote vs. 10 hr comparison, used at least two separate RNA preparations from each time-point, and we observed biological variation to be of the same order as experimental variation, with correlation coefficients in the range of 0.8-0.9 (data not shown).
Figure 1.
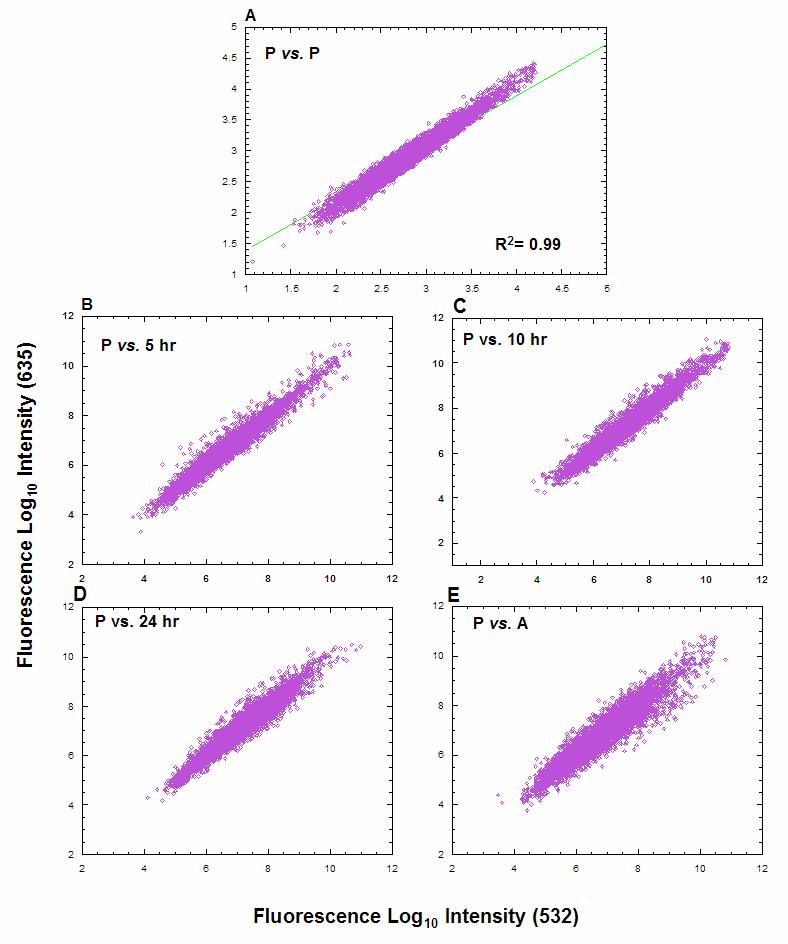
Pair-wise comparison of signal intensity on DNA microarrays hybridized with RNA from Leishmania donovani at different times after exposure to the differentiation signal. Each panel shows a log10 plot of Cy3 (532 nm) vs. Cy5 (635 nm) calibrated fluorescent response from representative hybridizations for (A) Cy3- vs. Cy5-labeled promastigotes (P) immediately before exposure to the differentiation signal, (B) promastigotes vs. parasites 5 hr after exposure to the differentiation signal, (C) promastigotes vs. 10 hr, (D) promastigotes vs. 24 hr and (E) promastigotes vs. axenic amastigotes (A).
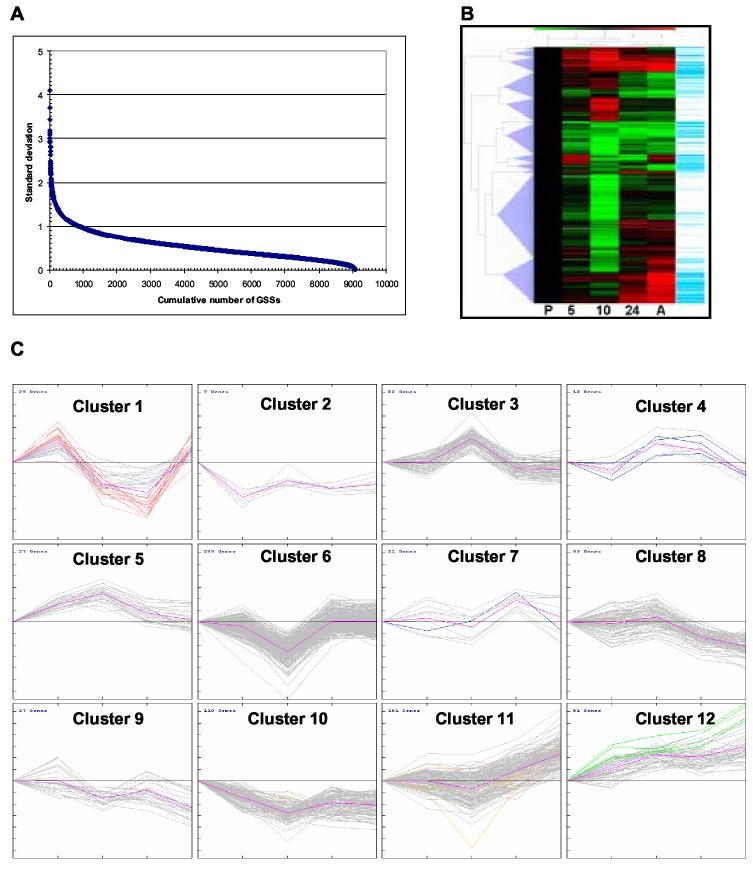
The data from replicate hybridizations were combined after normalization and statistical analysis of the data was carried out using a series of increasingly stringent criteria for differential expression (see Supplementary Methods). Of the 10,468 DNA elements, 1156 (11%) did not show enough signal at any time point to be evaluated, while 4844 (52%) of the remaining elements showed >2-fold change in signal compared to promastigotes at one or more time points, and 683 (7%) showed >5-fold change. Statistical analyses of the 9078 GSS elements with data from at least three time points revealed that 142 (1.6%) passed the most stringent criterion (p<0.5 in a pair-wise analyses with Bonferroni correction), while another 415 (4.5%) passed the more relaxed criteria (see Supplementary Methods). In order to focus on those genes that are the most highly regulated during differentiation, we used the TIGR MeV software package (see Materials and Methods) with a variance filter to include only those elements within the top tenth percentile of standard deviations of log2-transformed relative signal intensity (see Fig. 2A). All 907 of those GSS elements showed a 2-fold or more signal change for at least one time point, with 309 (34%) and 118 (13%) passing the less stringent and most stringent criteria, respectively. Hierarchical clustering (Fig. 2B) enabled us to distinguish 12 different expression profiles (Fig. 2C), which could be grouped into three categories: those up-regulated in amastigotes, those down-regulated in amastigotes, and those up- or down-regulated transiently during the differentiation process. Within each of these categories, we were able distinguish several sub-categories, based on the differences in their expression profiles. In addition, the 5 hr and 10 hr samples clustered together, as did the 24 hr and amastigote samples, indicating the greater relative similarity of gene expression at these time points. This supports our previous observation that morphological differentiation was essentially complete by 24 hr [19], although a number of gene expression changes were still observed between this time point and fully differentiated amastigotes.
