Abstract
Natural infection with simian immunodeficiency virus (SIV) is known to occur in the African green monkey (AGM). The actual onset of the disease has not been recognized in SIVagm infected AGM, and the precise reason for such apathogenicity in the AGM remains unclear. We reported previously that AGM peripheral CD4 lymphocytes underwent a peculiar differentiation from CD4+ to CD4− cells after in vitro activation, and we inferred that the AGM does not fall into a fatal immunodeficient state because of the generation of CD4− helper T cells in vivo. To evaluate this possibility, we examined the relationship between CD4 expression and helper T cell activity in the naturally infected AGM. We identified a healthy monkey almost lacking CD4 T cells in the periphery. This AGM showed no signs and symptoms of immunodeficiency and retained a helper T cell activity in antibody production comparable to those of CD4+ AGMs. In addition, SIVagm could be isolated from CD8+ lymphocytes in the CD4− AGM. These observations suggest that a unique host-virus adaptation has developed in the AGM, and may be helpful in explaining the fundamental reason for the apathogenicity occurring in this monkey.
Keywords: AIDS, apathogenicity, virus infection of CD8 cells
INTRODUCTION
Simian immunodeficiency viruses (SIV) can be classified into five genetically distinct groups: SIVagm (African green monkeys, AGMs) [1], SIVmnd (mandrills) [2,3], SIVsyk (Sykes' monkeys) [4], SIVsm (sooty mangabeys) [5,6] and SIVcpz (chimpanzees) [7,8]. These naturally infected primates remain healthy without the development of AIDS-like diseases. However, onset of the disease has been demonstrated in macaque monkeys experimentally inoculated with SIVsm or SIVagm, and these monkeys die of simian AIDS [9]. The above facts suggest that the issue of whether SIV is pathogenic or apathogenic is considerably influenced by the characteristics of the host-side factors.
AGMs have been classified into four subspecies: grivet monkeys (Cercopithecus aethiops), vervet monkeys (C. pygerythrus), tantalus monkeys (C. tantalus) and sabaeus monkeys (C. sabaeus), based on their phenotypic differences and geographical distributions [10]. All of the four subspecies are frequently infected with SIV in the wild [11–16], but none of these monkeys have been found to exhibit clinical signs of immunodeficiency. Immunological and virological studies performed to explain the apathogenicity observed in the AGM have revealed that neither humoral nor cellular immune responses were augmented in SIV-infected AGM [17], and although the mutation rate of SIVagm in vivo was comparable to that of HIV-1, the viral load in the peripheral blood remained low as noted in asymptomatic HIV-1 infected patients [18]. Furthermore, unlike in HIV-1 infected patients, the lymph nodes do not serve as a viral reservoir because the viral load in the lymph nodes of long-term infected AGMs was found to be similar to that in the peripheral blood mononuclear cells (PBMC) [19]. SIVagm can thus replicate in the AGM without activating immune responses of the host, but the level of infection remains relatively low throughout the lifetime of the AGM. In addition, neonatal AGMs have a higher percentage of circulating CD4 lymphocytes than adults; however, no differences in the in vitro replication kinetics of SIVagm in PBMC of adult or neonatal AGMs could be observed and none of the animals developed AIDS-like symptoms upon infection in vivo [20]. These observations imply that there must be a specific mechanism which permits the peaceful coexistence of SIVagm and its natural host. To elucidate the above mechanism, it is important to investigate the variations in helper T cell activity in SIV-infected AGMs and to compare the data with those for the HIV-human system.
One essential difference between the human and AGM immune systems is the mode of regulation of the CD4 and CD8α gene expressions in helper T cells. CD4+ lymphocytes also develop in the AGM thymus; however, mature peripheral CD4 cells coexpress the CD8α molecule and undergo a unique differentiation after lymphocyte activation, which results in a phenotypic conversion from CD4+ to CD4− cells [21]. Such CD4− CD8 helper T cells are resistant to SIV infection and the AGM may thus be able to survive with SIVagm due to host-virus adaptation, which has never been identified in the HIV-human system.
