Abstract
The close resemblance of MS to the animal model experimental autoimmune encephalomyelitis (EAE) has provided compelling data sustaining a pathogenic role of circulating T cells reactive against MBP. T cell antigen receptor (TCR) usage in EAE is commonly considered restricted; nevertheless, dynamic changes of TCR usage correlate with the course of EAE, resulting in a limited repertoire during early stages of disease activity followed by the recruitment of other T cells reactive against new determinants. Although a broader TCR repertoire mediates the response to MBP in humans, a restricted intra-individual heterogeneity may occur in some MS patients. In the present study we characterize the response to MBP in MS subjects with relapsing remitting disease from two sampling time points 12 months apart. MBP-specific T cell lines (TCL) were first generated from eight MS individuals and two healthy subjects. New TCL were obtained after 12 months from one control and three MS patients whose response, at the first time point, was directed against a single epitope. Interestingly, these three subjects had a stable and mild disease. Few TCL obtained at two time points from the MS individuals recognized the same immunodominant epitope and shared identical TCR Vβ sequences. In the control we could not detect a restriction of the repertoire. These findings suggest that in some MS patients with benign disease a predominant T cell response to a single determinant may be detectable at different moments and is mediated by clonally expanded populations.
Keywords: multiple sclerosis, T cell receptor, T cell lines, myelin basic protein
INTRODUCTION
MS is a chronic inflammatory disease of the central nervous system (CNS) characterized by perivenular mononuclear cell infiltrates and demyelination within the brain and spinal cord white matter [1]. Although the cause of MS remains unknown, clinical and experimental data support an autoimmune aetiology. Indirect evidence sustaining this hypothesis is the similarity of MS to experimental autoimmune encephalomyelitis (EAE), a disease induced in susceptible animals by immunization with myelin antigens [2]. EAE is mediated by class II-restricted CD4+ T cells reactive against MBP and other myelin proteins, and can also be induced by the adoptive transfer of MBP-specific T cells from one animal to another syngeneic one [3]. In inbred species, the T cell response to MBP has been shown to be directed against few epitopes and characterized by a restricted TCR usage [4]. This limited heterogeneity of T cell receptor (TCR) genes expressed by disease-inducing T cells has been exploited for therapeutic purposes, e.g. by means of antibodies directed against TCR carried on encephalitogenic T cells, or by ‘vaccination’ with synthetic TCR peptides [5, 6]. Nevertheless, a more heterogeneous reactivity to MBP and other myelin antigens is detected at later stages of EAE [7–9]. The response to MBP in MS subjects has been extensively studied and was observed to be diverse both in terms of epitope specificity and TCR usage [10–12]. Nevertheless, it has been reported that within the same individual a limited number of epitopes may be recognized [11, 13] and a restricted and predominant TCR repertoire may be detected [14, 15]. In monozygotic twins a restricted TCR usage was observed in individuals with a mild form of MS, while a broader heterogeneity correlated with disease severity [16]. In this study we examined the T cell response to MBP in healthy subjects and individuals with relapsing remitting (RR) MS during a 12-month period in order to characterize the dynamic changes of MBP-specific T cell populations over time.
MATERIALS AND METHODS
Antigens
MBP was purified from the white matter of human brain by standard methods according to Deibler et al. [17]. The antigen concentration in all experiments was 30 μg/ml. Overlapping peptides 20–24 amino acids long encompassing the whole MBP molecule were synthesized by a solid-phase method, purified by high performance liquid chromatography (HPLC) and used at 10 μg/ml concentration.
Subjects
At the first time point, eight RR MS patients with mild clinical course (EDSS ≤ 3·5) and two healthy individuals were selected for the study. MS subjects had a diagnosis of definite RR MS according to the Poser criteria [18], an average EDSS of 2·4 ± 1·1 and a disease duration of 8 ± 3 years. In order to study the response to MBP over time, we selected, after 12 months, one healthy control and three MS subjects whose response at the first time point was mainly directed against a single epitope. None of the three patients showed disease progression after 12 months (Table 1). Nevertheless, two patients had a mild relapse about 1 month before the second sampling, but they fully recovered without therapy.
Table 1.
