Abstract
HTLV-1 causes two distinct human diseases, HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) and adult T cell leukaemia/lymphoma (ATL). Persistently infected individuals carry a risk of < 1% of developing either disease. These basic epidemiological data imply that virus–host interactions, especially immunogenetic factors, influence the outcome of infection. Several studies showed that the HLA class II DR1 DQ5 haplotype is over-represented in HAM/TSP, but rare in ATL. Therefore, we selected four patients with HAM/TSP and one seronegative control who all carried the HLA DR1 DQ5 haplotype. We analysed the CD4+ T lymphocyte response against eight synthetic peptides of HTLV-1 envelope (env) glycoprotein gp21, a crucial target antigen in HAM/TSP. The first of two immunodominant epitopes corresponded to a domain of the HTLV-1 envelope protein which had previously been shown to be essential for HTLV-1 envelope function. The second immunodominant epitope overlapped a highly conserved sequence of the retroviral transmembrane envelope protein. DR1 (DRB1*0101)-restricted T lymphocytes were activated by the conserved peptide sequence in nanomolar concentrations. In contrast, this conserved sequence can also induce non-specific, cAMP-mediated immunosuppressive effects on T cells when added in micromolar concentrations to culture media, as shown by Haraguchi S, Good RA, James-Yarish M, Cianciolo GJ, Day NK, Proc Natl Acad Sci USA 1995; 92:5568–71. Hence, HTLV-1 env gp21 might exert either stimulating immunological or immunosuppressive effects in HTLV-1-infected individuals, depending on the level of its expression and the presence of HLA DRB1*0101.
Keywords: HTLV-1, T lymphocytes, epitope, HLA
INTRODUCTION
HTLV-1 infection is associated with two distinct diseases, a neoplastic disease, adult T cell leukaemia/lymphoma (ATL) [1], and a chronic inflammatory disease of the spinal cord, HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) [2, 3]. Either disease occurs in a small percentage of HTLV-1-infected individuals. The relatively rare occurrences of diseases in virus-infected humans and the different patterns of clinical diseases are thought to be linked to immunogenetic host factors. HAM/TSP patients suffer from spastic paraparesis and urinary symptoms, furthermore there is a frequent association with sicca syndrome, uveitis or arthritis. HAM/TSP is regarded as being an immunomediated disease.
Certain HLA antigens and haplotypes are segregated between patients with ATL and HAM/TSP [4–8]. The HLA haplotypes predominant in HAM/TSP are associated with a high immune responsiveness against HTLV-1 [4]. The most significant association between HAM/TSP and HLA is represented by HLA-DR1 (HLA DRB1*0101) [6, 8], which is known to be in strong linkage disequilibrium with HLA-DQ5 (HLA DQB1*0501).
HLA class II antigens have a strong effect on the epitope specificities of the CD4+ T cell immune response. To date, there is evidence of a significant CD4+ T cell response against HTLV-1 antigens [9–11], and also against central nervous system (CNS) autoantigen in HAM/TSP [12], but little is known about CD4+ T cell epitopes apart from one established epitope of HTLV-1 gp46 [9]. Furthermore, there is a lack of knowledge regarding the relation of particular HLA class II molecules to CD4+ T cell epitopes of HTLV-1 proteins. We found that there are far more CD4+ T cell epitopes in the transmembrane part of the HTLV-1 envelope (env) glycoprotein gp21 than in the extramembrane part of the envelope protein gp46, when we analysed the CD4+ T cell repertoire in peripheral blood mononuclear cells (PBMC) from HTLV-1−, ‘naive’ individuals with different HLA haplotypes [13]. Furthermore, in HAM/TSP, the B cell immune response against HTLV-1 is directed against a diversity of epitopes, both in the systemic and CNS compartment [14], but HTLV-1 gp21-specific antibodies were found frequently in those patients carrying HAM/TSP-associated HLA haplotypes [15].
Since B cell immune responses against peptide epitopes are controlled by the CD4+ T cell immune response [16], it is worthwhile to look for a particular association: the CD4+ T cell immune response against the HTLV-1 transmembrane envelope protein gp21, an important intrathecal target antigen [15] in HLA-DR1+ individuals who are at a higher risk of developing HAM/TSP [6, 8]. This study was designed to establish immunodominant CD4+ T cell epitopes of HTLV-1 gp21, to analyse their functional importance for HLA binding, and to correlate them to the B cell epitopes of HTLV-1 gp21 established previously [14, 15].
MATERIALS AND METHODS
Origin of lymphocytes and HLA typing
Four patients with HAM/TSP (diagnosed according to established guidelines [17]) and one control who was both HTLV-1 polymerase chain reaction (PCR)-negative and seronegative were selected, who all carried the HLA class II DR1 DQ5 haplotype. Three out of four HAM/TSP patients were homozygote for this haplotype.
