Abstract
A strong virus-specific CD4+ and CD8+ T lymphocyte response to hepatitis B virus (HBV) has been associated with viral clearance, but little is known about factors determining the individual's ability to mount such a T cell response. Recently a strong association between the HLA class II allele DR13 and a self-limited course of HBV infection has been described. In the present study of 33 patients with acute hepatitis B we show that individuals carrying HLA-DR13 mount a more vigorous CD4+ T cell response to HBV core (5706 ct/min (25th/75th percentile 3239 ct/min; 10 552 ct/min)) than patients without HLA-DR13 (1365 ct/min (490 ct/min; 5334 ct/min); P = 0.006). However, peptide epitopes aa 50–69, aa 61–85, and aa 81–105 were recognized most frequently by both patient groups. Moreover, among 14 HBV core-specific CD4+ T cell clones from two patients with HLA-DR13, only one T cell clone was HLA-DR13-restricted. Our data suggest that the beneficial effect of the HLA-DR13 alleles on the outcome of HBV infection could be explained by a more vigorous HBV core-specific CD4+ T cell response, which may either be due to more proficient antigen presentation by the HLA-DR13 molecules themselves or a linked polymorphism in a neighbouring immunoregulatory gene.
Keywords: CD4+ T lymphocytes, hepatitis B virus, immune response, viral infection
INTRODUCTION
Hepatitis B virus (HBV) infection is one of the most important chronic viral diseases in the world, yet the pathogenesis of chronic infection is still incompletely understood. Recently, an association of the HLA class II allele DRB1*1302 with a self-limited course of acute hepatitis B was reported in a large study from The Gambia [1] and was subsequently confirmed in a Caucasian population [2]. Whereas these observations provide strong evidence for a genetic variation in the ability to cope with HBV infection, it remains unknown whether this is due to the HLA-DR13 molecule itself or to a linkage disequilibrium of HLA-DR13 with another gene on the same chromosome. Since HLA class II molecules present antigens to CD4+ T cells and a strong HLA class II-restricted CD4+ T cell response to HBV core has been associated with viral clearance in acute hepatitis B [3,4], it was the aim of this study to elucidate the relation of HLA-DR13 to the HBc-specific CD4+ T cell response during viral elimination in acute hepatitis B.
PATIENTS AND METHODS
Patients
Thirty-two consecutive patients with chronic hepatitis B (defined by HBs and HBe antigen positivity and disease duration > 6 months) and 33 consecutive patients with acute hepatitis B (defined by new onset of liver disease, ALT > 1000 U/l, HBs, HBe antigen and anti-HBc-IgM positivity in first serum sample) as well as 208 healthy, unrelated controls were included in this study.
MHC typing
Typing for HLA-DRB1 alleles (low resolution) was performed using oligonucleotide hybridization with primers and oligonucleotides from the 11th and 12th International Histocompatibility Workshop [5,6] and a detection system as described elsewhere [7].
Recombinant protein and synthetic peptides
Recombinant HBV core protein (rHBc) obtained from bacterial extracts of Escherichia coli K 12 strain HB101 was purchased from Biogen (Geneva, Switzerland). Nine overlapping 25-mer peptides covering the entire HBV core sequence (aa 1–25, 20–45, 40–65, 60–85, 80–105, 101–125, 121–145, 141–165, 161–183) and an immunodominant peptide described previously (aa 50–69) [8] were synthesized by Multiple Peptide Systems (San Diego, CA). All peptides were purified to > 95% by high performance liquid chromatography (HPLC).
Peripheral blood mononuclear cell proliferation assay
Peripheral blood mononuclear cells (PBMC) were isolated on Ficoll–Isopaque gradients purchased from Pharmacia (Uppsala, Sweden) and washed four times in PBS. PBMC (5 × 104/well) were incubated in 96-well U-bottomed plates (Costar, Cambridge, MA) for 5 days in the presence of rHBc (1, 2 and 5 μg/ml) or peptides (10 μg/ml) in 150 μl RPMI 1640 medium (Gibco, Grand Island, NY) containing 2 mml-glutamine, 1 mm sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% human AB serum. The cell cultures were labelled by incubation for 16 h with 2 μCi 3H-thymidine (specific activity 80 mCi/mmol; Amersham, Aylesbury, UK). The cells were collected and washed on filters from Dunn (Asbach, Germany) using a cell harvester (Skatron, Sterling, VA), and the amount of radio label incorporated into DNA was estimated by a beta counter (LKB/Pharmacia). Triplicate cultures were assayed routinely and the results are expressed as mean ct/min. The stimulation index (SI) was calculated as the ratio between ct/min obtained in the presence of antigen and that obtained without antigen. An SI > 3 was considered significant. By separation experiments and FACS analysis it could be demonstrated that proliferation was confined to the CD4+ T cell subset [4,9].