Characteristics of individuals whose T cell lines (TCL) were obtained at the second time point
Establishment of MBP-specific T cell lines and proliferation assays
Long-term MBP-specific T cell lines (TCL) were generated from the peripheral blood of each individual using the ‘splitting well’ technique [19]. This technique allows isolation of clonal populations of MBP-specific T cells. Briefly, peripheral blood mononuclear cells (PBMC) were separated by Fycoll–Hypaque density gradient centrifugation (Pharmacia Biotech, Uppsala, Sweden), washed and seeded at 2 × 105 cells/well in 96-well U-bottomed plates (Corning, New York, NY) in the presence of MBP (30 μg/ml). Cells were grown in IL-2-rich medium (gift from Chiron-Eurocetus, Amsterdam, The Netherlands) and expanded by restimulation at 15-day intervals with autologous irradiated PBMC (1 × 105/well) as antigen-presenting cells (APC) pulsed with MBP (30 μg/ml). Antigen specificity of the TCL was determined by a standard 3H-thymidine incorporation assay after the third restimulation. Aliquots of the MBP-specific TCL (4 × 104) were plated out in duplicate in presence of irradiated autologous APC pulsed or not pulsed with antigen (for epitope specificity, APC were pulsed with MBP peptide at 10 μg/ml concentration). After 72 h, 0·5 μCi of 3H- thymidine (Amersham, Aylesbury, UK) was added to each well and incorporation measured after 16 h in a β scintillation counter. TCL were considered antigen-specific if the stimulation index (SI) was > 3.
Membrane phenotype analysis
Membrane phenotype of TCL was determined using a FACScan cytofluorometer (Becton Dickinson, San Jose, CA) by direct immunofluorescence with fluorescence dye-labelled MoAbs specific for CD3 (anti-Leu-4–PE; Becton Dickinson Immunocytometry Systems, San Jose, CA), TCR (anti-TCR αβ WT31-FITC; Becton Dickinson), CD4 (anti-CD4 Leu-3a–PE; Becton Dickinson) and CD8 (anti-CD8 Leu-2a–PE; Becton Dickinson).
RNA extraction, cDNA synthesis and linear polymerase chain reaction amplification
RNA was isolated from TCL using TRIzol according to the manufacturer's instructions (TRIzol; Gibco BRL, Gaithersburg, MD). First-strand cDNA was obtained by reverse-transcribing 1 μg of total RNA with random hexamers (Pharmacia Biotech) as primers and 5 U of M-MLV reverse transcriptase (Superscript II; Gibco) in a 30 μl reaction. cDNA was diluted to 300 μl and 24 10-μl aliquots were subjected to 30 cycles of polymerase chain reaction (PCR) amplification with a panel of Vβ primers [20, 21] in combination with a 3′ Cβ primer, each PCR cycle consisting of denaturation at 95°C for 60 s, annealing at 55°C for 60 s and extension at 72°C for 60 s. PCR products were visualized by electrophoresis on a 1.5% agarose gel stained with ethidium bromide, transferred to a nylon membrane (Hybond N; Amersham) according to standard methods and hybridized with a horseradish peroxidase (HRP)-labelled oligonucleotide probe specific for Cβ.
CDR3 size spectratyping
In order to determine the clonality of CDR3 junctional regions of those TCL expressing the same Vβ at different time points, an aliquot of the PCR product was subjected to CDR3 size spectratyping as described by Gorski and collegues [22]. Briefly, 5 μl of the amplicon were reamplified for 30 cycles with the same Vβ primer and a fluorescent Cβ primer, visualized on a denaturing polyacrylamide gel using an Applied Biosystem 373 DNA Sequencer (Perkin Elmer, Foster City, CA) and analysed with the software 672 GENESCAN (Perkin Elmer).
Inverse PCR
In some cases, TCL were analysed by inverse PCR [23]. Briefly, 1 μg of RNA isolated as previously described was reverse transcribed into first-strand cDNA using oligo (dT)12–18 (Pharmacia Biotech) and M-MLV reverse transcriptase (Superscript II; Gibco). Double-strand cDNA was synthesized using Escherichia coli DNA ligase (Gibco), E. coli DNA Polymerase I (Gibco) and E. coli RNase H (Gibco) according to standard methods and blunt ends were generated by digestion with T4 DNA polymerase (Gibco). After circularization with T4 DNA ligase (Gibco), an aliquot of cDNA was amplified for 35 cycles using Cβ primers annealing to the template in an outward orientation.
Cloning and sequencing of PCR products
Reverse transcriptase (RT)-PCR products were cloned into pMOS vector (Amersham) according to the manufacturer's instructions. After digestion with Not I and Sal I (Tib Molbiol, Genoa, Italy) inverse PCR products were resolved by electrophoresis, excised from the agarose gel and ligated into pBluescript II KS (-) vector (Stratagene, La Jolla, CA). Vectors containing RT and inverse PCR products were transformed into XL-1 Blue Supercompetent cells (Stratagene). Plasmid DNA was purified using Qiagen Spin Plasmid Miniprep (Qiagen, Hilden, Germany), hybridized with an HRP-labelled Cβ probe and sequenced with an Applied Biosystem 373 DNA Sequencer (Perkin Elmer).
Sequence analysis
Sequence computer analysis was performed using the software Lasergene for Windows (DNASTAR Inc., Madison, WI) and sequences were compared with nucleotide and amino acid databases using the BLAST algorithm of Genebank. Vβ gene names were assigned according to the criteria of the WHO-IUIS Nomenclature SubCommittee on TCR designation [24] and follow the classification of Arden et al. [25].