HLA antigens of HAM/TSP patients and the control individual were determined serologically by the standard National Institutes of Health (NIH) microcytotoxicity test using sera which were standardized to the criteria and nomenclature of the 11th International Histocompatibility Workshop. The linkage of HLA 5 locus haplotypes in HAM/TSP patients was confirmed by family HLA studies as described [4, 5]. In the seronegative control individual HLA phenotyping revealed a combination of HLA class II antigens, which also implicates the presence of the HLA class II DR1 DQ5 haplotype because of known linkage disequilibrium. Recently, our HLA haplotype data were confirmed and extended by HLA genotyping [8] using the PCR-restriction fragment length polymorphism (RFLP) method [18].
Microculture technique for selection of peptide-specific T cell lines
Eight synthetic peptides of the HTLV-1 gp21 amino acid sequence were provided by M. Nakamura (Kyushu University, Fukuoka, Japan) (Table 1). These peptides had been purified by high performance liquid chromatography (HPLC), and several had been used in serological studies, including our own studies of intrathecal antibody synthesis [14, 15]. Furthermore, overlapping 15 mer synthetic peptides were made using amide resin chemistry as described [13], based on the known sequence of HTLV-1 gp21. The latter peptides were used to identify the minimal essential HTLV-1 gp21 amino acid sequences to stimulate antigen-specific T cell proliferation.
Table 1.
Synthetic peptides of HTLV-1 env gp21 used to generate human T lymphocyte lines.
We used a T lymphocyte microculture approach which allows selection of antigen-specific T cell lines from PBMC and which was originally established to analyse the T lymphocyte repertoire against myelin basic protein in multiple sclerosis [19, 20]. Briefly, PBMC were isolated from peripheral blood using standard density gradient centrifugation. PBMC were suspended in RPMI maintenance medium which contained 5% inactivated (56°C for 30 min) human serum, penicillin as well as streptomycin. Cells were exposed to the mixture of eight HTLV-1 gp21 peptides (Table 1), each at a concentration of 1 μg/ml. The cell suspension was distributed in round-bottomed 96-well culture plates (Nunc, Roskilde, Denmark, or Iwaki, Funabashi, Japan) at 2.5 × 104, 5 × 104, 105, and 2 × 105 PBMC/well. For each experiment, at least 200 microcultures were set up and kept strictly separate. Four days after the start of culture, IL-2 (Takeda, Japan) was added to the microcultures to give a final concentration of 1 U/ml. Microcultures were supplemented with fresh medium containing IL-2 (final concentration 1 U/ml) every 2–4 days depending on culture growth.
On day 14 the cells in 96-well culture plates were centrifuged (Kubota cytocentrifuge; Tokyo, Japan) and washed once with PBS. After the last centrifugation step, PBS was replaced with RPMI maintenance medium. The eight gp21 peptides (each at a final concentration of 1 μg/ml RPMI maintenance medium) and antigen-presenting cells (APC), but no exogenous IL-2, were added to each microculture to restimulate T cells. DR1 DQ5-matched HTLV-1 PCR and seronegative donor PBMC treated by mitomycin C (2 × 105 PBMC/well) were used as APC. After another 4 days maintenance medium and exogenous IL-2 (final concentration 1 U/ml) were added. Fresh IL-2-containing medium was supplemented until day 28 of culture depending on lymphocyte proliferation.
On day 28 the cells in 96-well culture plates were centrifuged again and washed three times with PBS. After the last centrifugation step, PBS was replaced with maintenance medium without IL-2. Two suspensions of APC were prepared from mitomycin C-treated PBMC of the DR1 DQ5-matched HTLV-1−donor. One was preincubated for 1 h with the HTLV-1 gp21 peptides at final concentrations of 1 μg/ml for each peptide, while the other suspension acted as a control and was not exposed to peptides. The original washed microcultures were split into two wells in new microculture plates and cocultured with one or the other APC suspension. These duplicate microcultures were incubated for another 4 days and examined daily for microscopic evidence of antigen-specific cell proliferation, i.e. proliferation only in the presence of antigen. Microcultures which showed proliferation both with and without antigen were discarded. Precursor frequencies were estimated following established principles [21] as described previously [13]. According to the screening procedure, positive microcultures were selected, transferred to 24- or 12-well culture plates and further propagated in two to three cycles of restimulation in the presence of APC and antigen, followed by IL-2-driven proliferation (concentrations as described above).
T lymphocyte proliferation assay
After antigen-specific T cell lines had been washed three times in PBS, their specificities were assayed in the presence of APC and different peptides. Microcultures were subjected to the standard 3H-thymidine incorporation assay by pulsing with 1 μCi 3H-thymidine (specific activity 25 Ci/mmol) per well and harvesting after 16 h. In all experiments DR1 DQ5-matched HTLV-1−donor PBMC treated by mitomycin C (2 × 105 PBMC/well) were used as APC. Concentrations of antigens and MoAbs were as described in the figures and tables. The following anti-HLA antibodies were used: anti-HLA-ABC (Dako, Glostrup, Denmark), anti-HLA-DR, anti-HLA-DQw1/3 and anti-HLA-DP (Becton Dickinson, Mountain View, CA). Recombinant HTLV-1 p21 (amino acids 326–433; Intracel Corp., Cambridge, MA) was employed as a positive control. Experiments were performed in triplicate. Antigen-specific proliferation was regarded to be significant if the stimulation index (SI; mean of antigen-stimulated cultures/mean of control cultures without antigen) was > 2. The s.d. of triplicates was < 15% of the mean, except in control cultures (without antigen) showing a very low spontaneous proliferation.