Figure 2.
Patterns of gene expression changes during differentiation. (A) The mean fold-change in signal at each time-point during differentiation (relative to promastigotes) was averaged for the 9078 GSS elements with data from at least three time points and the standard deviation (y-axis) plotted against the cumulative number of GSSs (x-axis). (B) The 907 GSS showing the greatest standard deviation were clustered using the TMeV software package. The columns (from left to right) represent promastigotes (P), the 5, 10, 24 hr time-points after exposure to the differentiation signal and amastigotes (A), while the rows represent the fold-change values for each gene. Up-regulation is represented by red, down-regulation is represented by green and no change is represented by black. Genes showing statistically significant (by panel data or 2-group analysis) differences from promastigotes at one or more time-points are indicated by a blue line in the rightmost column. Clustered genes are indicated by the blue triangles to the left. (C) Expression graphs for each of the 12 clusters from panel B, plotting the log2 fold-change values (y-axis) at each of the five time-points (x-axis). The average for all genes in each cluster is shown by the magenta line. Orange and yellow lines indicate GSS elements containing TAS and THR sequences, green lines indicate GSSs containing snoRNA genes, and blue lines indicate GSSs containing kDNA sequence.
Two clusters (11 and 12) contained GSS elements that showed increased signal in amastigotes, which in most cases was statistically significant (Table III). The major difference between the clusters was that those in cluster 12 showed increased signal beginning at 5-10 hrs after exposure to the differentiation signal, while those in cluster 12 showed an increase beginning around 24 hr. A similar number of elements (237) showed decreased signal in amastigotes, but these were represented by four different clusters (2, 10, 9 and 8), which corresponded to decreased signal <5 hr, 5-10 hr, 10-24 hr, or >24 hr after exposure to the differentiation signal, respectively. The remaining clusters represented elements which showed a transient increase (130 GSSs) or decrease (299) in signal during differentiation, or both (29). Interestingly, the up-regulated GSSs fell into four clusters (5, 3, 4, and 7), with transient increases at 5-10 hr, 10 hr only, 10-24 hr and 24 hr only, respectively, while the down-regulated GSSs group into a single cluster (6) showing a transient decrease at 10 hr. A substantial fraction of the elements in clusters 3 and 6 showed transiently increases or decreases at 10 hr only that were not statistically significant, probably as a result of the small number of replicates performed at this time point.
Table III.
Patterns of gene expression during L. donovani differentiation.
| Lifecycle stage | Cluster | Description | Number of GSSs | Number of genesc | ||
|---|---|---|---|---|---|---|
| Totala | Significantb | |||||
| Amastigote | 12 | Up beginning at 5-10 hr | 51 | 44(17)d | 28(13)e | 112 (46) |
| 11 | Up beginning at 24 hr | 161 | 152(51) | 84(32) | ||
| Promastigote | 2 | Down beginning at 5 hr | 7 | 6(0) | 5(4) | 96 (44) |
| 10 | Down beginning at 5-10 hr | 110 | 86(13) | 32(11) | ||
| 9 | Down beginning at 10-24 hr | 27 | 27(7) | 14(7) | ||
| 8 | Down beginning at 24 hr | 93 | 67(16) | 45(22) | ||
| Transient | 5 | Up at 5 and 10 hr | 27 | 25(1) | 19(6) | 54 (8) |
| 3 | Up at 10 hr only | 82 | 28(0) | 31(1) | ||
| 4 | Up at 10 and 24 hr | 10 | 8(3) | 1(0) | ||
| 7 | Up at 24 hr only | 11 | 9(1) | 3(1) | ||
| 6 | Down at 10 hr only | 299 | 138(3) | 75(6) | ||
| 1 | Up at 5 hr, down at 10 and 24 hr, up in amastigotes | 29 | 28(6) | 7(2) | ||
| Total | 907 | 618(118) | 344(105) | |||
Number of GSS elements in each cluster
Number of GSS that showing statistical evidence for a change in signal at one or more time points (see Supplementary Methods)
Number of protein-coding genes represented by these GSSs, after removal of conflicts
The number in parenthesis indicates GSS elements showing p<0.05 by two-group analysis
The numbers in parenthesis indicates the number of genes showing confidence score of ++ or −− at one or more time points
In order to determine which gene(s) is represented by each GSS element, they were mapped to the annotated LmjF genome (Tables I and II) and, where possible, combined to provide the average expression profile for each protein-coding gene. This process also allowed us to remove GSS elements which showed conflicting expression profiles and assign a confidence score combining both the expression and mapping data (see Supplementary Methods). This analysis resulted in identification of 344 protein-coding genes showing potential regulation of gene expression during differentiation (Table III), of which 105 showed a high confidence score.
Genes with higher mRNA abundance in amastigotes
A total of 112 protein-coding genes showed higher expression in axenic amastigotes than in promastigotes (see Table S1), of which 28 were up-regulated within 5-10 hrs after the differentiation signal and 84 were not up-regulated until 24 hrs after exposure to the signal. In general, most genes showed relatively modest (2- to 10-fold) increases in mRNA level, and the changes in gene expression were greater for those genes that were up-regulated earlier during the differentiation process. However, four genes in the first cluster, and eight in the latter, showed substantial (10- to 46-fold) changes in gene expression. With a few exceptions, most genes up-regulated early in the differentiation process also showed continued increases in mRNA levels at later times. These genes include those encoding HASPA2, two SHERPs, a putative phosphatidylinositol 3-kinase, a Golgi transport protein, two ABC transporters, a putative choline/ethanolamine kinase, ascorbate-dependent peroxidase, a tryparedoxin-like protein, a putative ribonuclease, two heat shock proteins, two protein kinases and a number of hypothetical proteins. Those up-regulated later in differentiation include genes encoding several amastins, two dyneins, PPG4, PSA2, a putative ADP-ribosylation factor, four protein kinases, three protein phosphatases, a putative kinesin, a DNA replication licensing factor (minchromosome maintenance complex) subunit, argininosuccinate synthase, a mitochondrial helicase, fatty acyl CoA synthetase, glyceraldehyde 3-phosphate dehydrogenase, a putative lipase, two phosphoglycan-β-1,3-galactosyltransferases, superoxide dismutase, another tryparedoxin-like protein, a protein methyltransferase, three RNA binding proteins, two ABC transporters, an aminophospholipid translocase, an aquaporin, a phosphate permease, a phospholipidtranslocating P-type ATPase, as well as a number of hypothetical proteins.