RESULTS
Establishment of MBP-specific TCL: fine specificity and membrane phenotype
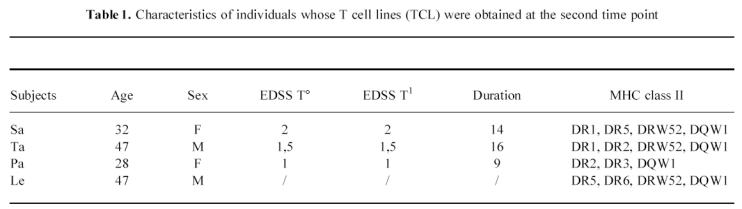
At the first time point 42 and eight long-term TCL were generated from the peripheral blood of eight MS patients and two healthy controls, respectively. The number of lines established from each individual ranged from 1 to 10, with an average of 5·2 TCL/subject in the MS patients and four TCL/subject in controls. All the TCL had a CD3+, CD4+, CD8−, TCRαβ+ membrane phenotype. Analysis of TCL fine specificity was determined by testing their reactivity against a panel of overlapping peptides encompassing the whole MBP molecule. All TCL tested (76%) were specific for the immunizing antigen. No significant difference of stimulation index (SI) was observed in the proliferation assays from MS subjects and controls (data not shown). Some lines recognizing MBP were specific for multiple contiguous peptides, while some others did not respond to any peptide, probably because of their specificity for epitopes spanning the cleavage regions. The response was mostly focused on peptides spanning amino acids 80–104, 142–172 and, in MS individuals, aa 16–38, a peptide rarely immunodominant in humans (Fig. 1). After 12 months we generated, respectively, three and 15 long-term TCL from one healthy control and three MS subjects. No disease progression was observed in any patient after 12 months. No significant change of epitope specificity was detected in TCL generated from MS individuals (Fig. 2). Only two lines specific for aa 80–104 were obtained from the third patient (Ta) before he dropped the study. Peptide 80–104 was the only one recognized by TCL established from the healthy donor at two time points (Table 2).
Fig. 1.
Epitope specificity of MBP-specific T cell lines (TCL) established from MS individuals (a) and healthy subjects (b). Some TCL from both groups recognized human MBP but did not respond to any peptide (nr). Ten lines were not tested in proliferation assay for epitope mapping.
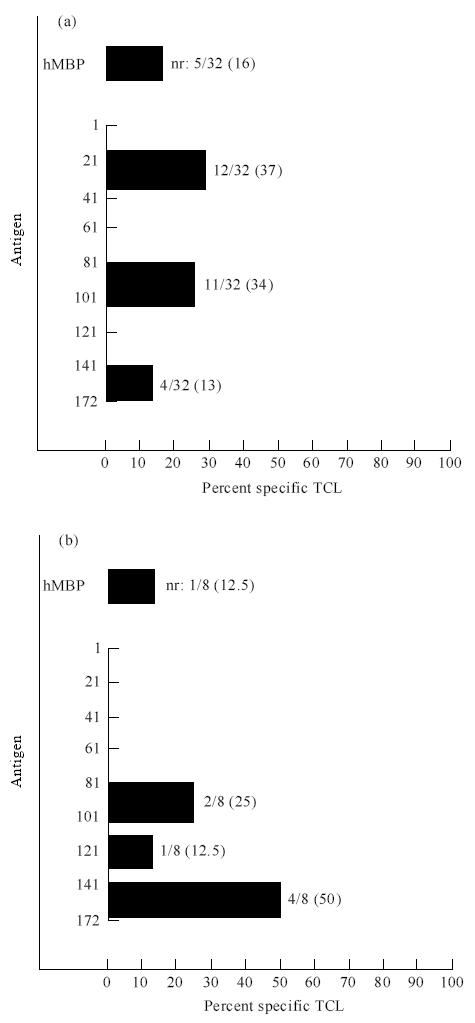
Fig. 2.
Peptide reactivities of MBP-specific T cell lines (TCL) generated from one MS patient (Sa). Most TCL recognized peptide 80–104 both at the first (a) and second time point (b). Two lines did not respond to any peptide (nr).