Search for homologies and phylogenetic analysis
In order to detect possible cross-reactivities with other proteins, the SWISSPROT database was searched for homologies of the newly defined HTLV-1 gp21 epitope sequences (using the server of the Computational Biochemistry Research Group, ETH Zürich, Switzerland). After finding retroviral homologous proteins, a multiple alignment was performed by selection of envelope sequences from different retroviral genera [22].
In order to find expressed homologous human sequences, a similarity search of the HTLV-1 gp21 amino acid sequences was performed using the TBLASTN algorithm and the Genebank database of expressed sequence tags, GeneBank EST (at the server of the NCBI, Bethesda, MD).
RESULTS
HTLV-1 env gp21-specific T cell lines
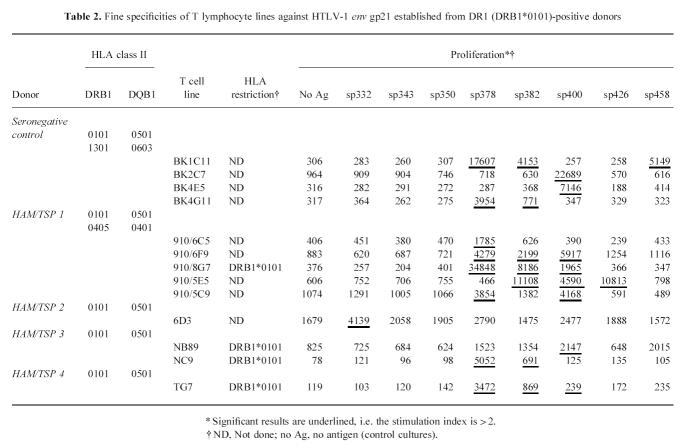
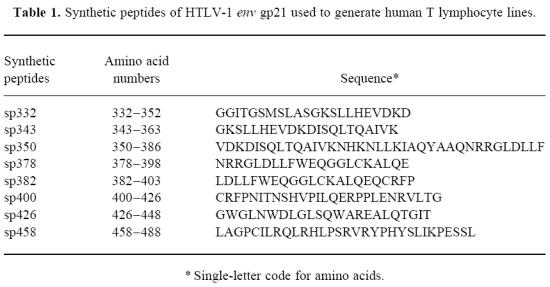
Using the T cell microculture approach and HTLV-1 non-infected, HLA-DR1 -DQ5-matched PBMC as APC, T lymphocyte lines were established against HTLV-1 gp21 synthetic peptides (Table 2). These T cell lines had the CD3+CD4+ phenotype (results not shown), as expected because of the selection procedure. In the presence of HLA-DR1 -DQ5-matched APC and antigen, proliferation of T cell lines could be blocked by a MoAb against HLA-DR, but not against other HLA class II or against HLA class I (as shown in a representative experiment in Fig. 1; summarized in Table 2). Under the conditions of the microculture approach, we estimated the precursor cell frequencies in the seronegative control and the non-homozygote HAM/TSP patient: 1 out of 106 PBMC and 1 out of 6 × 104 PBMC, respectively.
Table 2.
Fine specificities of T lymphocyte lines against HTLV-1 env gp21 established from DR1 (DRB1*0101)-positive donors
Fig. 1.
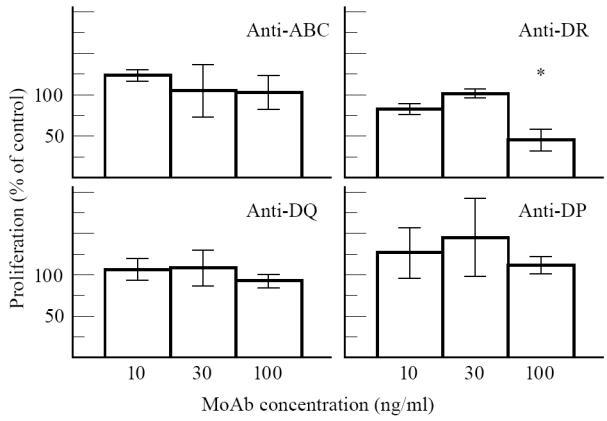
Inhibition of proliferation by anti-MHC MoAbs. In a representative experiment using T cell line 910/8G7, there was a significant inhibition of antigen-specific proliferation by anti-HLA-DR antibodies (*). Data are expressed as mean + s.d.
Fine specificities of T cell lines
Some T cell lines showed a proliferative response against more than one peptide, others against a single peptide (Table 2). Two specificities were most common: against the overlapping synthetic peptides sp378/sp382 and against sp400 (Table 2). These epitopes were further analysed using shorter synthetic peptides. An essential sequence of the sp378/sp382 epitope was 10 amino acids long (LFWEQGGLCK; Fig. 2), but T cell proliferation was greatly enhanced by a stretch of the N-terminal amino acids (Fig. 2b). A peptide lacking the leucine at the N-terminal end (FWEQGGLCKALQEQC) did not activate the T cell lines (data not shown). With respect to the sp400 epitope, the essential sequence consisted of 11 amino acids (RPPLENRVLTG; Fig. 3).