To evaluate the above possibility, we examined the relationship between CD4 expression and helper T cell activity in the naturally infected AGM. We identified an individual almost lacking CD4 T cells in the periphery, and this AGM retained a helper T cell activity in antibody production comparable to those of CD4+ AGMs. In addition, SIVagm could be isolated from CD8+ lymphocytes of this CD4− AGM. The findings obtained may be helpful in explaining the fundamental reason for the apathogenicity occurring in the AGM.
MATERIALS AND METHODS
Flow cytometry
Blood samples were collected from full-matured AGMs (vervet monkeys, C. pygerythrus) kept in individual cages at the Tsukuba Primate Center for Medical Science. The animals were wild-caught specimens, and 14 SIVagm seronegative monkeys and 14 seropositive monkeys were examined. PBMC were separated by the Ficoll (Pharmacia, Uppsala, Sweden) centrifugation method. The cells were stained with CD4 [phycoerythrin (PE)-Leu3a] and CD8α [fluorescein-isothiocyanate (FITC)-Leu2a] monoclonal antibodies (both from Becton-Dickinson, CA, USA) at 4°C for 20 min. After washing, the samples were fixed and analysed using a FACScan (Becton-Dickinson). Monkey CD3 (FITC-FN18, BIOsource International, CA, USA) and CD20 (PE-Leu16, Becton-Dickinson) antibodies were also employed.
To detect viral antigen, an anti-HIV gag monoclonal antibody (VAK4) [22] was employed. The cultured PBMC were fixed with 90% methanol overnight at − 20°C. After removal of the fixative, diluted VAK4 antibody was added to the cell pellets and the samples were incubated at 37°C for 2 h. Negative control cells were incubated with mouse immunoglobulin (Ig) G. The samples were washed, and then stained with FITC conjugated goat antimouse immunoglobulins antibody at 37°C for 2 h. After further washing, the intracellular expression of gag protein was analysed.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
The expression of CD4 mRNA in PBMC was investigated by the RT-PCR in an SIV seronegative monkey (Agm−) and three seropositive monkeys (Agm1–3). The total RNA was isolated from the PBMC using an RNeasy kit (QIAGEN, Chatsworth, CA, USA). Single stranded cDNA was synthesized with reverse transcriptase from 500 ng RNA. cDNA was then combined with sense and antisense primers in PCR buffer and the resultant solutions were subjected to 30 cycles of incubation, with each cycle consisting of denaturation for 1 min at 94°C, annealing for 2 min at 67°C (for β-actin) or at 55°C (for CD4), and extension for 2 min at 72°C. β-Actin mRNA was also amplified in each sample as an internal control. All samples were then subjected to 2% agarose gel electrophoresis and subsequent ethidium bromide staining. The primers employed in this study were: CD4-F, 5′-GTGGCACCTGG ACATGCAC-3′; CD4-R, 5′-GGTCAAAGGTGATCCAAGAC-3′; β-actin-F, 5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′; β-actin-R, 5′-CTAGAAGCATTGCGGTGGACGATGGAGGG-3′. The specificity and availability of above CD4 primers to detect AGM CD4 mRNA have been confirmed in our previous study [21].
Assay of helper activity
5×105 PBMC from Agm− and Agm1–3 were cultured in a 96-well U-bottom plate in a 200 μl volume of medium [RPMI 1640 (Gibco BRL, NY, USA)–10% foetal bovine serum] with or without 2μl of pokeweed mitogen (PWM, Gibco BRL) for 1 week at 37°C in a humidified 5% CO2 atmosphere. CD4 antibodies have been shown to inhibit in vitro several T lymphocyte functions, including helper activity for immunoglobulin production [23]. To confirm CD4-independent helper activity in Agm3, PBMC from Agm1–3 were also cultured with PWM in the presence of Nu-Th/i (Nichirei, Tokyo, Japan) antibody at final concentration of 1μg/ml. The antibody was previously freed of sodium azide by dialysis against phosphate-buffered saline. The IgG released into the culture supernatant was estimated by an indirect enzyme-linked immunosorbent assay. The values represent the mean of three wells.