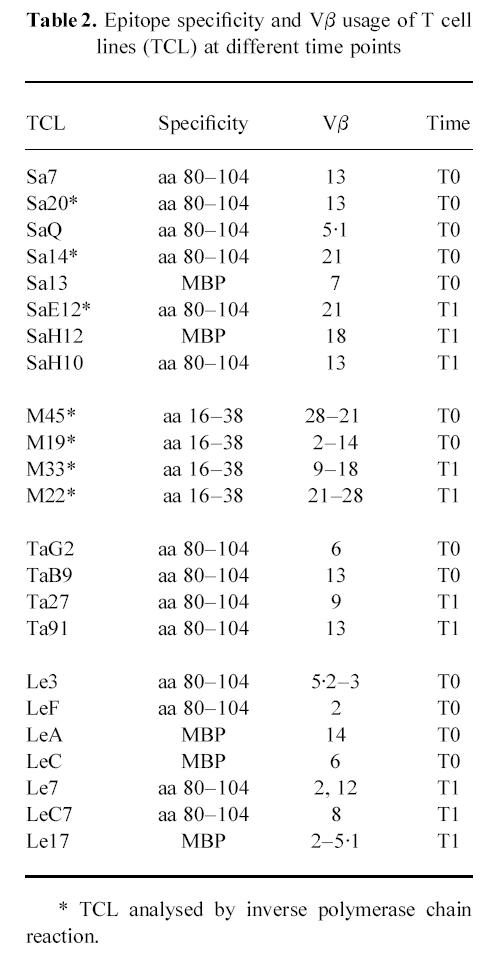
Table 2.
Epitope specificity and Vβ usage of T cell lines (TCL) at different time points
TCL TCR Vβ usage
Analysis of fine specificity and TCR usage of some MBP-specific TCL generated at the two time points is summarized in Table 2. RT and inverse PCR analysis showed an interindividual heterogeneity of TCR Vβ expression even in those lines recognizing the same epitope. This heterogeneous repertoire was observed both in MS individuals and in healthy subjects. Few TCL carried more than one Vβ, suggesting that the response to the MBP could have been oligoclonal rather than monoclonal even in the presence of single-epitope specificity. Interestingly, we observed that some TCL generated at T1 from all subjects expressed the same Vβ gene that was carried by TCL previously established from the same individual.
CDR3 spectratyping analysis of TCL expressing the same Vβ genes
In order to characterize the clonality of the response to MBP we performed CDR3 size spectratyping on TCL that were shown to express the same Vβ genes at the two time points. Single PCR bands originated from TCL utilizing the same V segment and migrating at the same molecular weight are likely to represent an identical rearrangement. The spectratype on a polyacrylamide gel of all the PCR products showed a single band, confirming the clonality of each rearrangement carried by the lines. Furthermore, in patient Sa the spectratype bands of the Vβ13 amplicons from lines Sa7 and SaH10 migrated at the same molecular weight, supporting the hypothesis that they represented CDR3 of the same length and possibly identical (Fig. 3a). In patient Ta and in the healthy subject, spectratype analysis, respectively, of Vβ13 (from TCL TaB9 and Ta91) and Vβ2 (LeF, Le7, Le17) PCR products revealed two bands migrating at slightly different molecular weights, suggesting different VDJ rearrangements (Fig. 3b).
Fig. 3.
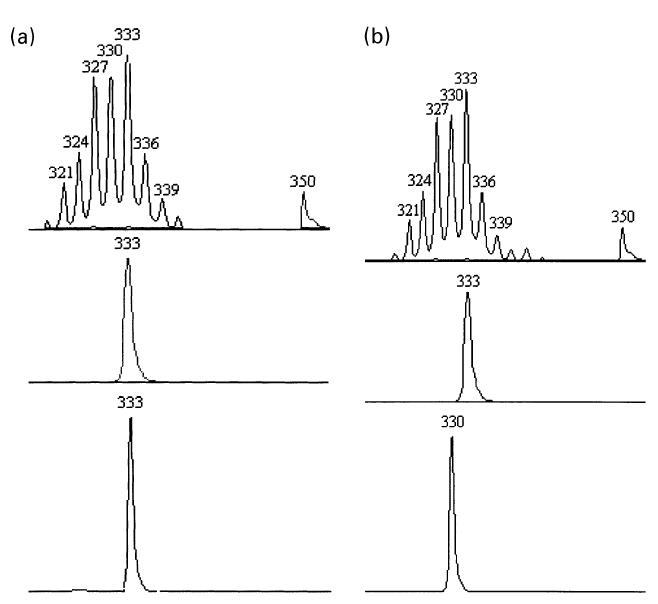
(a) Spectratyping analysis of Sa7 (bottom), SaH10 (centre) and peripheral blood mononuclear cell (PBMC) (top) Vβ13 T cell receptor (TCR) transcripts. One peak corresponding to a unique rearrangement migrates at the same expected size both for Sa7 and SaH10. Spectratyping of Vβ13 transcript from PBMC of the same patient is polyclonal, as shown by the bell-shape distribution of peaks in the top part of the figure. (b) Analysis of TaB9 (bottom), Ta91 (centre) and PBMC (top) Vβ13 TCR transcripts. One peak corresponding to a unique but diverse rearrangement migrates at slightly different sizes for Ta91 and TaB9. Spectratyping of Vβ13 transcript from PBMC of the same patient is polyclonal. The graph represents the intensity of fluorescence in arbitrary units as a function of polymerase chain reaction (PCR) product size in nucleotides and all the CDR3 peaks are 3 bp apart, suggestive of in-frame rearrangements.