Generation of T cell clones and specificity testing
Two million PBMC were stimulated with 3 μg/ml rHBc in 96-well U-bottomed plates as described above. On day 6 recombinant IL-2 was added to a final concentration of 15 U/ml (kindly provided by Boehringer, Mannheim, Germany). On day 10, cells were cloned by limiting dilution (0.5 cells/well) in the presence of 3 × 104 autologous, irradiated PBMC/well, 15 U/ml IL-2, and 2 μg/ml phytohaemagglutinin (PHA) from Murex Diagnostics (Dartford, UK). After 3–5 weeks growing clones were tested for specificity to rHBc and rHBc-specific clones were subsequently tested for peptide specificity. For this 1–5 × 104 clone cells were added to 3 × 104 autologous, irradiated PBMC with and without 2 μg/ml rHBc or 10 μg/ml peptide and cultured for 5 days. The proliferation assay was performed as described for PBMC. For expansion, T cell clones were stimulated every 3–5 weeks with irradiated autologous or allogeneic PBMC, 15 U/ml IL-2 and 2 μg/ml PHA.
FACS analyses
Triple immunofluorescence staining was performed on T cell clones with the following combinations of conjugated antibodies: CD3 (MT301–FITC, kindly provided by Dr E. P. Rieber, Institute for Immunology, Munich, Germany), CD4 (Leu-3a–PE; Becton Dickinson, Hamburg, Germany), CD8 (3B5-TRI-Colour; Medac, Hamburg, Germany) and CD25 (IL-2R1–FITC; Coulter, Hialeah, FL), HLA-DR (L243–PE; Becton Dickinson), and CD4 (S3,5-TRI-Colour; Medac). FACS analysis was performed with a FACScan from Becton Dickinson, as described previously [10].
HLA restriction
T cell clones were stimulated in the presence of irradiated autologous PBMC with rHBc and with 10 μl anti-DR (Cat. no. 7730), anti-DP (Cat. no. 7450) and anti-DQ (Cat. no. 7360) antibodies obtained from Becton Dickinson. Proliferation assays were performed as described above. For confirmation and identification of the exact HLA allele, the following well characterized B lymphoblastoid cell lines (kindly provided by Dr D. Schendel, Institute for Immunology, Munich, Germany) were used as antigen-presenting cells (APC) in proliferation assays: TEM (DRA*0101, DRB1*1401, DQA1*0101, DQB1*05031, DPA1*01, DPB1*0401), AMALA (DRA*0102, DRB1*1402, DQA1*0501, DQB1*0301, DPA1*01, DPB1*0402), CB6B (DRA*0101, DRB1*1301, DQA1*0103, DQB1*0603, DPA1*02021, DPB1*1901), SPO010 (DRA1*0101, DRB1*1101, DRB3*0202, DQA1*0102, DQB1*0502, DPA1*01, DPB1*02012). If only low numbers of clone cells were available HLA class II restriction was determined by FACS as previously described [11]. In that case HBc-specific CD4+ T cell clones (104 cells/well) were incubated with homozygous lymphoblastoid cell lines (3 × 104 cells/well) carrying one of the patients' HLA class II alleles for 20 h in the presence or absence of 2 μg/ml rHBc and stimulation was measured as an increase in CD25 expression in the CD4+ T cell population of rHBc-stimulated wells.
Statistical analysis
The χ2 test was used for the analysis of HLA-DR13 allele frequencies and for the response to individual HBV core peptide epitopes. Multiple comparisons were controlled by the method of Bonferroni. For the comparison of the strength of the HBc-specific CD4+ T cell response in different patient groups, non-parametric statistical analyses were performed using the Mann–Whitney U-test with a level of significance of 0.05. We first compared all assays performed in patients with HLA-DR13 with all assays performed in patients without HLA-DR13 with regard to total ct/min in the presence of rHBc minus control ct/min (Δct/min) and also with regard to SIs. Because in most assays an HBV core antigen concentration of 5 μg/ml yielded the strongest response, the results achieved with this concentration were used for all subsequent analyses. Since the number of proliferation assays differed in individual patients, we calculated the strength of the HBc-specific CD4+ T cell response of an individual patient as mean Δct/min or mean SI.