Fig. 2.
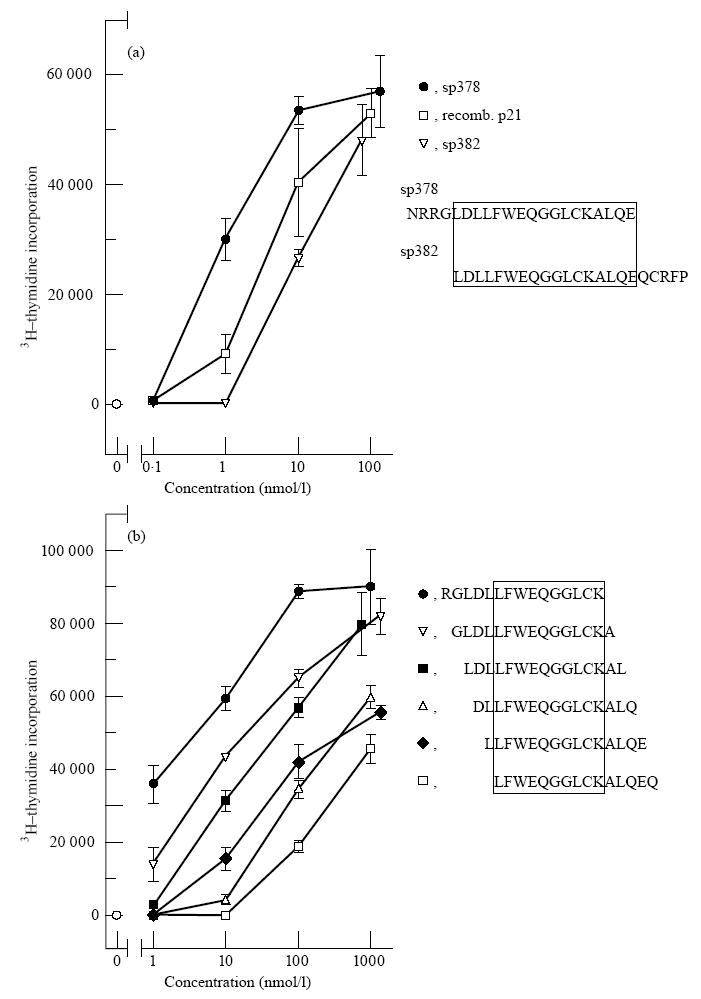
Proliferation of a HTLV-1 gp21 sp378/sp382-specific CD4+ T cell line (BK/1C11) depending on concentration and amino acid sequences of peptides. (a) Response to two long overlapping synthetic peptides, sp378 and sp382, which were used for selection of T cell lines, and to recombinant HTLV-1 p21. (b) Epitope analysis using short overlapping synthetic peptides with one amino acid shift. Values are expressed as mean + s.d. of triplicated data per point.
Fig. 3.
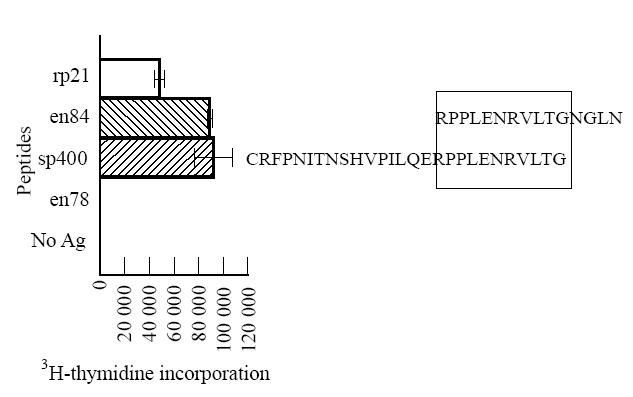
Analysis of epitope specificity of T cell line BK/2C7 for HTLV-1 gp21 using different overlapping peptides. Only recombinant p21 (rp21), peptides sp400 (amino acids 400–426) and en84 (amino acids 416–430) elicited a strong proliferative response, whereas all other peptides, including peptide sequences 396–410, 401–415, 406–420, 411–425, were negative (for clarity only one negative peptide is shown). Peptide concentrations were 1 μmolll. Data are expressed as mean + s.d.