Sequence analysis of TCR β-chain expressed by MBP-specific TCL
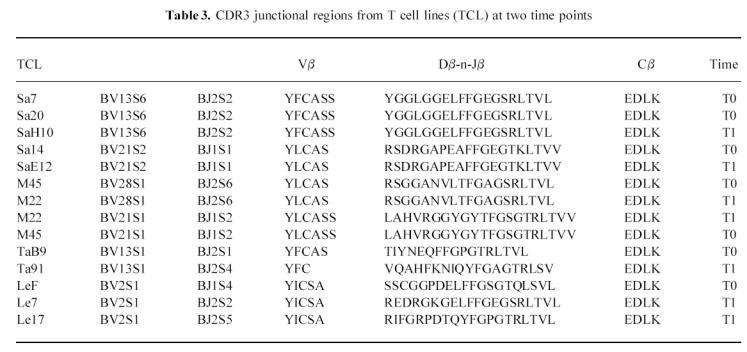
Sequence analysis of inverse PCR products and TCR transcripts subjected to CDR3 size spectratyping is shown in Table 3. The comparison of CDR3 sequences of TCL carrying Vβ13, generated from patient SA at two time points, revealed that Sa7, Sa20 and SaH10 expressed the same rearrangement BV13S6-n-D-n-BJ2S2. Two other lines isolated at T0 and T1 exhibited identical sequences (Sa14, SaE12). Similarly, identical rearrangements were obtained from TCL generated from patient Pa 12 months apart (M22, M45). In contrast, comparison of sequences from those TCL expressing the same Vβ but different CDR3 spectratypes showed different rearrangements, confirming that those TCL used a diverse β-chain repertoire. Remarkably, one of the sequences obtained from inverse PCR amplification of lines M22 and M45 specific for aa 16–38 did not show similarity > 75% with any of the Vβ described thus far. On-line comparison of this sequence with those in the nucleotide and protein databases revealed a high degree of similarity with a gene segment (BV28S1) located within the 685-kb TCR locus and yet undescribed at cDNA level.
Table 3.
CDR3 junctional regions from T cell lines (TCL) at two time points
DISCUSSION
Although the pathogenesis of MS still remains to be clarified, experimental data suggest that it may be caused by a failure in the central or peripheral tolerance induction leading autoreactive T cells escaping the process of negative selection [26]. EAE has provided compelling data on the mechanisms of organ-specific autoimmune disease. In this model it has been demonstrated that after initial priming with a single MBP determinant, intra- and intermolecular spreading of epitopes leads to the recruitment of a second wave of T cells with a different repertoire [27]. Thus, while EAE-inducing T cells utilize a very limited range of TCR genes in the recognition of single MBP determinants in the early stage of disease [4], a wider TCR repertoire has been observed in later phases [7–9]. This limited response has been exploited in successfully targeting the trimolecular complex TCR–MBP–MHC with specific immunotherapy [5, 6]. In humans, studies on the TCR repertoire used by T cells recognizing MBP have yielded conflicting data, supporting the idea that different individuals utilize a diverse repertoire in the recognition of MBP determinants [10–12]. Nevertheless, it has been observed that a limited usage of V genes within the same individual may arise from the clonal expansion of a limited T cell population [14, 15, 28]. Furthermore, restricted TCR rearrangements may be present in some MS individuals with a mild form of disease, while a more heterogeneous response to MBP could correlate with progression of disease [16]. We generated antigen-specific TCL from a group of MS individuals with RR disease and healthy subjects. The mean numbers of lines established from patients and controls were similar. Analogous results were obtained from other groups, confirming that autoreactive MBP-specific T cells are also present in the peripheral compartment of normal donors [19]. As in other reports, the response to MBP in some MS individuals clustered against a single epitope [11, 13].
