Abstract
Permanently transfected mouse cell lines which expressed different levels of the human autoantigen La/SS-B were infected with different strains of herpes simplex virus type 1, including the strains ANG, HSZP, 17syn+ and HFEM. During infection the localization of the human La protein was followed using an anti-La MoAb, which recognized only the human La protein but did not cross-react with either the endogenous mouse La protein or any viral encoded protein. After infection La protein was transported from the nucleus to the cytoplasm. The time course of translocation was dependent on the amount of human La protein expressed in the respective cell line. Moreover, acceleration of viral replication was dependent on the level of expression of human La protein, suggesting that La protein is a cellular factor that facilitates virus replication.
Keywords: autoimmunity, autoantigen, herpes simplex, La/SS-B
INTRODUCTION
Sera of patients with rheumatoid diseases such as systemic lupus erythematosus (SLE) or primary Sjögren's syndrome (pSS) contain autoantibodies to nuclear antigens including the autoantigen La (SS-B) [1]. The La protein was described to associate at least transiently with all primary RNA polymerase III transcripts including precursor molecules of ribosomal 5S RNA, tRNAs, some 4.5S RNAs, as well as a portion of the U1 and U6 snRNA [2–5]. Common to all primary RNA polymerase III transcripts is their 3′-terminal oligo (U)-tail, which is transcribed during the transcription termination step. These oligo (U)-tails were shown to be a binding site for the La protein [6]. Based on these findings a function of La protein in the transcription termination step of RNA polymerase III was proposed [7]. Besides the cellular small RNAs, La protein was found to associate with viral encoded small RNAs such as the adenovirus-encoded VA RNAs and the Epstein–Barr virus (EBV)-encoded EBER RNAs, thus forming ribonucleoprotein particles (RNPs) [8,9]. In addition to these viral encoded small RNPs, La protein was found associated with the leader RNAs of a series of viruses including the vesicular stomatitis virus (VSV), rabies viruses, poliovirus, HIV, influenza virus, rubella virus and hepatitis C virus [10–17]. Based on the association with viral genomes or mRNAs, a function of La protein in viral replication processes and/or in internal initiation of translation of uncapped viral mRNAs has been proposed.
Originally the protein involved in internal initiation of poliovirus mRNA was assumed to be encoded by the virus genome and proposed to be a dsRNA unwinding enzyme [15]. UV cross linking studies, however, showed that the protein is encoded by the host cell. The cross-linked cellular protein was characterized by sequencing and shown to be the La protein [16]. In line with a function in internal initiation of translation, La protein could be cross-linked not only to viral mRNAs but even to the translation initiation AUG start codon [18]. Moreover, it was described to be translocated from the nucleus to the cytoplasm after virus infections [18–23].
Similar to other mRNAs that are assumed to be regulated at the translational level, the poliovirus mRNAs contain a series of upstream open reading frames and a polypyrimidine stretch upstream of the translation initiation site. When a cDNA library, made from peripheral blood lymphocytes of an autoimmune patient, was screened for La cDNAs, similarly structured alternative La mRNA forms were identified [24–31]. In these La mRNA transcripts the exon 1 was replaced with alternative exons that were termed exon 1′ and 1′′. The 5′-start of the La mRNA isoforms varied in dependence on the analysed tissues and were found to influence the translational efficiency ([28,29]; to be published). Genomic analysis including an analysis using reporter gene fusion constructs [32] showed that the La mRNA isoforms were the result of a promoter switch in combination with alternative splicing pathways. Detailed analysis of expression showed that all La mRNA forms represent finally processed abundant cytoplasmic and translatable mRNAs.
Usually virus replication involves cellular proteins misusing their cellular function. As the unusual 5′-structure of the exon 1′ La mRNA isoform remembered to the structure of picorna virus mRNAs, it is tempting to speculate that one of the cellular functions of La protein could be an involvement in translation of its own exon 1′ La mRNAs. As herpes simplex virus type 1 (i) had been shown to cause a translocation of La protein from the nucleus to the cytoplasm, and (ii) is able to infect and replicate in a variety of species, we investigated whether virus infections may have different effects on La protein expression in murine cells permanently expressing the different human La mRNA forms.
Unexpectedly, during these studies we observed that herpes simplex type 1 replication appeared to be accelerated in dependence on the amount of human La protein expressed in the respective cell line.
MATERIALS AND METHODS
Materials
The following materials were obtained: Dulbecco's modified Eagle's medium (DMEM) from Gibco BRL Life Technologies (Eggenstein, Germany); anti-mouse IgG conjugated with Cy3 from Medac (Hamburg, Germany); anti-human IgG (γ-chain-specific) conjugated with alkaline phosphatase (A3150; 8 U/ml); protein A agarose from Sigma (St Louis, MO); 4-nitroblue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) from Roth (Karlsruhe, Germany); and PVDF-membrane (IPVH 000 10; pore size 0.45 μm) from Millipore (Bedford, MA).
Antibodies and sera
The anti-La MoAb SW5, which was found to be directed to the N-terminal domain of La protein [33], was originally described by Smith et al. [34]. The hybridoma supernatant was kindly provided by Professor Dr W. J. van Venrooij (University of Nijmegen, The Netherlands). The patient's anti-La antibody was prepared from the serum of the patient (Ma) [28,35]. Two antibodies to herpes simplex virus type 1 glycoproteins were obtained from Pharmingen (San Diego, CA). Both antibodies gave comparable results.