Homology analysis of HTLV-1 gp21 T cell epitopes
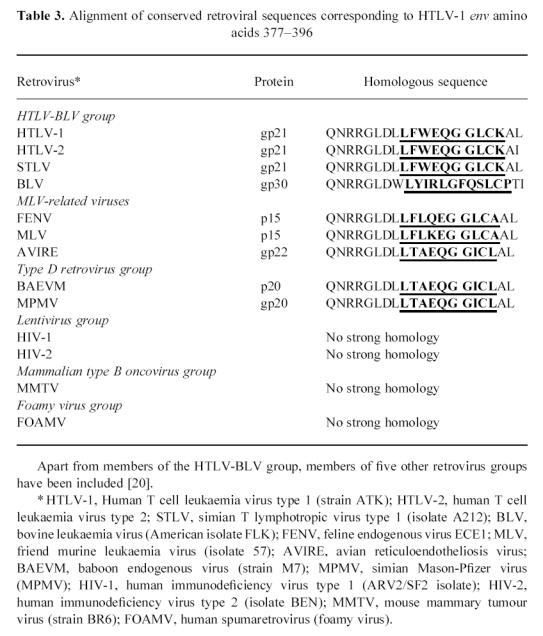
SWISSPROT database screening for homologies and subsequent phylogenetic analysis revealed that the sp378/sp382 epitope is localized in a highly conserved part of the retroviral envelope sequence, but there are no significant homologies to lentivirus, spumaretrovirus and MMTV envelope proteins (Table 3). The sp400 epitope has a very low homology to the same retroviral genera and BLV, but a high homology to HTLV-2 and STLV (results not shown).
Table 3.
Alignment of conserved retroviral sequences corresponding to HTLV-1 env amino acids 377–396
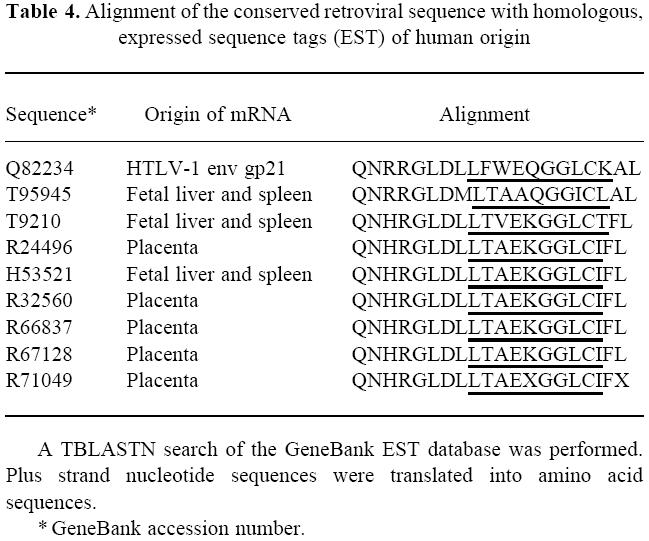
Furthermore, we searched the database of expressed human sequence tags (GeneBank EST database) for homologies with the conserved retroviral sequence which includes the sp378/sp382 epitope. Several human sequences could be identified which had previously been found to be expressed in human fetal tissues (Table 4).
Table 4.
Alignment of the conserved retroviral sequence with homologous, expressed sequence tags (EST) of human origin
DISCUSSION
In our study, we employed a microculture approach to select HTLV-1 transmembrane envelope protein gp21-specific CD4+ T cell lines from PBMC of four HTLV-1-infected and one noninfected individual who all carried the HLA class II DR1 DQ5 haplotype. Since HTLV-1 gp21-specific T cells could be isolated from the HTLV-1 non-infected donor, this T cell response appears to be part of the normal human T cell repertoire. T cell lines against the two common HTLV-1 gp21 epitopes were restricted by HLA-DR1 (HLA DRB1*0101). Thus, we report the immunodominant peptide motifs of HTLV-1 gp21 restricted by HLA-DR1 which have been established by T cell repertoire analysis. These peptide motifs were recognized by T cells when synthetic peptides bound to HLA were presented by APC to T cells, but also after antigen processing of a recombinant HTLV-1 p21 protein by APC (Figs 2a and 3).
The two immunodominant epitopes of HTLV-1 gp21 were further analysed by short overlapping synthetic peptides. Two unique T cell epitopes were defined (LFWEQGGLCK and RPPLENRVLTG) which showed special features compared with the established body of evidence regarding peptide sequences binding to HLA-DR1 (HLA DRB1*0101) [23–26]. In the epitope LFWEQGGLCK, three hydrophobic amino acids leucine, phenylalanine and tryptophan (underlined) are at the N-terminal end, which probably includes the main anchor position [23]. Furthermore, there are possible minor anchor positions five amino acids from the main anchor positions, i.e. two glycines [23]. We hypothesize that this epitope contains a strong HLA-DR1 binding motif. The enhancement of T cell proliferation by prolonging the peptide at the N-terminal end (Fig. 2b) might be related either to different folding of the longer peptide in water, or to enhanced binding to HLA-DR1 before or after proteolytic cleavage [26]. Next, we asked the question why this unique epitope is immunodominant although HTLV-1 gp21 contains a high number of possible HLA-DR1 binding sequences [13, 23]. Surprisingly, we found that this epitope corresponds to a highly conserved retroviral envelope sequence shared by many retroviruses, as shown in Table 3. Indeed it might be possible that the interaction of the retroviral envelope sequence, HLA and T cell receptor might be shaped by mammalian phylogeny. An immune response which is effective against a variety of retroviruses is likely to be conserved during phylogeny. There is another possibility: a similar sequence might be part of an endogenous retroviral sequence in the human genome. Indeed, two retroviral envelope sequences in Table 3 are derived from mammalian endogenous retroviruses. There is evidence that human cells express similar sequences (Table 4) which are probably derived from endogenous retroviral genes integrated in the human genome. To date it is unknown whether these sequences which are expressed in early life might induce tolerance or an autoreactive immune response in later life. Experimental studies on these endogenous conserved retroviral sequences need to be conducted to clarify their origin, expression in differentiated tissues and immunogenicity both in HTLV-1-infected and non-infected individuals.