In order to verify whether some changes of the response to MBP have occurred over time in those subjects with a response targeting a single epitope at T0, three and 15 TCL were generated after 12 months, respectively, from a normal donor and three MS subjects. Although a limited number of TCL was generated from each subject, a restricted response against a single MBP epitope was maintained after 12 months. Analysis of TCR Vβ gene usage showed that most TCL expressed diverse V elements in the recognition of MBP determinants. Different Vβs were exhibited also by TCL reactive against the same epitope. Similar results were obtained by other groups, confirming that the T cell response to MBP is characterized by an interindividual heterogeneity in TCR usage [10–12]. Nevertheless, few families were expressed more often than others. For example, Vβ13 was used by TCL specific for aa 80–104 generated in two individuals at different time points. In order to determine the clonality of lines carrying the same Vβ family, we performed CDR3 size spectratyping to see if each PCR product was migrating as a single band, possibly expression of a unique rearrangement [22]. Indeed, CDR3 hypervariable region arises from TCR gene rearrangement and represents the marker of the clonal origin of T lymphocytes [29]. In patient Sa spectratype bands of the Vβ13 PCR products migrating at the same molecular weight suggested that CDR3 regions were of the same length and probably identical. However, since the same CDR3 size can not exclude a different amino acid composition, we sequenced PCR products exhibiting the same spectratypes. Lines Sa7 and SaH10 established at different moments showed the same rearrangement that was observed also in line Sa20 analysed by inverse PCR. Indeed, the CDR3 sequences of four other independently derived TCL (Sa14, SaE12 from patient Sa and M45, M22 from patient Pa) were identical, suggesting the presence of stable and clonally expanded MBP-reactive T cell populations in vivo. In another MS patient and in the healthy subject, the spectratype analysis of those PCR products expressing the same Vβ gene (TaB9, Ta91, Le3, Le7) revealed two bands migrating at slightly different molecular weights, suggesting different VDJ rearrangements. Sequence analysis of this TCR transcript showed diverse hypervariable regions. These data, together with the presence of a conserved TCR repertoire in mildly affected MS individuals observed by another group [16], may indicate that a durable and restricted T cell response to MBP may be present in some subjects with a mild form of MS and could be the expression of a limited spreading of myelin determinants. The absence of a restricted TCR repertoire in the third patient with similar disease course might be due to the small number of TCL obtained before he received a pharmacological treatment, or to other factors, including MHC restriction and environmental exposure.
It is interesting to note that of those patients expressing clonally expanded and stable T cell populations, both had a mild clinical relapse a few weeks before new TCL were established. A similar observation was reported by another group [11]. In both cases no therapy was necessary and full recovery occurred within a few weeks. The persistence of clonally expanded and restricted T cell populations reacting against MBP in some MS patients right after the occurrence of a clinical relapse is noteworthy. Indeed, these findings may be consistent with the recent report that in SJL/J mice T cells specific to MBP can induce EAE after adoptive transfer but are not sufficient to determine epitope spreading [30]. Hence, other factors are probably necessary for acute CNS damage and release of previously cryptic determinants. It is possible that in MS individuals a few T cell populations recognizing a limited number of determinants may undergo clonal expansion following repeated antigen or superantigen stimulation [31, 32]. In some patients these cells may be kept initially under control by a regulatory circuitry [33]. Similarly to EAE, microbial superantigens and other environmental or genetic factors are likely to be responsible for the amplification of the autoimmune response followed by myelin destruction and consequential inflammatory spreading of new determinants [34, 35]. Thus, the presence of a restricted intra-individual response to MBP in some MS individuals with a mild clinical course may be exploited by tailoring early immunotherapies targeting selectively the recognition of the immunodominant epitope before the occurrence of spreading of determinants.
Finally, from sequence analysis of TCR rearrangements utilized by TCL reactive against MBP, we observed a high degree of homology between sequences obtained from two TCL generated in one patient and a Vβ gene segment located within the 685-kb TCR locus not yet described as cDNA [36]. Thus, the identity of this sequence obtained from MBP-specific TCL with a genomic sequence within the TCR locus proves that a novel Vβ family is used to recognize a rarely recognized MBP epitope in an MS patient (Uccelli et al., manuscript in preparation).
Acknowledgments
This study was funded by a grant from the Italian Society for Multiple Sclerosis (AISM). D.G. was supported by an Italian Society for Multiple Sclerosis fellowship. We wish to thank the Italian Centro Nazionale per le Ricerche (CNR) for financial support. We wish to thank Dr J. R. Oksenberg and Dr F. Manca for critical discussions and Dr L. Nobbio, Dr M. Seri and Dr C. Caroli for technical assistance.