Northern blot analyses were used to confirm the changes in the gene expression seen for LmjF23.1080, which encodes SHERP1 (Fig. 3A) and LmjF34.0070, which encodes the ascorbate-dependent peroxidase (Fig 3B, panel I). In both cases, agreement between the microarray and Northern blot results was good; with increased mRNA levels beginning by 5 hrs after exposure to the differentiation signal, and continuing to increase throughout differentiation. Indeed, up-regulation of SHERP1 mRNA could be seen as early as 1 hr after the signal, but interestingly never reached the level seen in stationary phase (metacyclic) promastigotes (Fig. 3A, lanes 1 and 2).
Figure 3.
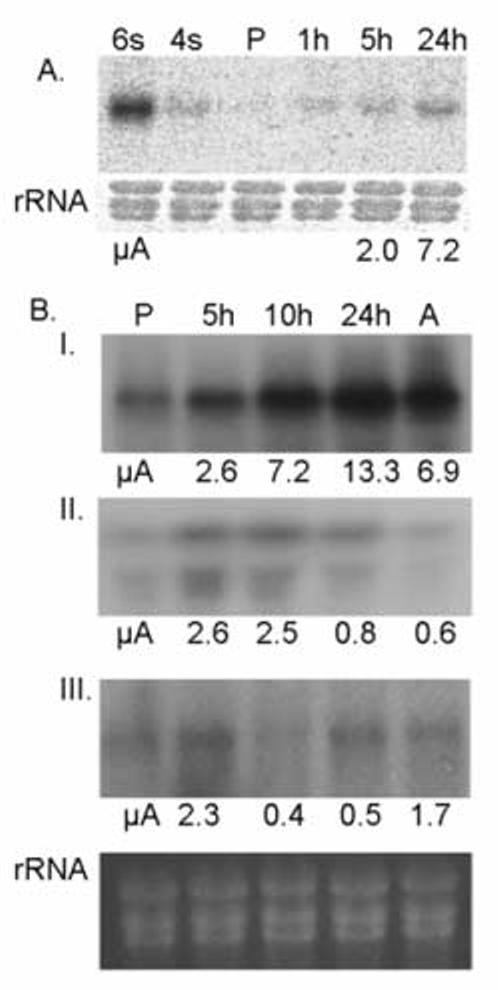
Northern analysis of differentially expressed genes. (A) RNA (10 μg in each lane) from metacyclic promastigotes maintained in stationary phase for 6 (6s) or 4 (4s) days; procyclic promastigotes in late-log phase (P), and 1, 5 and 24 hr after exposure to differentiation signal were hybridized with [32P]-labeled probe for SHERP1 (top panel). rRNA was visualized by ethidium bromide staining (bottom panel). (B) RNA from parasites at immediately before exposure to the differentiation signal (P), as well as 5, 10 and 24 hr after exposure, and axenic amastigotes (A) were hybridized with probes for LmjF34.0070 (top panel I), LmjF35.1200 (panel II) LmjF17.1090 (panel III). rRNAs are shown in the bottom panel. The fold-change values from the microarray results (μA) are indicated below each panel.
Genes with lower mRNA abundance in amastigotes
Cluster analysis of the microarray results revealed 96 genes that showed lower mRNA levels in amastigotes than promastigotes (Table S2), of which 37 were down-regulated within 5-10 hr after exposure to the differentiation signal, 14 beginning 10-24 hr after the signal, and 45 only at 24 hr or later. Once again most changes were relatively modest, but 24 genes showed 5-to 10-fold down-regulation. In contrast to the amastigote up-regulated genes described above, most of the genes that are down-regulated early during the differentiation process (clusters 2 and 9) showed rapid decreases in mRNA levels at 5 or 10 hrs, after which time they did not decline further. These genes include those encoding PSA2, HSP90 organizing protein, an amastin, a DNA topoisomerase IB subunit, an importin β-1 subunit, a pteridine transporter, paraflagellar rod protein 1D, an actin-like protein, a kinesin, a nucleolar GTP binding protein, the η subunit of T-complex protein 1, acetyl-CoA carboxylase, an RNA helicase, fatty acid elongase, mitochondrial proline oxidase, S-adenosylhomocysteine hydrolase, asparaginyl-tRNAsynthetase, a cysteine peptidase, putative GPI transamidase, a MAP kinase, ubiquitin activating enzyme E1, as well as several hypothetical proteins. Genes encoding dynein heavy chain, the γ and ϕ subunits of T-complex protein 1, enolase, a putative NADP-dependent sterol dehydrogenase, a pathothenate kinase subunit, translation elongation factor 2, HSP83, the α7 subunit of the proteasome, and five hypothetical proteins were down-regulated by 10-24 hrs after exposure to the differentiation signal, and generally continue to decrease slowly (cluster 9). Genes down-regulated only late in the differentiation process (cluster 8) include those encoding prostaglandin F2α synthase, a heat shock 90-related protein involved in lipophosphoglycan biosynthesis, two subunits of the mitochondrial DNA polymerase, an ABC transporter, mitochondrial HSP60, β-tubulin, actin, a CDC2-related protein kinase, a cyclin, an SMC (Structural Maintenance of Chromosomes) protein, a mitochondrial 3,2-trans-enoyl-CoA isomerase, an RNA helicase, a putative β-fructofuranosidase, a methylcrotonyl-CoA carboxylase subunit, ornithine decarboxylase, pyrroline-5-carboxylase, two peptidases, three protein kinases, a protein phosphatase, an N-myristoyl transferase, ribosomal protein L15 and a number of hypothetical proteins.