There is another interesting feature of this highly conserved peptide sequence. It has been found to be suppressive for several immune functions if added as a synthetic peptide in micromolar concentrations to the culture medium of mononuclear cells [27, 28]. This suppressive effect is non-specific and mediated via intracellular cAMP [28]. The HLA-DR1-restricted specific immune response against the epitope at concentrations in the nanomolar range (Fig. 2) might protect the individual from the deleterious effects of this HTLV-1 gp21 domain at much higher concentrations. Thus, the CD4+ T cell response might contribute to the protective effect of HLA-DR1 against ATL, since HLA-DR1 is very rare in patients with ATL [8].
The other immunodominant epitope RPPLENRVLTG lacks similarity to the established HLA-DR1 binding motif [23–26]. For this epitope it is very difficult to define the main anchor position for DR1 [23]. The sequence corresponds to another important domain of HTLV-1 gp21, a functional domain of the retrovirus as established by a peptide inhibition assay for syncytium formation [29]. Indeed, the function of this sequence might be important to make it the second immunodominant epitope. This sequence is conserved only in the HTLV and STLV retroviruses, but has a low homology to other retroviruses. To date, there is no evidence of a homologous endogenous retroviral sequence in the human genome.
At this stage of our studies of the CD4+ T cell response against HTLV-1 we used HTLV-1 gp21-specific CD4+ T cell lines cultured from PBMC to identify immunodominant epitopes (Table 2). In further experiments we need to select these lines into peptide-specific T cell clones in order to analyse their particular functions and to clarify if some of the clones are infected with HTLV-1.
In general, the functional and regulatory properties of CD4+ T cells are related to helper effects for B cells [16] and supportive effects for the cytotoxic T cell response [30]. The significance of ‘helper’ CD4+ T cells for the B cell immune response in HAM/TSP can be linked to our own studies of the systemic and intrathecal humoral immune response [14, 15]. Interestingly, the sequence sp382 contains an immunodominant T cell epitope and a frequently recognized B cell epitope [14, 15]. The sequence sp458 frequently binds antibodies in samples from HLA-DR1+ HAM/TSP patients [15], it does not contain an immunodominant T cell epitope restricted by HLA-DR1 (as shown), but two HLA-DR1-restricted CD4+ T cell epitopes have been identified in the proximity, i.e. amino acids 436–450 and 451–465 [13]. HTLV-1 gp21 is an important target antigen both for the T cell and the B cell immune response. The sites of immunodominant epitopes for the CD4+ T cell and B cell receptors tend to overlap or to be in close proximity. The same applies to HTLV-1 gp46 as shown by Jacobson et al. [9]. Using data from our previous studies [14, 15], we could compare the B cell and CD4+ T cell specificities in HAM/TSP patients 1 and 2. In HAM/TSP patient 1 the systemic and intrathecal humoral immune responses were directed against peptides sp332, sp350, sp382 and sp458, but only sp382 elicited a significant CD4+ T cell response. In HAM/TSP patient 2 there was a significant systemic humoral immune response only against peptide sp350, but the CD4+ T cell response was directed against sp332.
To date there is little knowledge of the interactions of B and T cell immune responses and of the distribution of B and T cell epitopes in viral infections. In measles [31] and influenza [32] virus infections there is evidence for adjacent or overlapping B and T cell epitopes. In a persisting viral infection, as in other secondary immune responses, B cells might act as highly efficient APC which present antigen, provide a second signal for T cell activation and secrete cytokines [33, 34]. Proximity of T cell and B cell epitopes in a protein antigen might facilitate the specific activation of both T cells and B cells for the same antigen [9, 13,31–34].
Regarding the cellular immune responses in HAM/TSP, recently the main research focus has been on HTLV-1 tax-specific CD8+ T cells [35–38]. The functional potential and data on high precursor frequencies of HTLV-1 tax-specific cytotoxic T cells (CTL) in HAM/TSP and asymptomatic carriers remain controversial (as reviewed in [38]). So far, there are no experimental data on the precursor frequencies and epitope specificities of HTLV-1 gp21-specific CD8+ CTL. Future studies should also evaluate CD8+ HTLV-1 gp21-specific T cells in HAM/TSP, as well as in asymptomatic HTLV-1 carriers and seronegative individuals.
Acknowledgments
B.K. was supported by a research grant from the Japanese Ministry of Education and in part by a fellowship of the European Commission. We thank K. Ikeda (MS TechnoSystems, Osaka, Japan) for support in making short synthetic peptides, and the Computational Biochemistry Research Group, ETH Zürich, for their valuable services and advice via the Internet.