Cells and viruses
Mouse NIH 3T3 cells were grown in DMEM containing 10% fetal calf serum (FCS) in a humidified CO2 atmosphere either in culture flasks for preparation of extracts or on coverslips for epifluorescence microscopy. A series of cell lines expressing either the human exon 1, exon 1′ or the human La gene had been recently prepared and shown to express different levels of human La protein ([31], see also Fig. 3). Two cell lines permanently expressed the exon 1 type of La mRNA. Five cell lines expressed the full length exon 1′ La mRNA form. In addition, a mouse 3T3 cell line was obtained that expressed all human La mRNA forms from the human La gene under its genuine promoter elements. Human La protein expression was highest in the mouse cell line expressing the human La gene using its genuine promoter. All cell lines expressing the human La mRNAs under control of thecytomegalovirus (CMV) promoter produced less human La protein. The level of human La protein was higher in cells expressing the exon 1 La mRNA form and reached the level close to the cell line expressing the human La gene. All exon 1′ La mRNA-expressing cell lines contained less human La protein. The differences in La protein content were not dependent on the construct or the promoter or the integration site, as all the different exon 1′ or all exon 1 La mRNA-expressing cells expressed comparable La protein levels. As control cells either untransfected cells or cells expressing neomycin resistance under control of the CMV promoter were used.
Fig. 3.
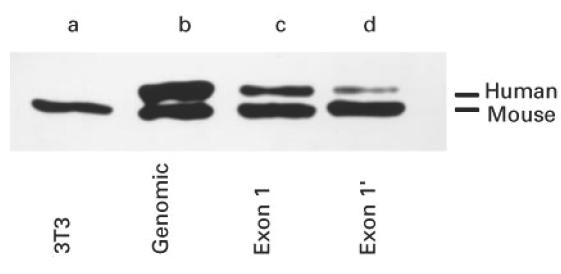
SDS–PAGE/immunoblotting analysis of mouse cell lines expressing the human La gene or either exon 1 or exon 1′ La mRNAs. Total extracts of mouse cells were analysed. Untransfected mouse 3T3 cells (3T3, lane a); cell lines expressing the human La gene (lane b), or human exon 1 La mRNA (lane c), or human exon 1′ La mRNA (lane d).
The cells were infected with herpes simplex virus type 1. We used the four different strains including ANG, HSZP, HFEM, and 17syn+ [36–38]. The cells were infected using different plaque-forming unit (PFU) ranges from 1 to 20 PFU per cell.
Epifluorescence microscopy
For immunofluorescence microscopy the cells were fixed with methanol containing 0.02% EGTA at −20°C for 1 h. Indirect immunofluorescence of cells with the anti-La MoAb SW5 was performed by incubating the fixed cells, which had been rehydrated for 5 min with PBS, with cell culture supernatant of hybridomas secreting the anti-La MoAb SW5 for at least 15 min. In the case of the MoAbs directed to herpes simplex virus surface proteins a dilution of 1:100 was used. The cells were washed with PBS (5 min) and the bound MoAbs were detected using Cy3-conjugated anti-mouse antibody. The incubation time for the secondary antibody was 15 min. The staining occurred at room temperature and the unbound secondary antibodies were removed by washing with PBS (twice, 5 min). The stained cells were mounted using PBS/glycerol (1:1 (v/v)) and analysed with a Zeiss Axiophot microscope equipped with epifluorescence optics. Photographs were taken using Kodak Ektachrome 400 film.
Preparation of total extracts
Total extracts were prepared by incubation with 350 μl SDS–PAGE sample solution (95°C). The lysed cells were heated for 5 min to 95°C and centrifuged at 14 000 g for 5 min at 4°C. Aliquots of 20 μl were used for SDS–PAGE.
SDS–PAGE/immunoblotting analysis
SDS–PAGE was performed according to Laemmli [39]. Transfer to PVDF membrane was performed according to Matsudaira [40]. Total extracts of mouse cells were analysed using the patient anti-La antibody (Ma). This anti-La antibody was shown to be useful to identify human La protein expressed in transfected cell lines besides endogenous mouse La protein [25,28,29].
Measurement of herpes virus DNA-dependent DNA polymerase activity
As described earlier, herpes simplex virus type 1 DNA replication depends on a viral encoded DNA polymerase [41]. Viral DNA polymerase activity can be estimated after inhibition of cellular DNA polymerase activities in the presence of 100 mm ammonium sulphate using a commercially available non-radioactive DNA polymerase assay provided by Boehringer (Boehringer, Mannheim, Germany). The ELISA was performed according to the supplier's instructions.
RESULTS AND DISCUSSION
Mouse 3T3 cells expressing permanently human La mRNA isoforms were infected with herpes simplex virus type 1 using the four different strains including ANG, HSZP, HFEM, and 17syn+. Due to alteration of the actin filament, herpes virus-infected cells usually alter their morphology. The virus strains HSZP, HFEM and ANG are herpes simplex viruses that finally cause giant cell formation, while the strain 17syn+ causes a rounding of the infected cells without formation of giant cells.
Analysis of localization of human La protein in virus-infected mouse 3T3 cells using epifluorescence microscopy
Untransfected and transfected mouse 3T3 cells were infected and fixed at various times post-infectionem (p.i.) including 1 h, 4 h, 8 h and 24 h p.i. The cells were stained with the anti-La MoAb SW5. The examples shown in Fig. 1 were taken for the strain ANG.