Proliferation assay to SIVagm antigens
106 PBMC from Agm3 were mixed together with 105 SIV-infected or uninfected CemX174 cells and cultured in a 96-well U-bottom plate in a 200μl volume of medium for 1 week at 37°C in a humidified 5% CO2 atmosphere. SIVagm-infected and uninfected CemX174 cells were pretreated with mitomycin C (25μg/ml, Kyowa-Hakko, Tokyo, Japan) at 37°C for 30 min and washed four times with medium before use. After the cultivation, 20μl of AlamarBlue solution (BIOsource International, Camarillo, CA, USA) was added to each well and further incubated for 3 h at 37°C. The absorbancy at 570 nm and 600 nm in each well was then determined. All experiments were carried out in triplicate and resultant data was expressed as mean percentage for PBMC cultured with SIVagm-infected CemX174 over control PBMC incubated with SIV-uninfected CemX174 cells.
Cell sorting and culture
PBMC from Agm2 and Agm3 were stained with PE-CD4, FITC-CD8 and PE-Cyanine5 (Cy5)-CD16 (3G8, Pharmingen, CA, USA) antibodies, and peripheral CD4+CD8lowCD16− (Agm2) and CD4−CD8lowCD16− (Agm3) cells were then sorted out aseptically with a FACSort cell sorter (Becton-Dickinson). The purity was > 99.5% for both fractions. The sorted cells were precultured in medium containing 5μg/ml of concanavalin A (ConA, Pharmacia) and 100 U/ml of human recombinant interleukin-2 (IL-2, Shionogi, Osaka, Japan) for 1 week at 37°C in a humidified 5% CO2 atmosphere. They were subsequently cocultured with human CD4+ cell lines, CemX174 or Molt4 clone8 cells, to isolate SIV. In addition, CD4+ and CD4−CD8low cells were collected from Agm3 PBMC for PCR assay to assess the influence of contamination of CD4+ cells. DNA was extracted from sorted 102–104 cells without cultivation and employed for nested-PCR.
PBMC derived from Agm− and Agm1 were cultured in medium containing ConA and IL-2 to examine the possibility that CD4 lymphocytes are eliminated by infection and replication of SIVagm. Half of the culture medium was replaced with fresh medium every 3–4 days. The CD3, CD4 and CD8 expressions of the cultured cells were analysed at 30 days of cultivation.
Nested PCR
The SIVagm env fragment containing the V3–V5 regions was amplified by the nested PCR. 500 ng DNA extracted from cultured cells was exposed to 100μl of PCR buffer containing 1 μm of sense (env-A: 5′-GAAGCTTGTGATAAAACATATTGGGAT-3′) and antisense (env-B: 5′-AGAGCTGTGACGCGGGCATTGAGG-3′) primers, 2.5 U of ExTaq polymerase (Takara, Kyoto, Japan) and 200μm of each dNTP. The resultant solution was subjected to 30 cycles of incubation, with each cycle consisting of denaturation for 1 min at 94°C, annealing for 2 min at 66°C and extension for 2 min at 72°C. The reaction mixtures were purified with a PCR purification kit (QIAGEN), and diluted solutions (1:10 in water) were then combined with inner sense (env-C: 5′-TGTAGGAGACCAGGAA ACA-3′) and antisense (env-D: 5′-GTGGGTGCAAAGCCAAT TGG-3′) primers in PCR reaction mixture. The second step of the PCR was performed by 30 amplification cycles consisting of denaturation for 1 min at 94°C, annealing for 2 min at 62°C and extension for 2 min at 72°C. The samples were then subjected to 2% agarose gel electrophoresis. DNA was transferred to a Biodyne plus membrane (Pall BioSupport Division, NY, USA) and hybridized with digoxigenin (DIG)-dUTP (Boehringer Mannheim, Germany) labelled probe containing the env region of SIVagm TYO-1. The chemiluminescent signal was detected with a DIG luminescent detection kit (Boehringer Mannheim).