RESULTS
HLA-DR13 is infrequent in patients with chronic hepatitis B
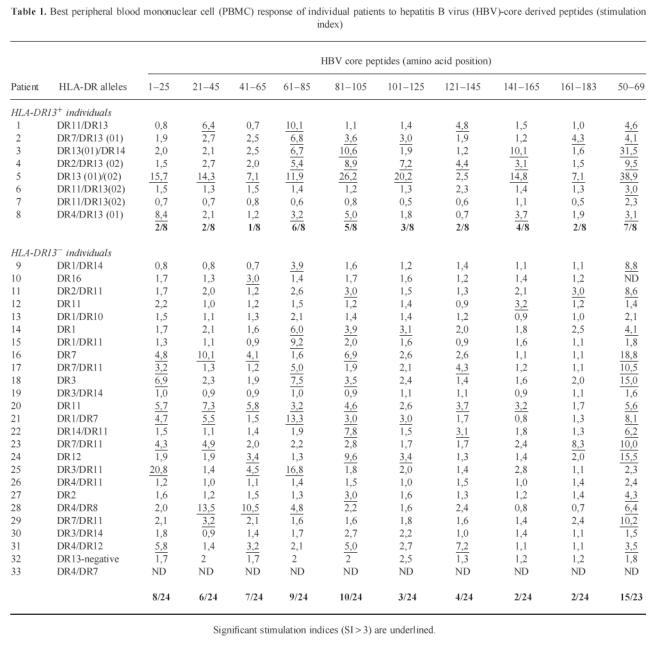
Thirty-two consecutive patients with chronic hepatitis B (cHB), 33 patients with acute, self-limited hepatitis B (aHB), and 208 healthy unrelated controls were typed for HLA class II by oligohybridization. Sixty of 208 healthy controls (28.8%) and 8/33 aHB patients (24.2%) carried a DR13 allele compared with only 1/32 cHB patients (χ2 test: healthy controls versus cHB P < 0.0019; aHB versus cHB P < 0.014). Moreover, the only cHB patient with HLA-DR13 was of Asian descent and was probably infected perinatally. Subtyping of the HLA-DR13 alleles was performed in seven patients; four patients were positive for HLA-DRB1*1302, one of those and three additional patients carried the HLA-DRB1*1301 allele (Table 1).
Table 1.
Best peripheral blood mononuclear cell (PBMC) response of individual patients to hepatitis B virus (HBV)-core derived peptides (stimulation index)
HLA-DR13 and HBV core-specific CD4+ T cell response
The proliferative response of PBMC to HBc was analysed in 33 patients with acute self-limited hepatitis B with regard to the presence or absence of HLA-DR13. All patients had been hospitalized for symptomatic aHB and were studied usually at weekly intervals until clinical recovery. No patient developed cHB. Ninety proliferation assays were performed, 23 in patients with HLA-DR13 (mean 2.9 assays per patient) and 67 in patients without HLA-DR13 (mean 2.7 assays per patient). For the quantitative analysis of the HBV core-specific CD4+ T cell response we used different approaches. First we analysed all individual proliferation assays with regard to total ct/min in the presence of HBV core minus control ct/min (Δct/min) and compared all observations in HLA-DR13+ patients with all observations in HLA-DR13− patients (Fig. 1a). The median proliferative response to HBV core in patients with HLA-DR13 was significantly higher (5706 ct/min (25th/75th percentile 3239 ct/min; 10 552 ct/min)) than in patients without HLA-DR13 (1365 ct/min (490 ct/min; 5334 ct/min); Mann–Whitney U-test; P = 0.006). The difference between the two groups was similar throughout the entire observation period (Fig. 2). No difference was found in the response to the non-specific T cell mitogen PHA that was determined simultaneously (78 896 ct/min (45 443 ct/min; 116 589 ct/min) versus 68 412 ct/min (32 439 ct/min; 121 997 ct/min); P = 0.51; Fig. 1b). If instead of Δct/min the SIs were compared between HLA-DR13+ and HLA-DR13− patients, the difference was also significant (median SI 8.3 versus 3.6; P = 0.019; Fig. 1c). Again, no difference was found in SIs for PHA (96.6 (52.5; 170.2) versus 102.6 (41.0; 223.8); P = 0.68; Fig. 1d). To avoid a bias due to the different number of assays that were performed in individual patients we calculated the mean Δct/min response and the mean SI per individual patient (Table 2), yielding a significant difference for the mean Δct/min (P = 0.017) but only a trend for the mean SI (P = 0.15). This was due to a significantly higher mean control ct/min value in patients with HLA-DR13 (1167 versus 696 ct/min, P = 0.036). Calculation of the average HBc-specific CD4+ T cell response for the other HLA-DR alleles (Table 3) revealed that indeed the strongest response was present for the HLA-DR13 group. However, there was a trend towards a weaker response in individuals carrying HLA-DR1 or HLA-DR3.
Fig. 1.