Transiently regulated genes
In addition to the genes described above, whose expression was permanently up- or down-regulated during differentiation, these analyses also identified 54 genes that were transiently up-regulated (Table S3), 75 that were transiently down-regulated (Table S4), and 7 that were initially up-regulated, then down-regulated, before returning to promastigote levels (or slightly higher; Table S5). A smaller proportion of these genes showed a high confidence score, probably due to the smaller number of replicates performed at the 10 hr time point. The transient increases in gene expression were generally modest, although 12 genes showed changes of 5- to 13-fold, and could be divided into 19 that occurred very early (5 hr) during differentiation (cluster 5); 31 that occurred at 10 hr after exposure to the differentiation signal (cluster 6); and 3 that occurred late (24 hr) during the differentiation process (cluster 7). Genes in the first group include those encoding nuclear transport factor, a putative 4-coumarate:CoA ligase, a putative acetylornithine deacetylases, an RNA helicase involved in pre-mRNA splicing, cyclophilin, three heat shock proteins, a MAP kinase, and several hypothetical proteins. Genes transiently up-regulated at 10 hr include those encoding an anion-transporting ATPase, a histone deacetylase, a putative lysophospholipase, an RNA binding protein, a heat shock protein, a protein kinase, ubiquitin fusion degradation protein, and numerous hypothetical proteins. A putative nucleoside transporter and two hypothetical protein genes are transiently up-regulated late in the differentiation process. Northern blot analyses confirmed transient up-regulation of expression for LmjF35.1200 (encoding the pre-mRNA splicing factor) by showing elevated mRNA levels at 5, 10 and possibly 24 hrs after the differentiation signal (Fig. 3B, panel II).
Conversely, almost all transiently down-regulated genes showed decreased mRNA levels only at 10 hrs after exposure to the differentiation signal (cluster 6), and in 36 cases this decrease was substantial (5- to 22-fold). These genes include those encoding PSA2, dihydrolipoamide dehydrogenase, dynein light chain, dynein heavy chain, two putative kinesins, phosphatidylinositol kinase, a putative helicase involved in mitochondrial DNA repair and recombination protein, a putative 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial aldehyde dehydrogenase, cytochrome p450 reductase, cytochrome b5, a galactofuranosyltransferase, a putative lathosterol oxidase, a putative phenylalanine-4-hydroxylase, a thiol-dependent reductase, a heat shock protein, a protein kinase, a protein phosphatase, a metallopeptidases, a ubiquitin hydrolase, a putative v-SNARE protein, three RNA binding proteins, an amino acid transporter, a clathrin coat assembly protein, a glycerol uptake protein, and a number of hypothetical proteins.
A small number of protein-coding genes were initially up-regulated 2- to 4- fold at 5 hr after exposure to the differentiation signal, subsequently down-regulated 2- to 4- fold at 10-24 hr, and finally returned to promastigote (or slightly higher) levels in amastigotes (see cluster 1 in Fig. 2C). These genes encode a putative cell cycle division protein, an RNA helicase, a ubiquitin hydrolase, and four hypothetical proteins. Northern blot analysis for LmjF17.1090 (encoding the ubiquitin hydrolase) confirmed that it is transiently up-regulated at 5 hr and down-regulated at 10 hr (Fig. 3B, panel III), although there is disagreement between the Northern blot and microarray data at the 24 hr time-point.
Other regulated RNAs
Of the 907 GSS elements that were used for the cluster analysis described above, 133 contained only intergenic regions. While most of these showed similar expression patterns to the adjacent protein-coding genes, suggesting that they represented mRNAs with extended 3′ UTRs (i.e. longer than the 500 nt used in our mapping); 55 showed conflicting patterns or clearly represented non-protein-coding RNAs (see Table S6). Seven GSS elements from the rRNA locus showed considerably (5- to 20-fold) lower signal beginning within 5 hr after exposure to the differentiation signal (cluster 10). It is not yet clear which particular transcripts within the rRNA region undergo this differentiation expression. Conversely, two GSS elements from the SL RNA locus show an increase in signal very late (in amastigotes) during the differentiation. However, the majority of GSSs representing the SL locus showed no change in signal during differentiation. Six GSS elements, representing a snoRNA cluster on chr35 (between LmjF35.4190 and LmjF35.4200), showed a dramatic (30- to 100-fold) progressive increase in signal beginning very early during differentiation (cluster 12), while two GSSs from this region showed a modest decrease (cluster 10), and one GSS showed little change. Two other snoRNA loci, on chr5 and chr26, appear to be transiently down-regulated during differentiation (cluster 6), with the latter also up-regulated in amastigotes. A single GSS representing the U2 snRNA and a tRNAAla on chr31 showed a substantial (2.7- to 6.5-fold) increase in signal beginning at 5 hr and continuing into amastigotes. We have not yet determined whether expression of both of these RNAs, or only one, is regulated during differentiation.
In addition to the annotated RNA genes described above, at least two other intergenic regions also appear to undergo regulation of RNA expression during differentiation. Three GSSs from a large intergenic region between LmjF34.2800 and LmjF34.2810 showed considerable (15- to 45-fold) increase in signal beginning 10-24 hrs after the differentiation signal, as did two GSSs from the intergenic region between LmjF31.1530 and LmjF31.1540. In addition, GSSs from several other large intergenic regions on chr18, 21, 23, 30, 31, and 33 also showed evidence of differential regulation, but the changes were more modest (see Table S6), and may represent false positives.
Interestingly, most of the GSSs containing telomeric or sub-telomeric sequences showed considerable changes in gene expression during differentiation (Table S7). Indeed, most of the GSS elements in cluster 1 (Fig. 2C) consisted of telomeric hexamer repeat (THR) and telomere associated sequences (TAS), and thus represented transcripts from the extreme ends of chromosomes, although it was not possible to determine from which chromosome(s) they were derived. These telomeric RNAs were initially up-regulated (4- to 12-fold) at 5 hr, followed by a substantial (8- to 27-fold) down-regulation at 10 and 24 hr, then a modest (2-to 6-fold) up-regulation in amastigotes. Another group of telomeric GSSs appears to be down-regulated slightly early in differentiation, then substantially (2- to 10-fold) up-regulated very late during differentiation (cluster 11). These GSSs contain one or more Leishmania Sub-Telomeric Repeat (LST-R) sequences [50] and are probably the right end of chr1 or chr22. Conversely, other LST-R-containing GSSs (from chr29 and chr22) show decreased signal beginning at 5 hr and continuing throughout differentiation (cluster 10). Interestingly, not all GSS elements containing LST-R sequences appear to shown differentiation expression. Finally, transcripts from another telomeric sequence (chr30R) were down-regulated late during the differentiation process (cluster 8).
Co-regulation of RNAs?