DNA sequence and phylogenetic analysis
In Agm2 and Agm3, the PCR derived SIVagm env fragments were cloned into pCR TRAP cloning vector (GenHunter, MA, USA) and then sequenced by the dye termination method employing a Perkin Elmer 377 sequencer (Perkin Elmer Biosystems, CA). The nucleotide sequences were determined in both strands at least twice. The sequences obtained were aligned using CLUSTALW [24]. The phylogenetic relationships among the SIVagm derived from velvet monkeys were estimated by the maximum likelihood method employing phylip (Phylogeny Inference Package version 3.5c) software, available on the Internet (URL http://evolution.genetics.washington.edu/phylip.html).
RESULTS
CD4 expression and helper activity of AGM PBMC
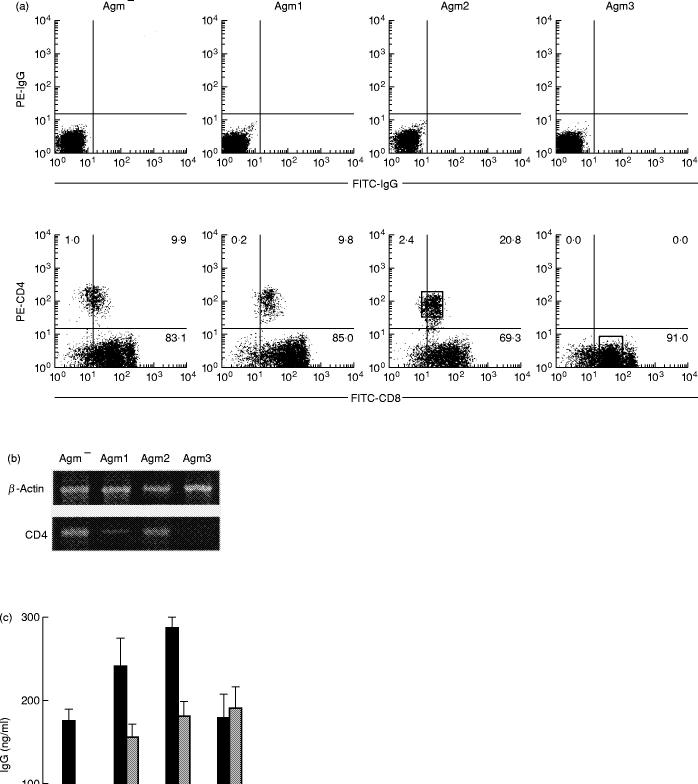
Figure 1(a) shows a two-colour flow cytometric profile of peripheral blood lymphocytes (PBL) stained with CD4 and CD8 antibodies. CD8 cells formed the major subset in the AGM and CD4 cells shared a low density of the CD8 antigen as reported previously [21]. The positive percentages of peripheral CD4 lymphocytes in the SIVagm seronegative and seropositive monkeys were 18.4 ± 9.0% (mean ± s.d., n = 14) and 12.1 ± 7.6%, respectively, and no significant difference observed (t-test). Interestingly, the PBL were virtually CD4− in Agm3, although this monkey remains healthy and no signs or symptoms of AIDS-like disease are observed. The lack of CD4 cells in Agm3 was confirmed by employing other monoclonal antibodies which recognize different epitopes from Leu3a, such as Nu-Th/i and our original simian CD4 antibody (U7b) [21] (data not shown). Figure 1(b) shows the results of the RT-PCR amplification of CD4 mRNA derived from freshly isolated PBMC. A PCR product with the predicted size (231 bp) could be identified in Agm−, Agm1 and Agm2 by the use of CD4 gene specific primers. On the other hand, CD4 gene expression could not be detected in the Agm3 PBMC.