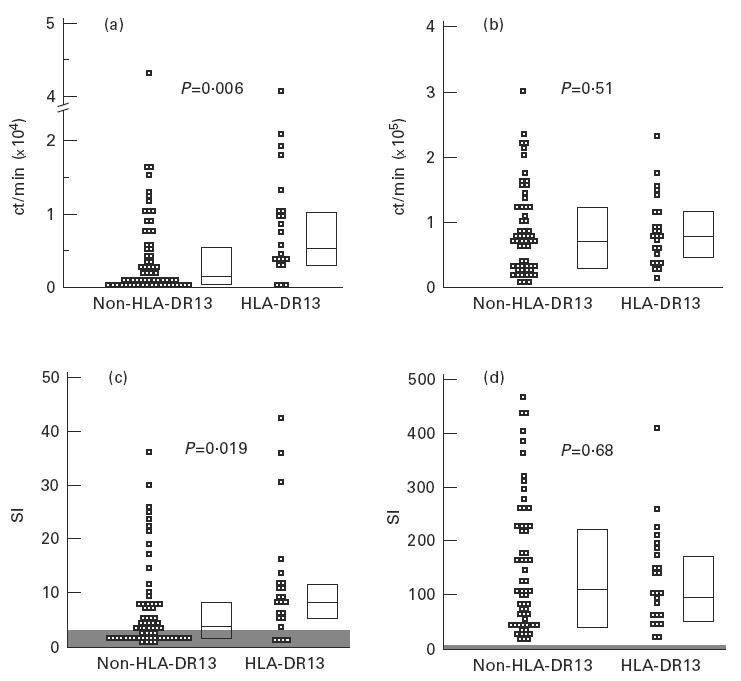
Patients with the HLA-DR13 allele show a significantly stronger peripheral blood mononuclear cell (PBMC) proliferation to recombinant hepatitis B virus (HBV) core protein (rHBc) than patients without HLA-DR13. All observations are shown as ct/min in the presence of rHBc minus control ct/min (Δct/min) (a) or as stimulation indices (SIs) (c). No difference is observed in the proliferative response to phytohaemagglutinin (b,d). Black bars represent the medians, the boxes enclose the 25th and 75th percentile. Shaded areas represent the level of significance for SIs (< 3). P values were calculated by Mann–Whitney U-test.
Fig. 2.
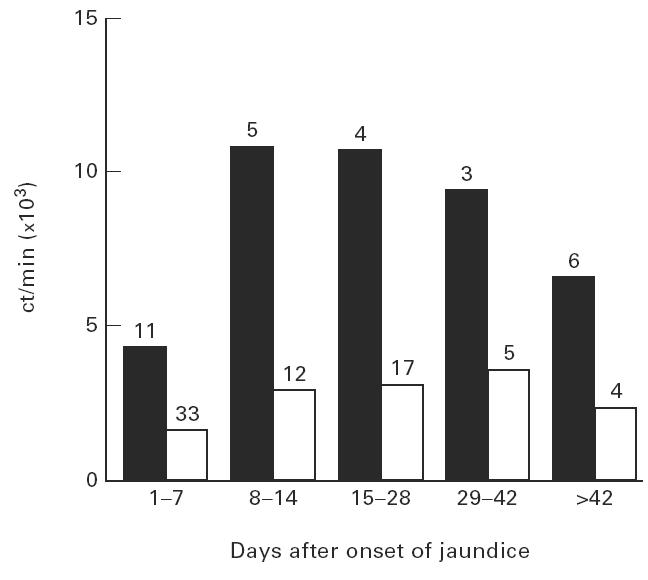
The difference in peripheral blood mononuclear cell (PBMC) response to recombinant hepatitis B virus (HBV) core protein (rHBc) in patients with HLA-DR13 compared with patients without HLA-DR13 is maintained throughout the observation period. The median Δct/min of patients with HLA-DR13 (▪) and of patients without HLA-DR13 (□) have been calculated for the first 2 weeks, weeks 3 and 4, weeks 5 and 6 and during follow up. The numbers on the bars represent the number of assays during that time period.
Table 2.
Analysis of hepatitis B virus (HBV) core-specific CD4+ T cell proliferative response in patients with or without HLA-DR13
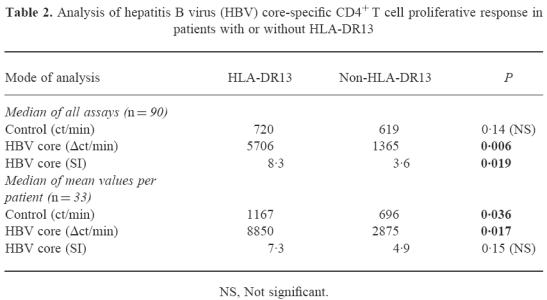
Table 3.