Fig. 1.
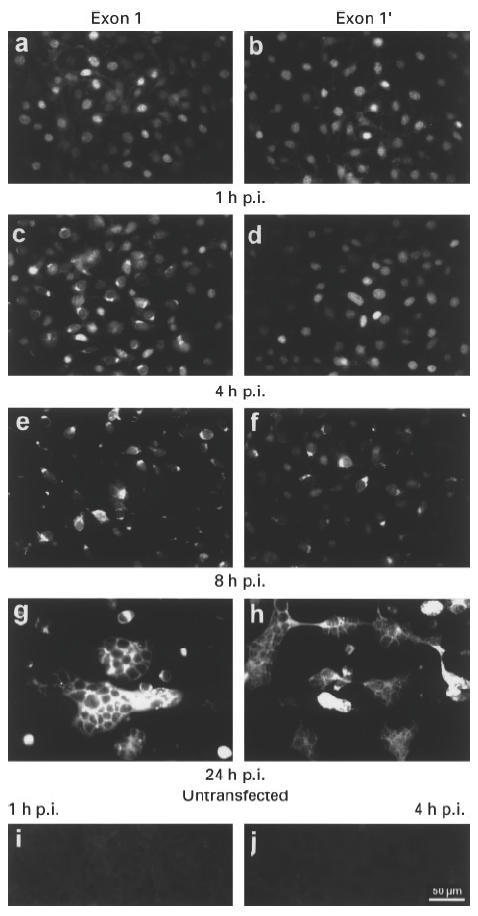
Epifluorescence analysis of mouse 3T3 cells permanently expressing human exon 1 (a,c,e,g) or 1′ La mRNAs (b,d,f,h). The cells were infected with herpes simplex virus type 1 strain ANG. (i,j) Untransfected mouse 3T3 cells. The cells were obtained 1 h post-infection (p.i.) (a,b,i) or 4 h p.i. (c,d,j), or 8 h p.i. (e,f) or 24 h p.i. (g,h).
The anti-La Moab SW5 did not stain the untransfected mouse 3T3 cells either before or after infection with herpes simplex virus type 1 (e.g. Fig. 1i, 1 h p.i.; Fig. 1j, 4 h p.i.). These results showed that the anti-La MoAb did not cross-react with any endogenous mouse La protein either before or during virus infection.
Fig. 4.
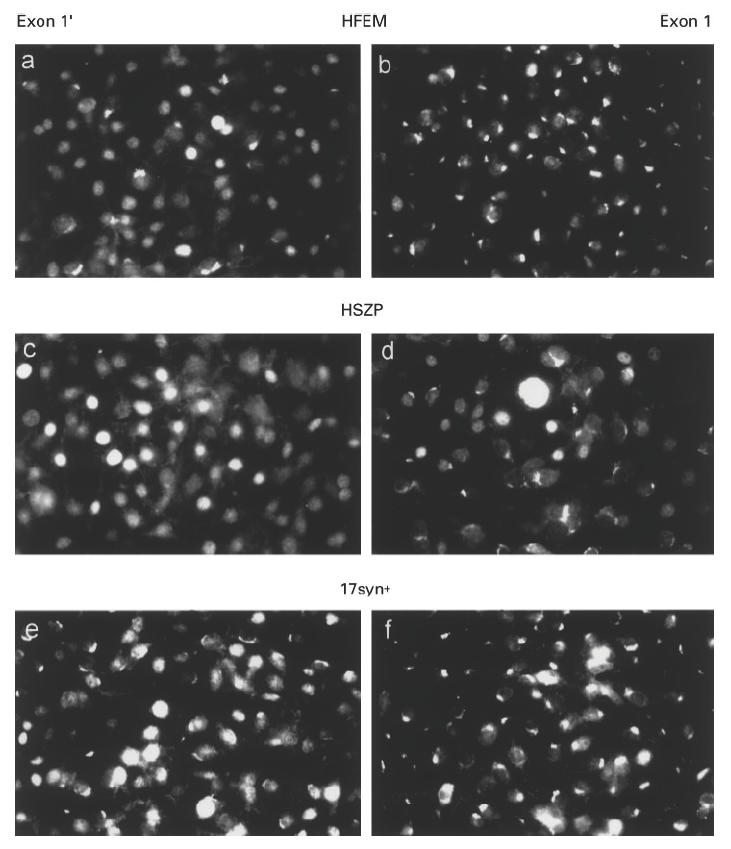
Epifluorescence analysis of mouse 3T3 cells permanently expressing human exon 1′ (a,c,e) or 1′ La mRNAs (b,d,f). The cells were infected with herpes simplex virus type 1 strain HFEM (a,b), strain HSZP (c,d) or strain 17syn+ (e,f). The cells were obtained 8 h p.i. (a,b,e,f) or 6 h p.i. (c,d).
In conclusion, the staining pattern of the cell lines expressing either the human exon 1 (Fig. 1a,c,e,g) or the exon 1′ (Fig. 1b,d,f,h) La mRNA must represent the localization of the human La protein in the transfected cells. As shown in Fig. 1a,b, 1 h p.i. both, the exon 1 and exon 1′ La mRNA-expressing cell lines gave a mostly nuclear staining. Four hours p.i. the nuclear staining pattern disappeared in a major portion of the exon 1 La mRNA-expressing cells, although some cells still displayed the nuclear staining (Fig. 1c). In contrast, most of the exon 1′ La mRNA-expressing cells still gave a nuclear staining pattern (Fig. 1d). Eight hours p.i. the exon 1′ La mRNA-expressing cell lines also displayed the cytoplasmic pattern (Fig. 1f), although some cells still gave a nuclear staining pattern. Giant cell formation was seen in both cell lines 24 h p.i. (Fig. 1g,h).