Analysis of the microarray data described above indicated that there was no statistically significant correlation between the expression profiles for adjacent genes, suggesting that most are not co-regulated. Unlike in most other eukaryotes, the protein-coding genes in Leishmania (and other trypanosomatids) are organized into large polycistronic gene clusters (PGCs) with tens-to-hundreds of adjacent genes [25,29] apparently transcribed from a single RNA polymerase II transcription initiation site [27,28]. Only eight of the 125 PGCs examined showed evidence for correlation of gene expression profiles that was significantly different (p<0.01) from that expected by chance (Table IV). However, even in these cases (with the exception of one PGC with only two genes) fewer than half of the genes were co-regulated at a single time-point.
Table IV.
Co-regulated genes in polycistronic gene clusters
| Down-regulated | Up-regulated | |||||||
|---|---|---|---|---|---|---|---|---|
| Polycistronic Gene Cluster |
5 hr | 10 hr | 24 hr | ama | 5 hr | 10 hr | 24 hr | ama |
| 6-3 | 1/2** | NS | 1/2* | 2/2*** | NS | NS | NS | NS |
| 8-3 | NS | NS | NS | NS | NS | NS | 4/13** | 4/13* |
| 9-1 | NS | NS | NS | NS | 4/39* | NS | NS | 10/39** |
| 11-1 | NS | NS | NS | 2/69* | NS | NS | 9/69* | 22/69*** |
| 26-1 | NS | NS | NS | NS | NS | 14/40** | 6/40* | NS |
| 28-3 | 4/29* | 10/29** | NS | NS | NS | NS | NS | NS |
| 29-2 | 3/25* | NS | NS | NS | NS | NS | 6/25** | NS |
| 31-1 | NS | 13/224* | NS | NS | NS | NS | NS | 39/224** |
| 35-2 | 7/61** | NS | NS | NS | NS | NS | NS | NS |
α<0.0001
α<0.001
α<0.05
NS – not significant
DISCUSSION
Promastigote-to-amastigote differentiation in Leishmania is a complex process, but the morphological changes that take place appear to be well-coordinated and regulated [19]. In other organisms, DNA microarray analysis has become a popular and powerful method for examining changes in gene expression during differentiation and/or adaptation to new growth conditions [52-57]. While this approach has been used previously to compare the different lifecycle stages (procyclics, metacyclics and amastigotes) of L. major [39-41], L. donovani [42], L. infantum [43], and L. mexicana [44], the work presented in this paper represents the first genome-wide analysis of changes in gene expression during the transition from promastigotes to amastigotes, a process which was impossible to follow until host-free differentiation systems had been developed. These experiments, especially the short time points, would be impossible to carry out using animal-derived parasites, and very difficult even using in vitro macrophage cultures.
The major new findings of this study are that expression of a substantial number of genes is transiently up- or down-regulated during differentiation and the permanent changes in genes expression occur at different times during differentiation. Thus, there appears to be an ordered progression of specific changes in gene expression after exposure of L. donovani promastigotes to the differentiation signal (37°C, pH 5.5), which eventually results in morphological and physiological differentiation to amastigotes that are adapted to intracellular growth in the macrophage phagolysosome.
Transient changes in gene expression during differentiation, development, or cell cycle progression have been described in several other organisms [54,57-59]. While we report here that 344 protein-coding genes are likely to be differentially regulated in terms of mRNA abundance during promastigote-to-amastigote differentiation, it is important to remember that these studies likely under-estimate the actual number of genes that are differentially regulated, since only about half of the predicted protein-coding genes are represented on our microarrays by GSS elements, and some of the GSSs overlap two or more genes. However, even the proportion of genes that change expression during L. donovani differentiation (∼9%) is substantially lower than that (40-86%) observed during development of the worm, fruit-fly or mouse [54,60,61] or the ∼75% of genes that are transiently activated during the 48-hour asexual intraerythocytic developmental cycle of Plasmodium falciparum, the causative agent of human malaria [57]. Indeed, the proportion is even lower than the changes seen during development of Dictyostelium [62] or during the cell cycles of the yeast [59] and HeLa cells [58], during which ∼15% of the total genome is periodically regulated. This relative dearth of gene expression changes at the level of transcript abundance probably reflects the lack of transcriptional control in the trypanosomatids [32,63].
Nevertheless, these studies revealed more differentially expressed genes than some previous microarray studies comparing Leishmania promastigotes and amastigotes, which reported only about 1-3% of the genes undergoing differential regulation in L. major [40] or L. mexicana [44], although another report claimed ∼35% of the genes surveyed showed differential expression between Leishmania promastigotes and amastigotes [41]. In this study, we find that ∼3% of the genes surveyed appeared to be up-regulated in amastigotes, relative to promastigotes, 2.5% are down-regulated, and ∼3.5% underwent transient changes during differentiation. It is likely that the inconsistencies between studies reflect differences in experimental protocols (such the use of axenic versus lesion-derived amastigotes and GSS or EST versus oligonucleotide probes), criteria used to define stage-regulated genes, and/or species-specific differences in differential gene expression. For example, we found the SHERP1 gene was up-regulated within 1 hr after exposure to the differentiation signal in L. donovani, and remained 10- to 40-fold higher in amastigotes than promastigotes (see Table S2 and Fig. 3A), whereas previous studies reported the increase to be metacyclic-specific with decreased expression in L. major lesion amastigotes [40,41,51]. This discrepancy may reflect a species-specific difference in SHERP expression or it may represent a difference between the axenic amastigotes used in our study and the lesion-derived amastigotes used by others. The L. mexicana study [44] found a number of differences in gene expression between lesion-derived and axenic amastigotes.
Interestingly, the majority (84/112) of genes that are up-regulated during differentiation do not change significantly until late (24 hours or more) in the differentiation process, when morphological differentiation is essentially complete. Conversely, a higher proportion (51/96) of those genes that are down-regulated (especially those change by more than 5-fold), show significant changes early in differentiation, suggesting that different pathways are involved in up-regulation and down-regulation. It is also interesting that there are more changes in gene expression at 10 hrs than at any of the other times. However, it is not yet clear whether this truly represents a critical time during differentiation or merely reflects a larger proportion of false positives due to the smaller number of replicates at this time-point.
Analysis of the stage-regulated genes by their Gene Ontology annotation showed that those that were down-regulated in amastigotes (i.e. up-regulated in promastigotes) were enriched for genes involved in cell growth and/or maintenance as well as cell motility, while those that were up-regulated in amastigotes were under-represented in genes involved in protein and other metabolism, but enriched for transporters and proteins of unknown function. These different distributions probably reflect the motility and higher growth rate of promastigotes and a larger number of uncharacterized amastigote-specific functions.