Comparison of mean Δct/min in response to hepatitis B virus (HBV) core according to the presence of individual HLA-DR alleles
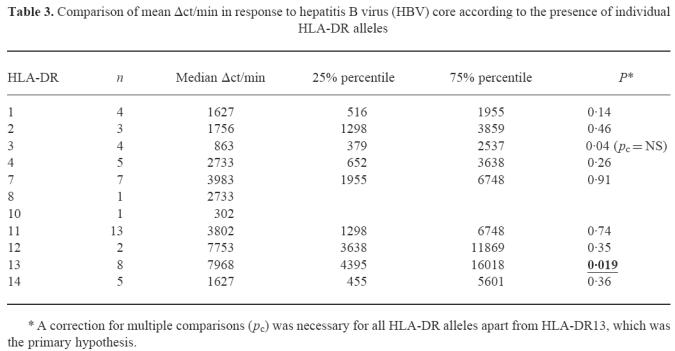
To define the role of individual CD4+ T helper cell epitopes we used overlapping 25-mer peptides and the previously described immunodominant epitope aa 50–69 [8] for stimulation of PBMC (Table 1). Patients carrying HLA-DR13 responded to an average of 2.38 ± 2.36 peptides per assay, whereas patients without HLA-DR13 recognized only 1.4 ± 1.77 peptides per assay (Mann–Whitney U-test, P = 0.057). Patients carrying HLA-DR13 most frequently responded in decreasing order of frequency to aa 50–69 (7/8), aa 61–85 (6/8), 81–105 (5/8), and 141–165 (4/8). While the first three epitopes were also most frequently recognized by patients without DR13 (by 16/23, 9/24, and 10/24, respectively), a response to aa 141–165 was found in only 2/24 HLA-DR13− patients (4/8 versus 2/24, χ2 test P = 0.009; Pcorr < 0.086). To clarify whether this trend was indeed due to the preferential presentation of aa 141–165 by HLA-DR13, we isolated three CD4+ T cell clones specific for that peptide from a patient carrying HLA-DR13 (DRB1*1301) and HLA-DR14 (DRB1*1401) (patient 3, Table 1), who responded strongly to peptide aa 141–165 (SI = 10.1). The minimal epitope for all three CD4+ T cell clones was determined by truncated peptides as aa 145–155 (data not shown) and all clones could be shown to be HLA-DR-restricted (Fig. 3). Using lymphoblastoid cell lines homozygous for the HLA class II locus as APC, the T cell clones could be stimulated by HLA-DR14 (DRB1*1401)-positive cell lines but not by HLA-DR13 (DRB1*1301)-positive cell lines (Fig. 4a–c). To further characterize the role of HLA-DR13 alleles in antigen presentation of HBV core epitopes we analysed the HLA restriction of another four clones from the same patient, three specific for aa 81–105 (minimal epitope for all three CD4+ T cell clones aa 93–103, data not shown) and one specific for aa 50–69 and seven HBV core-specific T cell clones from patient 2 who carried HLA-DR7 and HLA-DR13 (DRB1*1301). All aa 93–103-specific T cell clones were HLA-DR14-restricted (Fig. 4d–f) and the aa 50–69-specific T cell clone was stimulated neither by HLA-DR13 nor by HLA-DR14+ cell lines and is probably HLA-DQ- or HLA-DP-restricted (Fig. 4g). Six of seven HBV core-specific T cell clones from patient 2 were HLA-DR7-restricted and one was HLA-DR13 (DRB1*1301)-restricted and specific for aa 21–45 (data not shown).
Fig. 3.
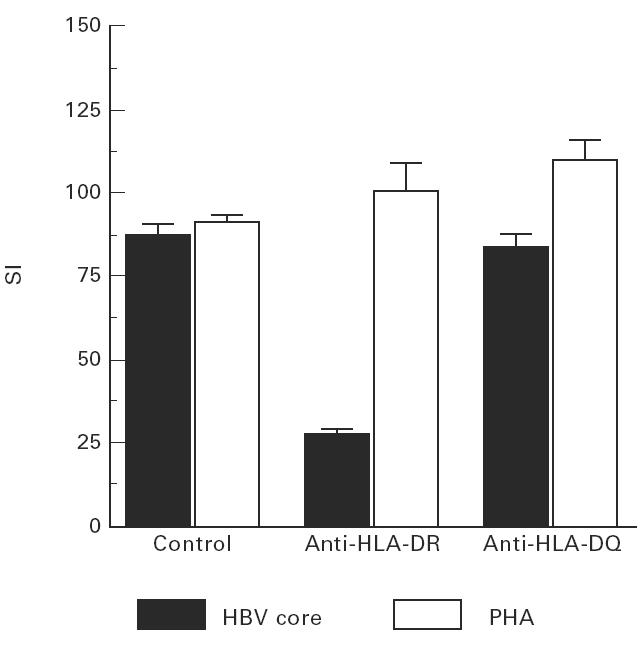
The activation of aa 141–165-specific CD4+ T cell clones can be inhibited by HLA-DR antibodies, whereas non-specific stimulation with phytohaemagglutinin (PHA) is not affected.
Fig. 4.