References
- 1.Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H. Adult T-cell leukemia: clinical and hematological features of 16 cases. Blood. 1977;50:481–92. [PubMed] [Google Scholar]
- 2.Osame M, Usuku K, Izumo S, et al. HTLV-I associated myelopathy. A new clinical entity. Lancet. 1986;1:1031–2. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 3.Gessain A, Barin F, Vernant JC, et al. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;2:407–10. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 4.Usuku K, Sonoda S, Osame M, et al. HLA haplotype-linked high immune responsiveness against HTLV-I in HTLV-I-associated myelopathy: comparison with adult T-cell leukemia/lymphoma. Ann Neurol. 1988;23(Suppl.):S143–50. doi: 10.1002/ana.410230733. [DOI] [PubMed] [Google Scholar]
- 5.Sonoda S, Yashiki S, Fujiyoshi T, et al. Immunogenetic factors involved in the pathogenesis of adult T-cell leukemia and HTLV-I- associated myelopathy. Gann Monograph Cancer Res. 1992;39:81–93. [Google Scholar]
- 6.Nishimura Y, Okubo R, Minato S, et al. A possible association between HLA and HTLV-1-associated myelopathy (HAM) in Japanese. Tissue Antigens. 1991;37:230–1. doi: 10.1111/j.1399-0039.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 7.Usuku K, Nishizawa M, Matsuki K, et al. Association of a particular amino acid sequence of the HLA DRβ1 chain with HTLV-I-associated myelopathy. Eur J Immunol. 1990;20:1603–16. doi: 10.1002/eji.1830200729. [DOI] [PubMed] [Google Scholar]
- 8.Sonoda S, Fujiyoshi T, Yashiki S. Immunogenetics of HTLV-I/II and associated diseases. J Acquir Immune Defic Syndr. 1996;13(Suppl. 1):S119–23. doi: 10.1097/00042560-199600001-00020. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson S, Reuben JS, Streilein RD, Palker TJ. Induction of CD4+, human T lymphotropic virus type-1-specific cytotoxic T lymphocytes from patients with HAM/TSP. Recognition of an immunogenic region of the gp46 envelope glycoprotein of human T lymphotropic virus type-1. J Immunol. 1991;146:1155–62. [PubMed] [Google Scholar]
- 10.Manca F, Li Pira G, Fenoglio D, et al. Recognition of human T-leukemia virus (HTLV-1) envelope by human CD4+ T-cell lines from HTLV-1 seronegative individuals: specificity and clonal heterogeneity. Blood. 1995;85:1547–54. [PubMed] [Google Scholar]
- 11.Katahira Y, Yashiki S, Fujiyoshi T, et al. In vitro induction of cytotoxic T lymphocytes against HTLV-I-infected T-cells from adult T-cell leukemia patients, asymptomatic HTLV-I carriers and seronegative healthy donors. Jpn J Cancer Res. 1995;86:21–27. doi: 10.1111/j.1349-7006.1995.tb02983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagai M, Yashiki S, Fujiyoshi T, et al. Characterization of a unique T-cell clone established from a patient with HAM/TSP which recognized HTLV- I-infected T-cell antigens as well as spinal cord tissue antigens. J Neuroimmunol. 1996;65:97–105. doi: 10.1016/0165-5728(96)00002-1. [DOI] [PubMed] [Google Scholar]
- 13.Yamano Y, Kitze B, Yashiki S, et al. Preferential recognition of HTLV-I gp21 envelope protein restricted by HLA-DRB1 alleles of HTLV-I-associated myelopathy/tropical spastic paraparesis (HAM/TSP) J Neuroimmunol. 1997;76:50–60. doi: 10.1016/s0165-5728(97)00041-6. [DOI] [PubMed] [Google Scholar]
- 14.Kitze B, Usuku K, Izumo S, et al. Diversity of intrathecal antibody synthesis against HTLV-I and its relation to HTLV-I associated myelopathy. J Neurol. 1996;243:393–400. doi: 10.1007/BF00868998. [DOI] [PubMed] [Google Scholar]
- 15.Kitze B, Usuku K, Yashiki S, et al. Intrathecal humoral immune response in HAM/TSP in relation to HLA haplotype analysis. Acta Neurol Scand. 1996;94:287–93. doi: 10.1111/j.1600-0404.1996.tb07067.x. [DOI] [PubMed] [Google Scholar]
- 16.Mosmann TR, Coffman RL. Th1 and Th2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–73. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 17.Osame M. Review of WHO Kagoshima Meeting and Diagnostic Guidelines for HAM/TSP. In: Blattner WA, editor. Human Retrovirology: HTLV. New York: Raven Press; 1990. pp. 191–7. [Google Scholar]
- 18.Ota M, Seki T, Fukushima H, Tsuji K, Inoko H. HLA-DRB1 genotyping by modified PCR-RELP method combined with group specific primers. Tissue Antigens. 1992;39:187–202. doi: 10.1111/j.1399-0039.1992.tb01935.x. [DOI] [PubMed] [Google Scholar]
- 19.Kitze B, Pette M, Rohrbach E, Städt D, Kappos L, Wekerle H. Myelin specific T-lymphocytes in multiple sclerosis patients and healthy individuals. J Neuroimmunol. 1988;20:237. doi: 10.1016/0165-5728(88)90166-x. [DOI] [PubMed] [Google Scholar]