Analysis of human La protein in virus-infected mouse 3T3 cells using SDS–PAGE/immunoblotting
La protein was described to be proteolytically sensitive and the N-terminal fragment, which contains the epitope recognized by the anti-La MoAb, does not contain a nuclear localization signal. In order to analyse whether the cytoplasmic staining pattern seen after virus infection was due to a degradation of human La protein, total extracts of cells obtained after virus infection were analysed by SDS–PAGE/immunoblotting. As shown in Fig. 2, when total extracts that were obtained from either uninfected cells (lanes a, b, d, g, j) or cells obtained 1 h p.i. (lanes c and e), 4 h p.i. (lanes f and h), or 8 h p.i. (lanes i and k) were analysed, no obvious degradation of the endogenous mouse or the transfected human La protein was observed. Thus the cytoplasmic staining observed after virus infection represented non-degraded human La protein.
Fig. 2.
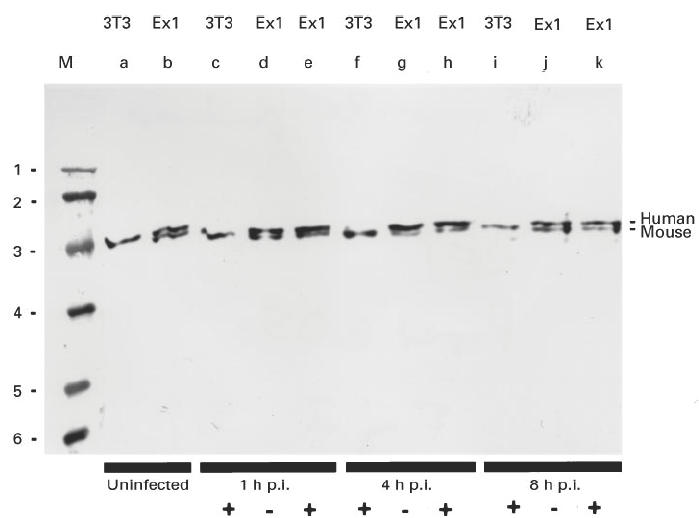
SDS–PAGE/immunoblotting analysis of mouse cells expressing the human exon 1 La mRNAs after infection with herpes simplex type 1. Total extracts of mouse cells were analysed. Untransfected mouse 3T3 cells (3T3, lanes a, c, f, i); cell lines expressing the exon 1 La mRNA under control of the cytomegalovirus (CMV) promoter (lanes b, d, e, g, h, j, k). The cells were infected with the herpes simplex virus type 1 strain ANG ((+), lanes c, e, f, h, i, k) or not infected ((–), lanes d, g, j). Cells were analysed 1 h (lanes c, d, e), 4 h (lanes f, g, h) or 8 h (lanes i, j, k) p.i. M, Marker, lane 1 = 97.4 kD, 2 = 66 kD, 3 = 43 kD, 4 = 31 kD, 5 = 21.5 kD, 6 = 14.5 kD.
Localization of human La protein in different permanently transfected mouse 3T3 cell lines after infection with different herpes virus strains
In an earlier approach a series of permanently transfected cell lines had been established. In addition to the cell lines mentioned above, an additional four cell lines had been established that permanently expressed the exon 1′ La mRNA form and a further cell line that permanently expressed the exon 1 La mRNA form. All exon 1′ La mRNA-expressing cell lines expressed comparable human La protein levels. The level was less than that of the cell lines expressing the human La gene or the exon 1 La mRNA form. The level of La protein expression was slightly less in the cell lines expressing the exon 1 La mRNA form than in the cell line expressing the human La gene [34]. A representative expression pattern is shown in Fig. 3 (lanes a–d).
In order to rule out that the observed effects shown in Fig. 1 were due to a unique property of a selected cell line or a certain virus strain, the experiments were repeated using different exon 1 or exon 1′ La mRNA-expressing cell lines. In addition, a cell line permanently expressing the neomycin resistance gene under the CMV promoter control was used as a further control.
Independently of the used cell lines or virus strains, the different onsets of the alteration of the staining pattern in exon 1 or exon 1′ expressing cell lines were seen. Selected examples for the virus strain HFEM (Fig. 4a,b), HSZP (Fig. 4c,d) or 17syn+ (Fig. 4e,f) are shown. A different onset of the alteration of the staining pattern was also seen when different cell densities or different PFU were used for infection (data not shown). In contrast, no difference was observed when the localization was analysed in untransfected 3T3 cells compared with 3T3 cells transfected with the neomycin resistance gene only (data not shown).
Taken together, these results indicate that either the different 5′-structure of the La mRNA forms or the different levels of human La protein were responsible for either a retardation or an acceleration of virus replication.
In order to resolve this question further studies were performed. In the first approach cells were infected with herpes simplex virus type 1 strain ANG and analysed by phase contrast. In the second approach cells were infected with the same virus strain but were stained with antibodies to viral glycoproteins and analysed by epifluorescence microscopy. The analysed glycoproteins belong to the late proteins, appearing in the late phase of herpes simplex virus infection. In the third approach the level of herpes virus DNA replicase activity was directly measured in extracts of herpes simplex virus-infected cells that were prepared at different times p.i.