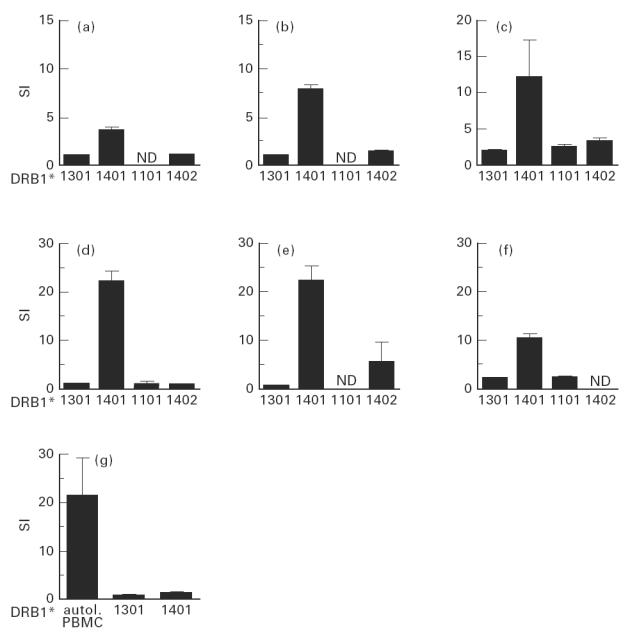
CD4+ T cell clones specific for aa 141–165 (a–c) or aa 93–103 (d–f) of hepatitis B virus (HBV) core respond to their specific peptide only if it is presented by HLA-DR14+ lymphoblastoid cell lines. A T cell clone specific for aa 50–69 (6g) only responds to its antigen presented by autologous peripheral blood mononuclear cells (PBMC), but not if presented by one of the patient's HLA-DR alleles.
DISCUSSION
Viral persistence in HBV infection is generally thought to be due to an inadequate antiviral T lymphocyte response. This concept is derived from animal models of viral infection and from the study of patients who spontaneously clear the virus and in whom a strong virus-specific CD4+ and CD8+ T lymphocyte response can be detected; in contrast, an HBV-specific T cell response is rarely found in chronic hepatitis B [4,8,12,13]. HBV-specific CD8+ T lymphocytes suppress viral replication in transgenic mice [14] and are therefore thought to be important antiviral effector cells. The relative contribution of HBV-specific CD4+ T lymphocytes is less well defined, although it can be expected that they are important regulators of the cellular and humoral antiviral immune defence. A recent study in a Gambian population has convincingly demonstrated an association between the HLA-DR allele DRB1*1302 and a self-limited course of acute hepatitis B [1]. The association seemed to hold true also for the other common HLA-DR13 allele, DRB1*1301, although the frequency of that allele was too small to achieve statistical significance. No association with HLA class I genes was detected. The low frequency of both DRB1*1301 and DRB1*1302 in patients with chronic hepatitis B was subsequently confirmed in a Caucasian population [2]. Basically, these studies demonstrate that there is a genetic dependence on the outcome of HBV infection, but at present it is unknown whether the HLA-DR13 molecule itself or a closely linked neighbouring gene mediates the protective effect.
In agreement with Thursz et al. [1] and Höhler et al. [2], we found a significantly lower frequency of the HLA-DR13 alleles in patients with chronic hepatitis B. In fact only one patient with chronic hepatitis B was positive for HLA-DR13 and this individual was of Asian descent and probably infected perinatally. It is important to mention that the frequency of HLA-DR13 in patients with acute hepatitis B was similar to healthy controls, indicating that the presence of HLA-DR13 does not seem to influence the susceptibility to clinical infection.
HLA class II molecules present viral peptides to CD4+ T lymphocytes, and in view of the correlation of a strong HBc-specific CD4+ T cell response with viral clearance during acute hepatitis B [3,4] it is tempting to speculate that the HLA-DR13 molecule could be more proficient for the presentation of HBV core epitopes. We quantified the strength of the HBc-specific CD4+ T cell response by serial proliferation assays during the symptomatic period of acute hepatitis B. Although the SI (i.e. ct/min in the presence of antigen ÷ control ct/min) is a well established parameter to detect antigen-specific proliferation, it is not known whether the SI is the best parameter to quantitatively compare different assays. For that reason all assays were analysed twice, with absolute ct/min minus controls (Δct/min) as well as with SI. When all observations were included into the analysis, both approaches yielded a significantly higher HBV core-specific CD4+ T cell response in HLA-DR13+ patients. Although all assays were performed before knowledge of the individuals' HLA-DR alleles, the fact that a different number of assays was performed in individual patients could have led to bias. A comparison of the mean Δct/min per patient confirmed the significantly stronger T cell response in HLA-DR13+ patients, whereas for the mean SI the difference fell short of statistical significance. This was due to a significantly higher mean control proliferation in patients with HLA-DR13 and may reflect either a higher level of in vivo activation of PBMC in those patients or a greater propensity for proliferation and/or 3H-thymidine uptake. The latter two hypotheses, however, are unlikely because proliferation, i.e. 3H-thymidine uptake in response to PHA, did not differ between the two groups.