Fig. 1.
(a) CD4 and CD8 expressions in simian immunodeficiency virus (SIV) seronegative (Agm−) and seropositive (Agm1–3) monkey peripheral blood lymphocytes (PBL). Two-colour flow cytometric analysis was performed with forward-and right-angle scatter gates set on the lymphocyte fraction. 104 cells were counted. Negative control cells were stained with control phycoerythrin- (PE) and fluorescein-isothiocyanate- (FITC) immunoglobulin (Ig) G antibodies. The sorting gates were set on the rectangles as indicated. (b) CD4 gene expression in peripheral blood mononuclear cells (PBMC). The total RNA was isolated from PBMC and the expressions of mRNA were analysed by the reverse transcriptase-polymerase chain reaction. (c) pokeweed mitogen (PWM)-induced antibody production in the African green monkey (AGM). PBMC were cultured in medium with (closed bars) or without (open bars) PWM for 1 week. In Agm1–3, the inhibitory effect for PWM-induced antibody synthesis by CD4 antibody was also examined (grey bars). The IgG released into the culture supernatant was estimated by an indirect enzyme-linked immunosorbent assay. (d) Proliferative response of Agm3 PBMC against SIV-uninfected (open bar) and SIV-infected (closed bar) CemX174 cells. The cell proliferation was estimated by AlamarBlue assay and the response of control PBMC incubated with SIV-negative CemX174 cells was represented as 100%.
We next determined the helper activities in PWM induced IgG production in these monkeys (Fig. 1c). CD20+ B cells amounted to 6–8% in these specimens. Antibody production was clearly observed in Agm1 and Agm2 as well as the seronegative Agm− and the anti-CD4 antibody inhibited the immunoglobulin production induced by PWM. The PBMC of Agm3 also displayed significant helper activity despite the lack of detectable CD4+ cells and the anti-CD4 antibody was not able to reduce immunoglobulin production.
The next experiment was performed to determine whether or not the specific immune response against SIV antigens was maintained in Agm3 PBMC (Fig. 1d). The proliferative response of Agm3 PBMC to SIV-infected CemX174 cells was significantly higher than the background response to SIV-uninfected CemX174 cells (P < 0.05, t-test).
SIV infection in peripheral T lymphocytes
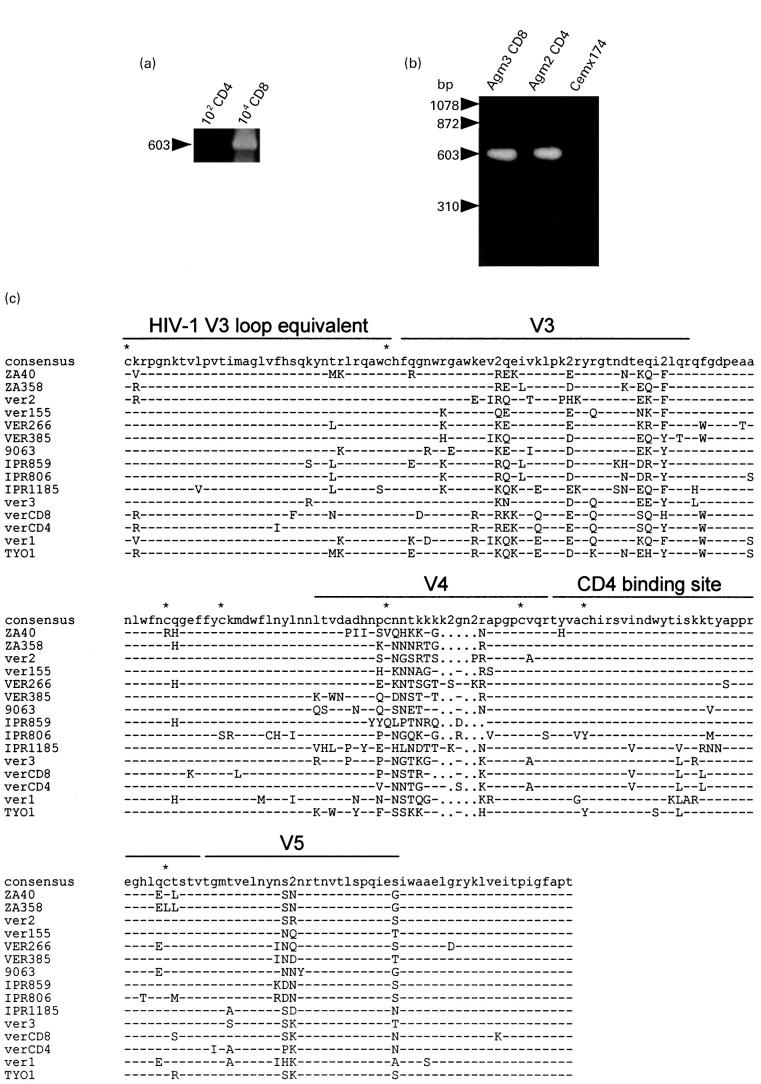