Acceleration of rounding up of virus-infected cells in dependence on the level of human La protein expression
The cells were infected with the herpes simplex type 1 strain ANG. Infection with the virus caused an alteration of the shape of the cells from the fibroblast morphology (e.g. Fig. 5a) to rounded cells (e.g. Fig. 5c). All the images shown in Fig. 5 were obtained 5 h p.i., but comparable differences were also seen at earlier stages p.i. and using different virus strains. As shown in Fig. 5, the infection course was slowest in the mouse cells lacking additional human La protein (Fig. 5a) and accelerated in the cells expressing the highest amount of human La protein (Fig. 5c,d). Compared with the untransfected 3T3 cells (Fig. 5a) the effect appeared earlier in cells expressing the exon 1′ La mRNA (Fig. 5b), but less if compared with the cell line expressing the human La gene (Fig. 5d) or the exon 1 La mRNA (Fig. 5c).
Fig. 5.
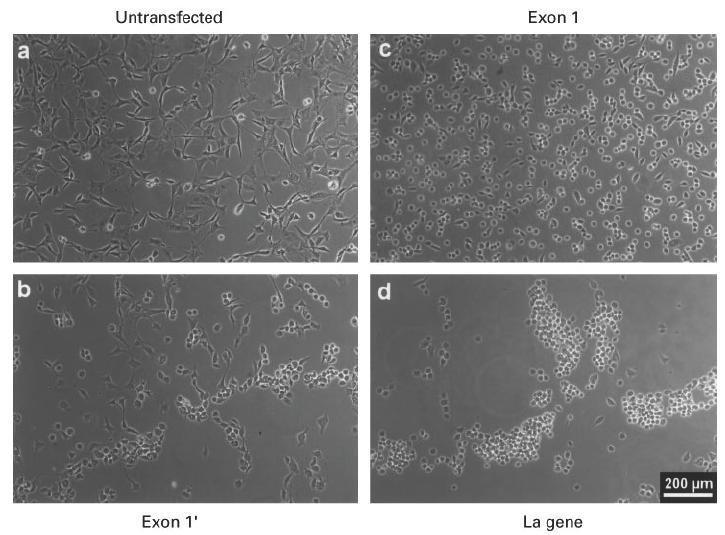
Phase contrast analysis of mouse 3T3 cells after herpes simplex type 1 virus infection. The cells were infected with herpes simplex virus type 1 strain ANG. Untransfected mouse 3T3 cells (a), or cells expressing the exon 1′ under cytomegalovirus (CMV) promoter control (b), or cells expressing the human La gene under control of the genuine promoter elements (d), or cells expressing the exon 1 La mRNA under CMV promoter control (c) were fixed 5 h p.i. and phase contrast images were taken.
Acceleration of appearance of viral glycoproteins in dependence on the level of human La protein expression
Cells were infected with the herpes simplex type 1 strain ANG. The cells were fixed at different times p.i. and stained with two commercially available MoAbs to herpes simplex virus glycoproteins. The MoAbs did not stain uninfected cells (data not shown). After infection, first a perinuclear staining appeared. The perinuclear staining was similar to the pattern of antibodies to Golgi. Later in infection, the whole cells were stained. The appearance of the respective pattern was dependent on the analysed cell line. All the images shown in Fig. 6 were obtained 6 h p.i. As shown in Fig. 6a, stained cells could hardly be detected in the case of untransfected mouse 3T3. In the case of the human exon 1′ La mRNA-expressing cell line, a higher number of cells displayed the perinuclear staining pattern (Fig. 6b). The amount of stained cells was larger in the case of the human exon 1 La mRNA-expressing cell line (Fig. 6c). At the same time all the human La gene-expressing cells already displayed an intense staining of the whole cell.
Fig. 6.
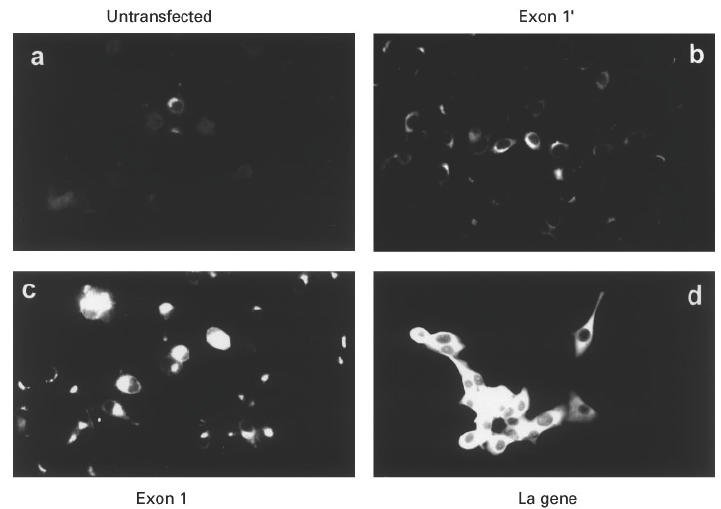
Epifluorescence analysis of cells stained with a MoAb to a surface protein of herpes simplex virus type 1. The cells were infected with herpes simplex virus strain ANG. Six hours p.i. the cells were stained with the MoAb HSV1-06 (see Materials and Methods). (a) Untransfected mouse 3T3 cells. (b) Cells transfected with the exon 1′ construct. (c) Cells transfected with the exon 1 construct. (d) Cells transfected with the human La gene.