In the second part of the study we tried to elucidate whether the HLA-DR13 molecule itself is more proficient in the presentation of HBV core-derived peptides to CD4+ T lymphocytes. Patients with HLA-DR13 on average tended to respond to more individual HBV core peptides at a given time point, supporting the hypothesis that HLA-DR13 molecules are able to present more HBV core-derived CD4+ T cell epitopes than other HLA-DR molecules. However, the three immunodominant peptides were the same in the two groups of patients. Moreover, for the only peptide that was preferentially recognized by HLA-DR13+ individuals, we could not demonstrate restriction to HLA-DR13 at a T cell clonal level. Similarly, among another 11 HBV core-specific CD4+ T cell clones from two different HLA-DR13+ patients only one clone was HLA-DR13-restricted and this clone was specific for the peptide epitope aa 21–45 which was recognized at a similar frequency by HLA-DR13+ and HLA-DR13− patients. Thus we failed to demonstrate clear evidence for a more efficient presentation of HBV core peptides to T cells by the HLA-DR13 molecules themselves. Alternatively, the data may suggest that a genetic polymorphism in an immunoregulatory gene that is in linkage disequilibrium with the HLA-DR13 alleles may facilitate the more vigorous HBV core-specific CD4+ T cell response by influencing T cell activation and/or cytokine production. This hypothesis is also supported by previous observations in other viral infections that HLA-DR13+ individuals are less susceptible to human papillomavirus-associated cervical cancer [15,16] and may run a more favourable clinical course in HIV infection [17,18] and hepatitis C virus infection [19]. The MHC class II and the neighbouring MHC class III region are characterized by a high density of genes, many of which have important immunoregulatory properties. Polymorphisms in the tumour necrosis factor-alpha promotor [1], the heat shock protein (hsp)70-1 promotor region [20] and the TAP2 gene [21], which is crucial for antigen presentation to cytotoxic T cells, are all in linkage disequilibrium with the HLA-DR13 alleles. Future studies have to investigate whether one of these polymorphisms or a yet unidentified immunoregulatory gene is possibly associated with a more successful antiviral immune response.
In conclusion, our data suggest that patients with HLA-DR13 can mount a more vigorous CD4+ T cell response to HBV core antigen during acute HBV infection and that progression to chronic hepatitis B is rare in that group. From our analysis of the HBV core-specific CD4+ T cell response this may be due to either a more proficient presentation of HBV core-derived peptides by the HLA-DR13 molecules themselves or a linked polymorphism in a neighbouring immunoregulatory gene. Further characterization of the HLA-DR13-restricted, HBV core-specific CD4+ T cell response and/or identification of a responsible genetic polymorphism could significantly contribute to our understanding of the pathogenesis of chronic HBV infection.
Acknowledgments
Financial support for this work was given by the Wilhelm-Sander-Stiftung (Grant No. 94.072.1) and the Deutsche Forschungsgemeinschaft (SFB 217). We thank Frau Jutta Döhrmann, Frau Carola Steiger, and Frau Carmen Amsel for excellent technical assistance.
REFERENCES
- 1.Thursz MR, Kwiatkowski D, Allsopp CEM, Greenwood BM, Thomas HC, Hill AVS. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995;332:1065–9. doi: 10.1056/NEJM199504203321604. [DOI] [PubMed] [Google Scholar]
- 2.Höhler T, Gerken G, Notghi A, et al. HLA DRB1*1301 and *1302 protect against chronic hepatitis B. J Hepatol. 1997;26:503–7. doi: 10.1016/s0168-8278(97)80414-x. [DOI] [PubMed] [Google Scholar]
- 3.Ferrari C, Penna A, Bertoletti A, et al. Cellular immune response to hepatitis B virus encoded antigens in acute and chronic hepatitis B virus infection. J Immunol. 1990;145:3442–9. [PubMed] [Google Scholar]
- 4.Jung M-C, Diepolder HM, Spengler U, et al. Activation of a heterogenous hepatitis B (HB) core and e antigen-specific CD4+ T-cell population during seroconversion to anti-HBe and anti-HBs in hepatitis B virus infection. J Virol. 1995;69:3358–68. doi: 10.1128/jvi.69.6.3358-3368.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura A, Sasazuki T. Eleventh International Hisocompatibility Workshop reference protocol for the HLA DNA typing technique. In: Tsuji K, Aizawa M, Sasazuki T, editors. HLA 1991. Oxford–Tokyo: Oxford University Press; 1992. pp. 397–419. [Google Scholar]