- 20.Pette M, Fujita K, Kitze B, et al. Myelin basic protein specific T lymphocyte lines from MS patients and healthy individuals. Neurol. 1990;40:1770–6. doi: 10.1212/wnl.40.11.1770. [DOI] [PubMed] [Google Scholar]
- 21.Lefkovits I, Waldmann H. Cambridge: Cambridge University Press; 1979. Limiting dilution analysis of cells in the immune system; pp. 38–82. [Google Scholar]
- 22.Coffin JM. Retroviridae. In: Francki RIB, Fauquet CM, Knudson DL, Brown F, editors. Classification and nomenclature of viruses—Fifth report of the international committee on taxonomy of viruses. New York: Springer; 1991. pp. 290–9. [Google Scholar]
- 23.Hammer J, Nagy ZA, Sinigaglia F. Rules governing peptide-class II MHC molecule interactions. Behring Inst Mitt. 1994;94:124–32. [PubMed] [Google Scholar]
- 24.Hill CM, Liu A, Marshall KW, et al. Exploration of requirements for peptide binding to HLA DRB1*0101 and DRB1*0401. J Immunol. 1994;152:2890–8. [PubMed] [Google Scholar]
- 25.Brown JH, Jardetzky TS, Gorga JC, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA- DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 26.Engelhard VH. Structure of peptides associated with class I and class II MHC molecules. Annu Rev Immunol. 1994;12:181–207. doi: 10.1146/annurev.iy.12.040194.001145. [DOI] [PubMed] [Google Scholar]
- 27.Haraguchi S, Good RA, James-Yarish M, Cianciolo GJ, Day NK. Differential modulation of Th1- and Th2-related cytokine mRNA expression by a synthetic peptide homologous to a conserved domain within retroviral envelope protein. Proc Natl Acad Sci USA. 1995;92:3611–5. doi: 10.1073/pnas.92.8.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haraguchi S, Good RA, James-Yarish M, Cianciolo GJ, Day NK. Induction of intracellular cAMP by a synthetic retroviral envelope peptide: a possible mechanism of immunopathogenesis in retroviral infections. Proc Natl Acad Sci USA. 1995;92:5568–71. doi: 10.1073/pnas.92.12.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sagara Y, Inoue Y, Shiraki H, Jinno A, Hoshino H, Maeda Y. Identification and mapping of functional domains on human T-cell lymphotropic virus type 1 envelope proteins by using synthetic peptides. J Virol. 1996;70:1564–9. doi: 10.1128/jvi.70.3.1564-1569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stuhler G, Walden P. Collaboration of helper and cytotoxic T lymphocytes. Eur J Immunol. 1993;23:2279–86. doi: 10.1002/eji.1830230934. [DOI] [PubMed] [Google Scholar]
- 31.Muller CP, Handtmann D, Brons NHC, et al. Analysis of antibody response to the measles virus using synthetic peptides of the fusion protein: evidence of non-random pairing of T and B cell epitopes. Virus Res. 1993;30:271–80. doi: 10.1016/0168-1702(93)90095-5. [DOI] [PubMed] [Google Scholar]
- 32.Graham CM, Barnett BC, Hartlmeyer I, et al. The structural requirements for class II (I-Ad)-restricted T cell recognition of influenza hemagglutinin: B cell epitopes define T cell epitopes. Eur J Immunol. 1989;19:523–8. doi: 10.1002/eji.1830190317. [DOI] [PubMed] [Google Scholar]
- 33.Lanzavecchia A. Antigen-specific interaction between T and B cells. Nature. 1985;314:537–9. doi: 10.1038/314537a0. [DOI] [PubMed] [Google Scholar]
- 34.Pistoia V. Production of cytokines by human B cells in health and disease. Immunol Today. 1997;18:343–50. doi: 10.1016/s0167-5699(97)01080-3. [DOI] [PubMed] [Google Scholar]
- 35.Elovaara I, Koenig S, Brewah AY, Woods RM, Lehky T, Jacobson S. High human T cell lymphotropic virus type 1 (HTLV-1)-specific precursor cytotoxic T lymphocyte frequencies in patients with HTLV-1-associated neurological disease. J Exp Med. 1993;177:1567–73. doi: 10.1084/jem.177.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elovaara I, Utz U, Smith L, Jacobson S. Limited T cell receptor usage by HTLV-I tax-specific, HLA class I restricted cytotoxic T lymphocytes from patients with HTLV-I associated neurological disease. J Neuroimmunol. 1995;63:47–53. doi: 10.1016/0165-5728(95)00129-8. [DOI] [PubMed] [Google Scholar]
- 37.Daenke S, Kermode AG, Hall SE, et al. High activated and memory cytotoxic T-cell responses to HTLV-1 in healthy carriers and patients with tropical spastic paraparesis. Virol. 1996;217:139–46. doi: 10.1006/viro.1996.0101. [DOI] [PubMed] [Google Scholar]
- 38.Bangham CRM, Kermode AG, Hall SE, Daenke S. The cytotoxic T-lymphocyte response to HTLV-I: the main determinant of disease? Sem Virol. 1996;7:41–48. [Google Scholar]