Acceleration of appearance of viral DNA replication in dependence on the level of human La protein expression
In addition to the cell biology approaches, herpes viral DNA replication activity was directly measured in extracts of infected cells expressing different levels of human La protein. Besides the 3T3 cell line expressing the human La gene, a neomycin-resistant mouse 3T3 cell line and a further exon 1′ La mRNA-expressing cell line was used. Extracts of cells were harvested at different times p.i., including 2 h, 4 h, 6 h and 8 h p.i. Viral DNA polymerase activity was measured using a commercially available ELISA system. Cellular DNA polymerase activity was inhibited by the addition of 100 mm ammonium sulphate according to the supplier. As expected, the onset of viral DNA polymerase activity started between 4 h and 6 h p.i. (Fig. 7). Six hours and 8 h p.i. the increase of viral DNA polymerase activity was highest in the cell line expressing the human La gene, medium in the cell line expressing the exon 1′ La mRNA and lowest in the neomycin-resistant 3T3 cell line.
Fig. 7.
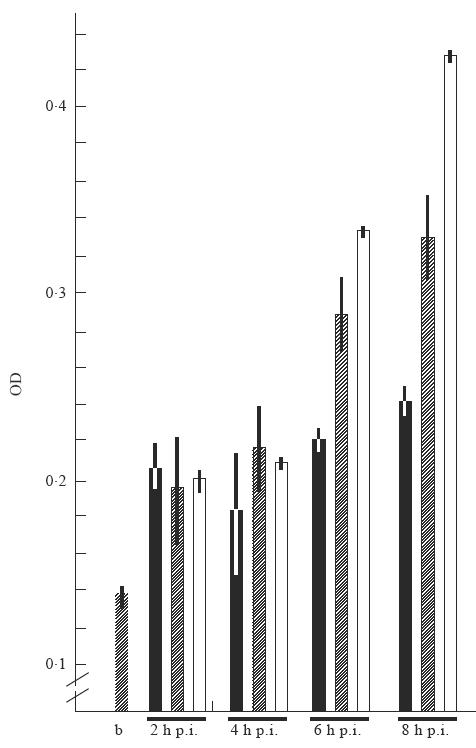
Estimation of herpes virus DNA polymerase activity. Viral DNA polymerase activity was measured in transfected cell lines 2 h, 4 h, 6 h and 8 h post-infection (p.i.). Viral DNA polymerase activity was estimated using a commercially available ELISA system (see Materials and Methods). b, hatched bar, activity of Klenow enzyme in the presence of ammonium sulphate; ▪, neomycin-resistant cell line; hatched bars, exon 1′ La mRNA-expressing cell line; □, cell line expressing the human La gene. OD, Optical density. Error bars of two assays are given.
Taken together, these results strongly indicate that the infection course was accelerated in the transfected mouse 3T3 cells in dependence on the level of expression of human La protein. Bearing in mind that (i) untransfected and neomycin-resistant cell lines displayed no difference, (ii) the transcription of the exon 1 and exon 1′ La mRNA-expressing cell lines was under control of the same CMV promoter while exon 1 and exon 1′ La mRNA expression in the genomically transfected cell line was under control of the genuine promoter elements, the observed effects might not be related to effects at the transcriptional level. As the acceleration coincides with the translocation to the cytoplasm, these data strongly support the idea that La protein does not only play a nuclear role in transcription/termination of RNA polymerase III, but may also have an additional cytoplasmic function, at least in virus-infected cells.
It is well established that herpes simplex virus is able to infect a wide variety of cell lines, including from different species, e.g. human cell lines, green African monkey kidney cells (Vero and CV1 cells), mouse 3T3 cells and Chinese hamster ovary (CHO) cells. Although herpes simplex virus can replicate in a series of different species, differences in replication are known. For example, herpes virus can infect CHO cells but does not replicate in them (e.g. [21]). The presented data point to a species-specific effect of human La protein. In this context it is of interest to mention that all La proteins isolated so far contain species-specific differences due to species-specific inserts [42]. However, future studies are required to examine if these species-specific differences are responsible for the observed increase of replication in mouse cells expressing the human La protein.
At present we also do not know the mechanism for the acceleration of herpes simplex virus type I replication and whether it is a more general effect. As mentioned above, La protein binds to a series of viral encoded dsRNAs both in vitro and in vivo. Moreover, La protein was able to interact with the double-strand RNA binding protein kinase (DAI, also known as PKR) in vitro and inhibited the function of DAI in vitro [43]. DAI is known to be one key enzyme in the interferon pathway activated by dsRNA and thereby protecting cells against virus infection. La protein has been shown to contain dsRNA binding motifs related to that of DAI. Moreover, it was able to unwind dsRNA [35,43,44]. Furthermore, it was able to alleviate the translational repression by the 5′-leader sequence of HIV type 1 mRNA [13]. Thus the observed translocation of La protein after virus infection may have two advantages for viruses: (i) the translation of unusual structured mRNAs by the mechanism of internal initiation; and (ii) bypassing the protecting mechanisms dependent on dsRNA.
Acknowledgments
This work was supported by grants given to M.B. by the Deutsche Forschungsgemeinschaft DFG (Ba1145/4–2) and the VW-Stiftung. M.B. was a recipient of a professorship granted by the ‘Hermann-and-Lilly Schilling Stiftung’ in the ‘Stifterverband für die Deutsche Wissenschaft’.