- 6.Bodmer JG, Marsh SGE, Albert ED, et al. Nomenclature for factors of the HLA system, 1996. Eur J Immunogenet. 1997;24:105–51. [PubMed] [Google Scholar]
- 7.Nevinny-Stickel C, Bettinotti MP, Andreas A, Hinzpeter M, Mühlegger K, Schmitz G, Albert ED. Nonradioactive HLA class II typing using polymerase chain reaction and digoxigenin-11-2′-3′-dideoxy-uridinetriphosphate labeled oligonucleotide probes. Hum Immunol. 1991;31:7–13. doi: 10.1016/0198-8859(91)90042-8. [DOI] [PubMed] [Google Scholar]
- 8.Ferrari C, Bertoletti A, Penna A, et al. Identification of immunodominant T cell epitopes of the hepatitis B virus nucleocapsid antigen. J Clin Invest. 1991;88:214–22. doi: 10.1172/JCI115280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diepolder HM, Jung MC, Wierenga E, et al. Anergic TH1 clones specific for hepatitis B virus (HBV) core peptides are inhibitory to other HBV core-specific CD4+ T cells in vitro. J Virol. 1996;70:7540–8. doi: 10.1128/jvi.70.11.7540-7548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruber R, Reiter C, Riethmüller G. Triple immunofluorescence flow cytometry, using whole blood, of CD4+ and CD8+ lymphocytes expressing CD45RO and CD45RA. J Immunol Methods. 1993;163:173–9. doi: 10.1016/0022-1759(93)90120-v. [DOI] [PubMed] [Google Scholar]
- 11.Diepolder HM, Gerlach JT, Zachoval R, et al. Immunodominant CD4+ T cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol. 1997;71:6011–9. doi: 10.1128/jvi.71.8.6011-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marinos G, Torre F, Chokshi S, et al. Induction of T-helper cell response to hepatitis B core antigen in chronic hepatitis B. A major factor in activation of the host immune response to the hepatitis B virus. Hepatology. 1995;22:1040–9. doi: 10.1016/0270-9139(95)90607-x. [DOI] [PubMed] [Google Scholar]
- 13.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 14.Chisari FV. Hepatitis B virus transgenic mice: insights into the virus and the disease. Hepatology. 1995;22:1316–25. doi: 10.1016/0270-9139(95)90645-2. [DOI] [PubMed] [Google Scholar]
- 15.Sastre Garau X, Loste MN, Vincent Salomon A, et al. Decreased frequency of HLA-DRB1(*)13 alleles in Frenchwomen with HPV-positive carcinoma of the cervix. Int J Cancer. 1996;69:159–64. doi: 10.1002/(SICI)1097-0215(19960621)69:3<159::AID-IJC1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 16.Apple RJ, Erlich HA, Klitz W, Manos MM, Becker TM, Wheeler CM. HLA DR-DQ associations with cervical carcinoma show papillomavirus-type specificity. Nat Genet. 1994;6:157–62. doi: 10.1038/ng0294-157. [DOI] [PubMed] [Google Scholar]
- 17.Itescu S, Rose S, Dwyer E, Winchester R. Certain HLA-DR5 and -DR6 major histocompatibility complex class II alleles are associated with a CD8 lymphocytic host response to human immunodeficiency virus type 1 characterized by low lymphocyte viral strain heterogeneity and slow disease progression. Proc Natl Acad Sci USA. 1994;91:11472–6. doi: 10.1073/pnas.91.24.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winchester R, Chen Y, Rose S, Selby J, Borkowsky W. Major histocompatibility complex class II DR alleles DRB1*1501 and those encoding HLA-DR13 are preferentially associated with a diminution in maternally transmitted human immunodeficiency virus 1 infection in different ethnic groups: determination by an automated sequence-based typing method. Proc Natl Acad Sci USA. 1995;92:12374–8. doi: 10.1073/pnas.92.26.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuzushita N, Hayashi N, Katayama K, et al. Increased frequency of HLA DR13 in hepatitis C virus carriers with persistently normal ALT levels. J Med Virol. 1996;48:1–7. doi: 10.1002/(SICI)1096-9071(199601)48:1<1::AID-JMV1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 20.Cascino I, D'Alfonso S, Cappello N, Giordano M, Pugliese A, Awdeh Z, Alper CA, Richardi PM. Gametic association of HSP70-1 promoter region alleles and their inclusion in extended HLA haplotypes. Tissue Antigens. 1993;42:62–66. doi: 10.1111/j.1399-0039.1993.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 21.Ronningen KS, Undlien DE, Ploski R, et al. Linkage disequilibrium between TAP2 variants and HLA class II alleles; no primary association between TAP2 variants and insulin-dependent diabetes mellitus. Eur J Immunol. 1993;23:1050–6. doi: 10.1002/eji.1830230511. [DOI] [PubMed] [Google Scholar]