The differentiation process in Leishmania is likely to involve one or more signal transduction pathways which trigger changes in gene expression and involve a cascade of molecular interactions. In other organisms, these pathways often involve protein kinase/phosphatase cascades that results in transcriptional up- or down-regulation of specific effector genes. While these studies will not identify the earliest components of these signal transduction pathways, since these are likely to involve changes in protein turn-over and/or phosphorylation state, they should reveal components of more downstream effector pathways. In particular, those genes whose expression is transiently up- or down-regulated during differentiation would be likely candidates for secondary components involved in up-regulating expression of amastigote-specific genes. These may include the protein kinase genes (LmjF36.0860, and LmjF11.0510) that are transiently up- and down-regulated, respectively, at 5-10 hrs, as well as the heat shock proteins (encoded by LmjF30.2490, LmjF17.0040 and LmjF33.0360) transiently up-regulated early in the differentiation process.
Our analyses provide little evidence for co-regulation of genes within the same polycistronic gene cluster, suggesting that the genes are regulated individually. This is consistent with the finding that Leishmania (and other trypanosomatids) appear not to regulate gene expression at the level of transcription, relying instead on changes in mRNA stability and translation [32]. The presence of several RNA binding proteins and helicases among those regulated during the differentiation process also supports this hypothesis. Interestingly, a number of up-regulated genes (mostly amastins, but also HSP100) contain 3′ UTR sequences similar to those implicated in stage-specific changes in gene expression in L. infantum, even though this sequence appears to be involved in translational control [64]. Nevertheless, the transient up-regulation of a histone deacetylase gene (LmjF21.0680) at 10 hrs is provocative in suggesting possible changes in chromatin structure, which may lead to alterations in the rate of global or specific transcription.
In addition to the changes in protein-coding gene expression, we also observed substantial (20- to 100-fold) changes in RNA concentration from a number of non-coding and/or intergenic regions. At least one of the rRNAs appears to be down-regulated during differentiation, perhaps reflecting a generalized decrease in protein synthesis, while several snoRNAs are up- or down-regulated, presumably resulting in amastigote-specific RNA modifications. Transcripts from at least two intergenic regions (LmjF34.2800-LmjF34.2810 and LmjF31.1530-LmjF31.1540) are also up-regulated in amastigotes. Characterization of these RNAs has not yet been completed, but some are adjacent to short degenerate non-LTR retrotransposon-like sequences.
Transcripts containing telomere-associated (THR and TAS) sequences were more abundant in amastigotes than promastigotes, as shown previously [40]. Interestingly, these transcripts were apparently up-regulated at 5 hours, then substantially down-regulated at 10 and 24 hours. Due to the repetitive nature of these sequences, it is not clear whether this apparent differentiation-associated regulation occurs at all telomeres, or only a sub-set. However, subtelomeric repetitive sequences on chr1 and/or chr22 appear to be up-regulated throughout differentiation, while those on chr20 and chr2/29 appear to be down-regulated. The results are confirmed by a recent publication showing that sub-telomeric non-coding RNAs are developmentally up-regulated in L. infantum amastigotes [65]. The significance of these changes in telomeric transcript abundance is not yet clear.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Stephen Beverley (Washington University) for the kind gift of the LmjF GSS clones and end-sequences (GenBank Accession numbers AQ843743-AQ853356, AQ901732-AQ902705 and AQ911373-AQ912039), the SBRI genome sequencing team for additional end-sequencing, Aaron Leland for his technical assistance with RNA labeling, and Dr. Deborah Smith for her gift of the pDA-PET 33/7 plasmid. This work was supported by PHS grant AI47234 to PJM from the National Institutes of Health, Grant T24-86-1 from the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases and grant 2003237 from the U.S.-Israel Binational Foundation.
Abbreviations
- (LdoS)
L. donovani MHOM/SD/00/1SR
- (PGC)
Polycistronic Gene Cluster
- (GSS)
Genome Survey Sequence
- (THR)
Telomere hexamer repeat
- (TAS)
Telomere associated sequence
- (PSA2)
promastigotes surface antigen
- (HSP90)
Heat shock protein 90
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chang KP, Dwyer DM. Multiplication of human parasite Leishmania donovani in the phagolysosomes of hamster macrophages in vitro. Science. 1976;193:678–80. doi: 10.1126/science.948742. [DOI] [PubMed] [Google Scholar]
- 2.Killick-Kendrick R. Biology of Leishmania in phlebotime sand flies. In: Lumsden WHR, Evans DA, editors. Biology of Kinetoplastidae. Academic Press; New York: 1979. pp. 395–449. [Google Scholar]
- 3.Zilberstein D, Shapira M. The role of pH and temperature in the development of Leishmania parasites. Annu Rev Microbiol. 1994;48:449–70. doi: 10.1146/annurev.mi.48.100194.002313. [DOI] [PubMed] [Google Scholar]
- 4.Burchmore RJ, Barrett MP. Life in vacuoles--nutrient acquisition by Leishmania amastigotes. Int J Parasitol. 2001;31:1311–20. doi: 10.1016/s0020-7519(01)00259-4. [DOI] [PubMed] [Google Scholar]
- 5.Mukkada AJ, Meade JC, Glaser TA, Bonventre PF. Enhanced metabolism of Leishmania donovani amastigotes at acid pH: an adaptation for intracellular growth. Science. 1985;229:1099–101. doi: 10.1126/science.4035350. [DOI] [PubMed] [Google Scholar]
- 6.Mazareb S, Fu ZY, Zilberstein D. Developmental regulation of proline transport in Leishmania donovani. Exp Parasitol. 1999;91:341–8. doi: 10.1006/expr.1998.4391. [DOI] [PubMed] [Google Scholar]
- 7.Bates PA. Characterization of developmentally-regulated nucleases in promastigotes and amastigotes of Leishmania mexicana. FEMS Microbiol Lett. 1993;107:53–8. doi: 10.1016/0378-1097(93)90353-4. [DOI] [PubMed] [Google Scholar]