References
- 1.Tan EM. Antinuclear antibodies: diagnostic markers for autoimmune diseases and probes for cell biology. Adv Immunol. 1989;44:93–151. doi: 10.1016/s0065-2776(08)60641-0. [DOI] [PubMed] [Google Scholar]
- 2.Hendrick JP, Wolin S, Rinke J, Lerner M, Steitz JA. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: Further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol. 1981;12:1138–49. doi: 10.1128/mcb.1.12.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madore SJ, Wieben ED, Pederson T. Eukaryotic small ribonucleoproteins. Anti-La human autoantibodies react with U1 RNA–protein complexes. J Biol Chem. 1984;259:1929–33. [PubMed] [Google Scholar]
- 4.Rinke J, Steitz JA. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucl Acids Res. 1985;13:2617–29. doi: 10.1093/nar/13.7.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pruijn GJM, Slobbe RL, van Venroooij WJ. Structure and function of La and Ro RNPs. Mol Biol Rep. 1990;14:43–48. doi: 10.1007/BF00360410. [DOI] [PubMed] [Google Scholar]
- 6.Stefano JE. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–54. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 7.Gottlieb E, Steitz JA. Function of the mammalian La protein: evidence for its activity in transcription termination by RNA polymerase III. EMBO J. 1989;8:841–50. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francoer M, Mathews M. Interaction between VA RNA and the lupus antigen La: formation of a ribonucleoprotein particle in vitro. Proc Natl Acad Sci USA. 1982;79:6772–6. doi: 10.1073/pnas.79.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lerner MR, Andrews NC, Miller G, Steitz JA. Two small RNAs encoded by Epstein–Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1981;78:805–9. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilusz JM, Kurilla G, Keene JD. A host protein (La) binds to a unique species of minus-sense leader RNA during replication of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1983;80:5827–31. doi: 10.1073/pnas.80.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurilla MG, Cabradilla CD, Holloway BP, Keene JD. Nucleotide sequence and host La protein interactions of rabies virus leader RNA. J Virol. 1984;50:773–8. doi: 10.1128/jvi.50.3.773-778.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park YW, Katze MG. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J Biol Chem. 1995;270:28433–9. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 13.Svitkin YV, Pause A, Sonenberg NJ. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J Virol. 1994;68:7001–7. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pogue GP, Hofmann J, Duncan R, Best JM, Etherington J, Sontheimer RD, Nakhasi NL. Autoantigens interact with cis-acting elements of rubella virus RNA. J Virol. 1996;70:6269–77. doi: 10.1128/jvi.70.9.6269-6277.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meerovitch K, Pelletier J, Sonenberg NJ. A cellular protein that binds to the 5′-non-coding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989;3:1026–34. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- 16.Meerovitch K, Svitkin YV, Lee HS, et al. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulo lysate. J Virol. 1993;67:3798–807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali N, Siddiqui A. The La antigen binds 5′-noncoding region of hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–54. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBratney S, Sarnow P. Evidence for involvement of trans-acting factors in selection of the AUG start codon during eukaryotic translational initiation. Mol Cell Biol. 1996;16:3523–34. doi: 10.1128/mcb.16.7.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachmann M, Pfeifer K, Schröder HC, Müller WEG. The nucleocytoplasmic shuttling of the La antigen in CV-1 cells. Mol Biol Rep. 1987;12:239–40. [Google Scholar]
- 20.Bachmann M, Schröder HC, Falke D, Müller WEG. Alteration of the intracellular localization of the La protein compared with the localization of U snRNPs. Cell Biol Int Rep. 1988;12:765–89. doi: 10.1016/0309-1651(88)90088-4. [DOI] [PubMed] [Google Scholar]
- 21.Bachmann M, Falke D, Schröder HC, Müller WEG. Intracellular distribution of the La-antigen in CV-1 cells after herpes simplex type 1 infection compared with the localization of U snRNPs. J Gen Virol. 1989;70:881–9. doi: 10.1099/0022-1317-70-4-881. [DOI] [PubMed] [Google Scholar]
- 22.Bachmann M, Pfeifer K, Schröder HC, Müller WEG. The La antigen shuttles between the nucleus and the cytoplasm in CV-1 cells. Mol Cell Biochem. 1989;85:103–14. doi: 10.1007/BF00577106. [DOI] [PubMed] [Google Scholar]
- 23.Baboonian C, Venables PJW, Booth J, Williams DG, Roffe LM, Maini RN. Virus infection induces redistribution and membrane localization of the nuclear antigen La (SS-B): a possible mechanism for autoimmunity. Clin Exp Immunol. 1989;78:454–9. [PMC free article] [PubMed] [Google Scholar]