To examine the distribution of SIVagm provirus in the peripheral lymphocytes of Agm3, CD4 and CD8 cells were isolated and proviral DNA was analysed by nested PCR. As illustrated in Fig. 2(a), PCR product with the predicted size could be identified in 104 CD8 cells whereas no PCR product was detected in 102 CD4 cells. Since the purity of CD8 cell fraction was > 99.5%, it is unlikely that the contamination of CD4 cells would have yield a PCR positive reaction, even if the contaminated cells were all CD4 lymphocytes. To isolate SIVagm, purified CD8 T lymphocytes from Agm3 were cocultured with CD4 cell lines, such as CemX174 and Molt4 clone8. CD4 T lymphocytes from Agm2 were also used for virus isolation. After cultivation for 1 month, a cytopathic effect (CPE) became obvious in the cultured cells. By Southern blotting analysis, the env gene fragment of SIVagm could be detected in the CemX174 cells cocultured with CD4 lymphocytes (Fig. 2b). In addition, the env gene fragment was recognized in the CemX174 cells cocultured with CD8 T lymphocytes. Similar results were obtained for Molt4 clone8 cells (data not shown). These findings indicated that SIVagm was distributed in the peripheral CD8 T lymphocytes of Agm3. SIVagm in such CD8 lymphocytes maintained an infectious ability to CD4 cell lines.
Fig. 2.
(a) Detection of proviral DNA in CD4 and CD8 subsets in Agm3. DNA was extracted from sorted 102–104 cells and employed for nested-polymerase chain reaction (PCR). (b) Detection of SIVagm provirus by the PCR and Southern blotting in Cemx174 cells cocultured with peripheral CD4 or CD8 cells from African green monkeys. A negative reaction was confirmed in the DNA sample of CemX174 cells cultured alone. (c) Multiple alignment of deduced amino acid sequences of SIV isolated from vervet monkeys. PCR derived env nucleotide sequences from Agm2 CD4 cells (verCD4) and Agm3 CD8 cells (verCD8) were translated and aligned with previously reported SIV env sequences. Dashes indicate sequence identity with the consensus sequence, while dots represent gaps introduced to optimize the alignment. ‘2’ in the consensus sequence denotes sites in which two amino acids were shared in viruses at the same frequency. Asterisks indicate cysteine residues. V3, V4 and V5 designate variable regions. The putative CD4 binding site and HIV-1 V3 loop equivalent were as described previously [16,26]. The sequences of the references were obtained from the DNA data bank of Japan (DDBJ): ZA40 (AF015906), ZA358 (AF015905), IPR806 (AF015907), IPR859 (AF015908) and IPR1185 (AF015809) [30]; ver1 (U04003) and ver2 (U04004) [31]; ver155 (M29975) [32]; VER266 (U10896) and VER385 (U10898) [33]; 9063 (L40990) [34]; ver3 (M30931) [35]; and TYO1 (X07805) [1]. 9063 was isolated from a pig-tailed macaque inoculated with SIVagm90.