- 8.Charest H, Zhang WW, Matlashewski G. The developmental expression of Leishmania donovani A2 amastigote-specific genes is post-transcriptionally mediated and involves elements located in the 3′-untranslated region. J Biol Chem. 1996;271:17081–90. doi: 10.1074/jbc.271.29.17081. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W-W, Matlashewski G. Loss of virulence in Leishmania donovani deficient in anamastigote-specific protein, A2. Proc Natl Acad Sci U S A. 1997;94:8807–11. doi: 10.1073/pnas.94.16.8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubel A, Krobitsch S, Horauf A, Clos J. Leishmania major Hsp100 is required chiefly in the mammalian stage of the parasite. Mol Cell Biol. 1997;17:5987–95. doi: 10.1128/mcb.17.10.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiese M, Gorcke I. Homologues of LMPK, a mitogen-activated protein kinase from Leishmania mexicana, in different Leishmania species. Med Microbiol Immunol (Berl ) 2001;190:19–22. doi: 10.1007/s004300100072. [DOI] [PubMed] [Google Scholar]
- 12.Handman E, Osborn AH, Symons F, Van Driel R, Cappai R. The Leishmania promastigote surface antigen 2 complex is differentially expressed during the parasite life cycle. Mol Biochem Parasitol. 1995;74:189–200. doi: 10.1016/0166-6851(95)02500-6. [DOI] [PubMed] [Google Scholar]
- 13.McConville MJ, Blackwell JM. Developmental changes in the glycosylated phosphatidylinositols of Leishmania donovani. Characterization of the promastigote and amastigote glycolipids. J Biol Chem. 1991;266:15170–9. [PubMed] [Google Scholar]
- 14.Turco SJ, Sacks DL. Expression of a stage-specific lipophosphoglycan in Leishmania major amastigotes. Mol Biochem Parasitol. 1991;45:91–9. doi: 10.1016/0166-6851(91)90030-a. [DOI] [PubMed] [Google Scholar]
- 15.Saar Y, Ransford A, Waldman E, et al. Characterization of developmentally-regulated activities in axenic amastigotes of Leishmania donovani. Mol Biochem Parasitol. 1998;95:9–20. doi: 10.1016/s0166-6851(98)00062-0. [DOI] [PubMed] [Google Scholar]
- 16.Gupta N, Goyal N, Rastogi AK. In vitro cultivation and characterization of axenic amastigotes of Leishmania. Trends Parasitol. 2001;17:150–3. doi: 10.1016/s1471-4922(00)01811-0. [DOI] [PubMed] [Google Scholar]
- 17.Somanna A, Mundodi V, Gedamu L. In vitro cultivation and characterization of Leishmania chagasi amastigote-like forms. Acta Trop. 2002;83:37–42. doi: 10.1016/s0001-706x(02)00054-2. [DOI] [PubMed] [Google Scholar]
- 18.Debrabant A, Joshi MB, Pimenta PF, Dwyer DM. Generation of Leishmania donovani axenic amastigotes: their growth and biological characteristics. Int J Parasitol. 2004;34:205–17. doi: 10.1016/j.ijpara.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Barak E, Amin-Spector S, Gerliak E, Goyard S, Holland N, Zilberstein D. Differentiation of Leishmania donovani in host-free system: analysis of signal perception and response. Mol Biochem Parasitol. 2005;141:99–108. doi: 10.1016/j.molbiopara.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Mengeling BJ, Zilberstein D, Turco SJ. Biosynthesis of Leishmania lipophosphoglycan: solubilization and partial characterization of the initiating mannosylphosphoryltransferase. Glycobiology. 1997;7:847–53. doi: 10.1093/glycob/7.6.847. [DOI] [PubMed] [Google Scholar]
- 21.Shaked-Mishan P, Ulrich N, Ephros M, Zilberstein D. Novel Intracellular SbV reducing activity correlates with antimony susceptibility in Leishmania donovani. J Biol Chem. 2001;276:3971–6. doi: 10.1074/jbc.M005423200. [DOI] [PubMed] [Google Scholar]
- 22.Bente M, Harder S, Wiesgigl M, et al. Developmentally induced changes of the proteome in the protozoan parasite Leishmania donovani. Proteomics. 2003;3:1811–29. doi: 10.1002/pmic.200300462. [DOI] [PubMed] [Google Scholar]
- 23.Goyard S, Segawa H, Gordon J, Showalter M, Duncan R, Turco SJ, Beverley SM. An in vitro system for developmental and genetic studies of Leishmania donovani phosphoglycans. Mol Biochem Parasitol. 2003;130:31–42. doi: 10.1016/s0166-6851(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 24.Ephros M, Bitnun A, Shaked P, Waldman E, Zilberstein D. Stage-specific activity of pentavalent antimony against Leishmania donovani axenic amastigotes. Antimicrob Agents Chemother. 1999;43:278–82. doi: 10.1128/aac.43.2.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myler PJ, Audleman L, deVos T, et al. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc Natl Acad Sci U S A. 1999;96:2902–6. doi: 10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Worthey E, Martinez-Calvillo S, Schnaufer A, et al. Leishmania major chromosome 3 contains two long “convergent” polycistronic gene clusters separated by a tRNA gene. Nucl Acids Res. 2003;31:4201–10. doi: 10.1093/nar/gkg469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Calvillo S, Yan S, Nguyen D, Fox M, Stuart KD, Myler PJ. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol Cell. 2003;11:1291–9. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Calvillo S, Nguyen D, Stuart K, Myler PJ. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot Cell. 2004;3:506–17. doi: 10.1128/EC.3.2.506-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivens AC, Peacock CS, Worthey EA, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–42. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry K, Agabian N. mRNA processing in the Trypanosomatidae. Experientia. 1991;47:118–28. doi: 10.1007/BF01945412. [DOI] [PubMed] [Google Scholar]
- 31.LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 32.Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–8. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aly R, Argaman M, Halman S, Shapira M. A regulatory role for the 5′ and 3′ untranslated regions in differential expression of hsp83 in Leishmania. Nucl Acids Res. 1994;22:2922–9. doi: 10.1093/nar/22.15.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beetham JK, Myung KS, McCoy JJ, Donelson JE. Glycoprotein 46 mRNA abundance is post-transcriptionally regulated during development of Leishmania chagasi promastigotes to an infectious form. J Biol Chem. 1997;273:17360–6. doi: 10.1074/jbc.272.28.17360. [DOI] [PubMed] [Google Scholar]
- 35.Burchmore RJ, Landfear SM. Differential regulation of multiple glucose transporter genes in Leishmania mexicana. J Biol Chem. 1998;273:29118–26. doi: 10.1074/jbc.273.44.29118. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y, El Fakhry Y, Sereno D, Tamar S, Papadopoulou B. A new developmentally regulated gene family in Leishmania amastigotes encoding a homolog of amastin surface proteins. Mol Biochem Parasitol. 2000;110:345–57. doi: 10.1016/s0166-6851(00)00290-5. [DOI] [PubMed] [Google Scholar]