- 24.Tröster H, Metzger TE, Semsei I, Schwemmle M, Winterpacht A, Zabel B, Bachmann M. One gene, two transcripts: isolation of an alternative transcript encoding for the autoantigen La/SS-B from a cDNA library of a patient with primary Sjögren's syndrome. J Exp Med. 1994;180:2059–67. doi: 10.1084/jem.180.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bachmann M, Tröster H, Bartsch H, Grölz D. A frame shift mutation in a hot spot region of the nuclear autoantigen La (SS-B) J Autoimmun. 1996;9:747–56. doi: 10.1006/jaut.1996.0097. [DOI] [PubMed] [Google Scholar]
- 26.Hilker M, Tröster H, Grölz D, Hake U, Bachmann M. The autoantigen La/SS-B: analysis of the expression of alternatively spliced La mRNA isoforms. Cell Tis Res. 1996;284:383–9. doi: 10.1007/s004410050599. [DOI] [PubMed] [Google Scholar]
- 27.Bachmann M, Hilker M, Grölz D, Tellmann G, Hake U, Kater L, deWilde P, Tröster H. Different La/SS-B mRNA isoforms are expressed in salivary gland tissue of patients with primary Sjögren's syndrome. J Autoimmun. 1996;9:757–66. doi: 10.1006/jaut.1996.0098. [DOI] [PubMed] [Google Scholar]
- 28.Grölz D, Bachmann M. The nuclear autoantigen La/SS-B: one gene three functional La mRNAs. Biochem J. 1997;323:151–8. doi: 10.1042/bj3230151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grölz D, Laubinger J, Wilmer F, Tröster H, Bachmann M. The nuclear autoantigen La/SS-B: transfection analysis of expression of mRNA isoforms. J Biol Chem. 1997;272:12076–82. doi: 10.1074/jbc.272.18.12076. [DOI] [PubMed] [Google Scholar]
- 30.Grölz D, Bartsch H, Tröster H, Bachmann M. The nuclear autoantigen La/SS-B: mapping and sequencing of the gene and the three retropseudogenes. Gene. 1997;191:23–29. doi: 10.1016/s0378-1119(97)00023-1. [DOI] [PubMed] [Google Scholar]
- 31.Grölz D, Bachmann M. An altered distribution of the autoantigen La/SS-B when translated from a La mRNA isoform. Exp Cell Res. 1997;234:329–35. doi: 10.1006/excr.1997.3608. [DOI] [PubMed] [Google Scholar]
- 32.Grölz D, Tröster H, Semsei I, Bachmann M. Analysis of expression of the gene encoding for the nuclear autoantigen La/SS-B using reportergene constructs. Biochem Biophys Acta. doi: 10.1016/s0167-4781(97)00201-7. in press. [DOI] [PubMed] [Google Scholar]
- 33.Pruijn GJM, Thijssen JPH, Smith PR, Williams DG, van Venrooij WJ. Anti-La monoclonal antibodies recognizing epitopes within the RNA-binding domain of the La protein show differential capacities to immunoprecipitate RNA-associated La protein. Eur J Biochem. 1995;232:611–9. doi: 10.1111/j.1432-1033.1995.611zz.x. [DOI] [PubMed] [Google Scholar]
- 34.Smith PR, Williams DJ, Venables PJW, Maini RN. Monoclonal antibodies to the Sjögrens's syndrome associated antigen SS-B (La) J Immunol Methods. 1985;77:63–69. doi: 10.1016/0022-1759(85)90184-x. [DOI] [PubMed] [Google Scholar]
- 35.Bachmann M, Pfeifer K, Schröder HC, Müller WEG. Characterization of the autoantigen La as a nucleic acid dependent ATPase/dATPase with melting properties. Cell. 1990;60:85–93. doi: 10.1016/0092-8674(90)90718-t. [DOI] [PubMed] [Google Scholar]
- 36.Lingen M, Seck T, Dehoust U, Weise K, Falke D. Amino acid substitutions in glycoprotein D mediate the ability of fusion from without-positive herpes simplex virus type 1 strains to penetrate at 4 degrees and in the presence of soluble glycoprotein D. Intervirology. 1995;38:283–9. doi: 10.1159/000150452. [DOI] [PubMed] [Google Scholar]
- 37.Lingen M, Seck T, Weise K, Falke D. Two mutations in gB-1 and gD-1 of herpes simplex virus type 1 are involved in the fusion from without phenotype in different cell types. Virus Genes. 1996;3:221–8. doi: 10.1007/BF00366982. [DOI] [PubMed] [Google Scholar]
- 38.Seck T, Koch O, Lingen M, Falke D. Influence of glycoproteins B, C and D on the conversion of virus to cell attachment from heparin sensitivity to resistance. Acta Virologica. 1996;40:179–5. [PubMed] [Google Scholar]
- 39.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–8. [PubMed] [Google Scholar]
- 41.Müller WEG, Falke D, Zahn RK. DNA-dependent DNA polymerase pattern in noninfected and herpesvirus infected rabbit kidney cells. Virusforschung. 1973;42:278–84. doi: 10.1007/BF01265653. [DOI] [PubMed] [Google Scholar]
- 42.Semsei I, Tröster H, Schwemmle M, Bachmann M. Cloning of the autoantigen La/SS-B from a rat cDNA library: detection of species specific variations. Gene. 1993;126:265–8. doi: 10.1016/0378-1119(93)90378-g. [DOI] [PubMed] [Google Scholar]
- 43.Xiao Q, Sharp TV, Jeffrey IW, James MC, Pruijn GJM, van Venrooij WJ, Clemens MJ. The La antigen inhibits the activation of the interferon-inducible protein kinase PKR by sequestering and unwinding double-stranded RNA. Nucl Acids Res. 1994;22:2512–8. doi: 10.1093/nar/22.13.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hühn P, Pruijn GJM, van Venrooij WJ, Bachmann M. Characterization of the autoantigen La/SS-B as dsRNA unwinding enzyme associated with a RNA cleavage activity. Nucleic Acids Res. 1997;25:410–6. doi: 10.1093/nar/25.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]