References
- 1.Prineas JW. The neuropathology of multiple sclerosis. In: Koetsier JC, editor. Handbook of clinical neurology. Vol. 3. Amsterdam: Elsevier; 1985. pp. 213–58. [Google Scholar]
- 2.Alvord EC, Kies MW, Suckling AJ. Experimental allergic encephalomyelitis: a useful model for multiple sclerosis. Prog Clin Biol Res. 1984;146:523–54. [PubMed] [Google Scholar]
- 3.Pettinelli DB, McFarlin DE. Adoptive transfer of experimental allergic encephalomyelitis in SJL/J mice after in vitro activation of lymph node cells by myelin basic protein: requirement for Lyt 1+ 2− T-lymphocytes. J Immunol. 1981;127:1420–3. [PubMed] [Google Scholar]
- 4.Burns FR, Li RX, Shen L, Offner H, Chou K, Vandenbark A, Haber-Katz E. Both rat and mouse T-cell receptors specific for the encephalitogenic determinant of myelin basic protein use similar Vα and Vβ chain genes even though the major histocompatibility complex and encephalitogenic determinants being recognized are different. J Exp Med. 1989;169:27–39. doi: 10.1084/jem.169.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urban JL, Kumar V, Kono DH, et al. Restricted use of the T cell receptor V genes in murine autoimmune encephalomyelitis raises the possibility for antibody therapy. Cell. 1988;54:577–92. doi: 10.1016/0092-8674(88)90079-7. [DOI] [PubMed] [Google Scholar]
- 6.Acha-Orbea H, Mitchell DJ, Timmermann L, Wraith DC, Tausch GS. Limited heterogeneity of T cell receptors from lymphocytes mediating autoimmune encephalomyelitis allows specific immune intervention. Cell. 1988;54:263–73. doi: 10.1016/0092-8674(88)90558-2. [DOI] [PubMed] [Google Scholar]
- 7.Mor F, Cohen IR. Shifts in the epitopes of myelin basic protein recognized by Lewis rat T cells before, during and after the induction of experimental autoimmune encephalomyelitis. J Clin Invest. 1993;92:2199–206. doi: 10.1172/JCI116822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehmann PV, Sercarz EE, Forsthuber T, Dayan CM, Gammon G. Determinant spreading and the dynamics of the autoimmune T-cell repertoire. Immunol Today. 1993;14:203–8. doi: 10.1016/0167-5699(93)90163-F. [DOI] [PubMed] [Google Scholar]
- 9.McRae BL, Vanderlugt CL, Dal Canto MC, Miller SD. Functional evidence for epitope spreading in the relapsing pathology of experimental autoimmune encephalomyelitis. J Exp Med. 1995;182:75–85. doi: 10.1084/jem.182.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin R, Utz U, Colligan JE, et al. Diversity in fine specificity and T cell receptor usage of the human CD4+ cytotoxic T cell response specific for the immunodominant myelinic basic protein peptide 87–106. J Immunol. 1992;148:1359–66. [PubMed] [Google Scholar]
- 11.Meinl E, Weber F, Drexel K, et al. Myelin basic protein-specific T lymphocyte repertoire in multiple sclerosis: complexity of the response and dominance of nested epitopes due to recruitment of multiple T cell clones. J Clin Invest. 1993;92:2633–43. doi: 10.1172/JCI116879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafler DA, Saadeh MG, Kuchroo KV, Milford E, Steinman L. TCR usage in human experimental demyelinating disease. Immunol Today. 1996;17:152–9. doi: 10.1016/0167-5699(96)80611-6. [DOI] [PubMed] [Google Scholar]
- 13.Salvetti M, Ristori G, D'Amato M, Buttinelli C, Falcone M, Fieschi C, Wekerle H, Pozzilli C. Predominant and stable T cell response to regions of myelin basic protein can be detected in individual patients with multiple sclerosis. Eur J Immunol. 1993;23:1232–9. doi: 10.1002/eji.1830230606. [DOI] [PubMed] [Google Scholar]
- 14.Wucherpfennig KW, Zhang J, Witek C, Matsui M, Modabber Y, Ota K, Hafler DA. Clonal expansion and persistence of human T cells specific for an immunodominant myelin basic protein peptide. J Immunol. 1994;152:5581–92. [PubMed] [Google Scholar]
- 15.Vandevyver C, Mertens N, van den Elsen P, Medaer R, Raus J, Zhang J. Clonal expansion of myelin basic protein-reactive T-cells in patients with multiple sclerosis: restricted T-cell receptor V gene rearrangements and CDR3 sequence. Eur J Immunol. 1995;25:958–68. doi: 10.1002/eji.1830250416. [DOI] [PubMed] [Google Scholar]
- 16.Utz U, Brooks JA, McFarland HF, Martin R, Biddison WE. Heterogeneity of T-cell receptor α-chain complementarity-determining region 3 in myelin basic protein-specific T cells increases with severity of multiple sclerosis. Proc Natl Acad Sci USA. 1994;91:5567–71. doi: 10.1073/pnas.91.12.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deibler G, Martensson RE, Kies MW. Large sale preparation of myelin basic protein from central nervous tissue of several mammalian species. Prep Biochem. 1972;2:139–65. doi: 10.1080/00327487208061467. [DOI] [PubMed] [Google Scholar]