- 37.Brittingham A, Miller MA, Donelson JE, Wilson ME. Regulation of GP63 mRNA stability in promastigotes of virulent and attenuated Leishmania chagasi. Mol Biochem Parasitol. 2001;112:51–9. doi: 10.1016/s0166-6851(00)00346-7. [DOI] [PubMed] [Google Scholar]
- 38.Papadopoulou B, Huang X, Boucher N, McNicoll F. Stage-specific regulation of gene expression in Leishmania. ASM News. 2003;69:282–8. [Google Scholar]
- 39.Saxena A, Worthey EA, Yan S, Leland A, Stuart KD, Myler PJ. Evaluation of differential gene expression in Leishmania major Friedlin procyclics and metacyclics using DNA microarray analysis. Mol Biochem Parasitol. 2003;129:103–14. doi: 10.1016/s0166-6851(03)00100-2. [DOI] [PubMed] [Google Scholar]
- 40.Akopyants NS, Matlib RS, Bukanova EN, Smeds MR, Brownstein BH, Stormo GD, Beverley SM. Expression profiling using random genomic DNA microarrays identifies differentially expressed genes associated with three major developmental stages of the protozoan parasite Leishmania major. Mol Biochem Parasitol. 2004;136:71–86. doi: 10.1016/j.molbiopara.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Almeida R, Gilmartin BJ, McCann SH, et al. Expression profiling of the Leishmania life cycle: cDNA arrays identify developmentally regulated genes present but not annotated in the genome. Mol Biochem Parasitol. 2004;136:87–100. doi: 10.1016/j.molbiopara.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Duncan RC, Salotra P, Goyal N, Akopyants NS, Beverley SM, Nakhasi HL. The application of gene expression microarray technology to kinetoplastid research. Curr Mol Med. 2004;4:611–21. doi: 10.2174/1566524043360221. [DOI] [PubMed] [Google Scholar]
- 43.McNicoll F, Drummelsmith J, Muller M, Madore E, Boilard N, Ouellette M, Papadopoulou B. A combined proteomic and transcriptomic approach to the study of stage differentiation in Leishmania infantum. Proteomics. 2006;6:3567–81. doi: 10.1002/pmic.200500853. [DOI] [PubMed] [Google Scholar]
- 44.Holzer TR, McMaster WR, Forney JD. Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol Biochem Parasitol. 2006;146:198–218. doi: 10.1016/j.molbiopara.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 45.Thomas JG, Olson JM, Tapscott SJ, Zhao LP. An efficient and robust statistical modeling approach to discover differentially expressed genes using genomic expression profiles. Genome Res. 2001;11:1227–36. doi: 10.1101/gr.165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. BioTech. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 47.Akopyants NS, Clifton SW, Martin J, et al. A survey of the Leishmania major Friedlin strain V1 genome by shotgun sequencing: a resource for DNA microarrays and expression profiling. Mol Biochem Parasitol. 2001;113:337–40. doi: 10.1016/s0166-6851(01)00227-4. [DOI] [PubMed] [Google Scholar]
- 48.Wilson R, Ainscough R, Anderson K, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–8. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 49.Chiurillo MA, Beck A, deVos T, Myler PJ, Stuart K, Ramirez JL. Cloning and characterization of Leishmania donovani telomeres. Exp Parasitol. 2000;94:248–58. doi: 10.1006/expr.2000.4499. [DOI] [PubMed] [Google Scholar]
- 50.Sunkin SM, Kiser P, Myler PJ, Stuart KD. The size difference between Leishmania major Friedlin chromosome one homologues is localized to sub-telomeric repeats at one chromosomal end. Mol Biochem Parasitol. 2000;109:1–15. doi: 10.1016/s0166-6851(00)00215-2. [DOI] [PubMed] [Google Scholar]
- 51.Knuepfer E, Stierhof YD, McKean PG, Smith DF. Characterization of a differentially expressed protein that shows an unusual localization to intracellular membranes in Leishmania major. Biochem J. 2001;356:335–44. doi: 10.1042/0264-6021:3560335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith L, Greenfield A. DNA microarrays and development. Hum Mol Genet. 2003;12(Spec No 1):R1–R8. doi: 10.1093/hmg/ddg053. [DOI] [PubMed] [Google Scholar]
- 53.Jiang M, Ryu J, Kiraly M, Duke K, Reinke V, Kim SK. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:218–23. doi: 10.1073/pnas.011520898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner RA, Tabibiazar R, Liao A, Quertermous T. Genome-wide expression dynamics during mouse embryonic development reveal similarities to Drosophila development. Dev Biol. 2005;288:595–611. doi: 10.1016/j.ydbio.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 55.Maeda M, Sakamoto H, Iranfar N, et al. Changing patterns of gene expression in Dictyostelium prestalk cell subtypes recognized by in situ hybridization with genes from microarray analyses. Eukaryot Cell. 2003;2:627–37. doi: 10.1128/EC.2.3.627-637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–6. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 57.Bozdech Z, Llinas M, Pulliam B, Wong E, Zhu J, DeRisi J. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLOS. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitfield ML, Sherlock G, Saldanha AJ, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spellman PT, Sherlock G, Zhang MQ, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–97. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim SK, Lund J, Kiraly M, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–92. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 61.Arbeitman MN, Furlong EE, Imam F, et al. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–5. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- 62.Sasik R, Iranfar N, Hwa T, Loomis WF. Extracting transcriptional events from temporal gene expression patterns during Dictyostelium development. Bioinformatics. 2002;18:61–6. doi: 10.1093/bioinformatics/18.1.61. [DOI] [PubMed] [Google Scholar]
- 63.Campbell DA, Thomas S, Sturm N. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect. 2003;5:1231–40. doi: 10.1016/j.micinf.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 64.Boucher N, Wu Y, Dumas C, Dube M, Sereno D, Breton M, Papadopoulou B. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J Biol Chem. 2002;277:19511–20. doi: 10.1074/jbc.M200500200. [DOI] [PubMed] [Google Scholar]
- 65.Dumas C, Chow C, Muller M, Papadopoulou B. A novel class of developmentally regulated non-coding RNAs In Leishmania. Eukaryot Cell. 2006 doi: 10.1128/EC.00147-06. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.