- 18.Poser CM, Paty DW, Scheinberg L, et al. New diagnostic criteria for multiple sclerosis: guidelines for research protocols. Ann Neurol. 1983;13:227–31. doi: 10.1002/ana.410130302. [DOI] [PubMed] [Google Scholar]
- 19.Pette M, Fujita K, Kitze B, Whitaker JN, Albert E, Kappos L, Wekerle H. Myelin basic protein specific T lymphocyte lines from MS patients and healthy individuals. Neurology. 1990;40:1770–6. doi: 10.1212/wnl.40.11.1770. [DOI] [PubMed] [Google Scholar]
- 20.Oksenberg JR, Panzara MA, Begovich AB, et al. Selection for T-cell receptor Vb-Db-Jb gene rearrangements with specificity for a myelin basic protein peptide in brain lesions of multiple sclerosis. Nature. 1993;362:68–70. doi: 10.1038/362068a0. [DOI] [PubMed] [Google Scholar]
- 21.Usuku K, Joshi N, Hatem CJJ, Alper CA, Schoenfeld DA, Hauser SL. The human T-cell receptor β-chain repertoire: longitudinal fluctuations and assessment in MHC matched populations. Immunogenetics. 1993;38:193–8. doi: 10.1007/BF00211519. [DOI] [PubMed] [Google Scholar]
- 22.Gorski J, Yassai M, Zhu X, Kissela B, Keever C, Flomenberg N. Circulating T cell repertoire complexity in normal individuals and bone marrow recipients analyzed by CDR3 spectratyping: correlation with the immune status. J Immunol. 1994;152:5109–19. [PubMed] [Google Scholar]
- 23.Uematsu Y. A novel and rapid cloning method for the T-cell receptor variable region sequences. Immunogenetics. 1991;34:174–8. doi: 10.1007/BF00205820. [DOI] [PubMed] [Google Scholar]
- 24.WHO-IUIS. Nomenclature Sub-Committee on TCR designation. Nomenclature for T-cell receptor (TCR) gene segments of the immune system. Immunogenetics. 1995;42:451–3. doi: 10.1007/BF00172175. [DOI] [PubMed] [Google Scholar]
- 25.Arden B, Clarck SP, Kabelitz D, Mak TW. Human T-cell receptor variable gene segment families. Immunogenetics. 1995;42:455–500. doi: 10.1007/BF00172176. [DOI] [PubMed] [Google Scholar]
- 26.Steinman L. Escape from ‘horror autotoxicus’: pathogenesis and treatment of autoimmune disease. Cell. 1995;80:7–10. doi: 10.1016/0092-8674(95)90443-3. [DOI] [PubMed] [Google Scholar]
- 27.Lehmann PV, Forsthuber T, Miller A, Sercarz EE. Spreading of T cell autoimmunity to cryptic determinants of an autoantigen. Nature. 1992;358:155–7. doi: 10.1038/358155a0. [DOI] [PubMed] [Google Scholar]
- 28.Hohlfeld R, Meinl E, Weber F, et al. The role of autoimmune T lymphocytes in the pathogenesis of multiple sclerosis. Neurology. 1995;45:33–38. doi: 10.1212/wnl.45.6_suppl_6.s33. [DOI] [PubMed] [Google Scholar]
- 29.Chothia C, Boswell DR, Lesk AM. The outline structure of the T-cell αβ receptor. EMBO J. 1988;7:3745–55. doi: 10.1002/j.1460-2075.1988.tb03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voskuhl RR, Wesley Farris IIR, Nagasato K, McFarland HF, Dubois Dalcq M. Epitope spreading occurs in active but not passive EAE induced by myelin basic protein. J Neuroimmunol. 1996;70:103–11. doi: 10.1016/s0165-5728(96)00054-9. [DOI] [PubMed] [Google Scholar]
- 31.Allegretta M, Nicklas JA, Sriram S, Albertini RJ. T-cells responsive to myelin basic protein in patients with multiple sclerosis. Science. 1990;247:718–21. doi: 10.1126/science.1689076. [DOI] [PubMed] [Google Scholar]
- 32.Wucherpfennig KW, Strominger JL. Molecular mimicry in the T-cell mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar V, Tabibiazar R, Geysen HM, Sercarz EE. Immunodominant framework region 3 peptide from TCR Vβ8·2 chain controls murine experimental autoimmune encephalomyelitis. J Immunol. 1995;154:1941–50. [PubMed] [Google Scholar]
- 34.Brocke S, Gaur A, Piercy C, Gautam A, Gijbels K, Fathman GC, Steinman L. Induction of relapsing paralysis in experimental autoimmune encephalomyelitis by bacterial superantigen. Nature. 1993;365:642–4. doi: 10.1038/365642a0. [DOI] [PubMed] [Google Scholar]
- 35.Goverman J, Woods A, Larson L, Weiner LP, Hood L, Zeller DM. Transgenic mice that express a myelin basic protein-specific T cell receptor develop spontaneous autoimmunity. Cell. 1993;72:551–60. doi: 10.1016/0092-8674(93)90074-z. [DOI] [PubMed] [Google Scholar]
- 36.Rowen L, Koop BF, Hood L. The complete 685-kilobase DNA sequence of the human β T cell receptor locus. Science. 1996;272:1755–62. doi: 10.1126/science.272.5269.1755. [DOI] [PubMed] [Google Scholar]