Sequence analysis of proviral DNA
PCR products of proviral DNA from CemX174 cells cocultured with Agm2 CD4 cells and Agm3 CD8 cells were cloned and sequenced. The deduced amino acid sequences of the env region from the new isolates were compared with the corresponding sequences of previously reported SIVagm from vervet monkeys [25–30] (Fig. 2c).
In the external glycoprotein of SIVagm, five conserved and five variable regions could be distinguished. The putative CD4 binding site is located in the fourth-conserved region. The deduced amino acid sequences of verCD4 derived from CD4 lymphocytes and verCD8 from CD8 lymphocytes displayed strong similarities with those of other SIVagm obtained from vervet monkeys. In verCD4 and verCD8, eight cysteine residues between the HIV-1 V3 equivalent and V5 regions were conserved. The V4 region of verCD8 was shorter than that of verCD4 by two amino acids. Only one amino acid substitution was recognized in the putative CD4 binding site between verCD4 and verCD8. Phylogenetic analysis indicated that verCD4 was most closely related to verCD8 (Fig. 3).
Fig. 3.
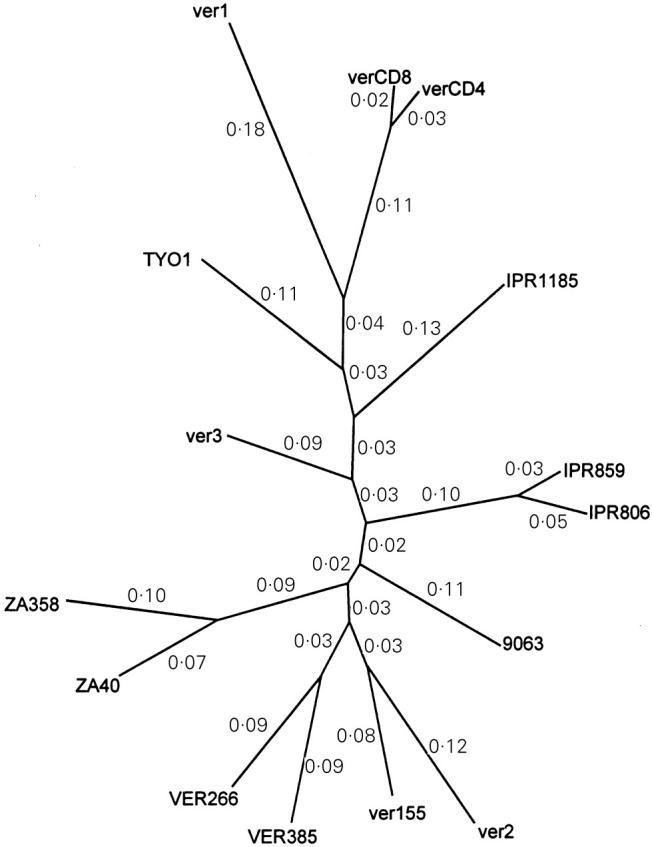
Unrooted phylogenetic tree of SIVagm isolated from vervet monkeys. Phylogenetic relationships were estimated by the maximum likelihood method. The values in the figure indicate the rate of divergence.
CD4 expression and SIV infection in cultured cells
In Agm3, CD4− CD8 lymphocytes were infected with SIVagm. This finding demonstrated that SIVagm could also be harbored in CD4−, CD8+ lymphocytes as reported previously in other SIVs [31,32] and HIV [33–35]. However, the reason for the loss of CD4 lymphocytes observed in Agm3 remains unclear. CD4 lymphocytes could be eliminated by infection and replication of SIVagm. To examine this possibility, PBMC from Agm1 and Agm− were cultured with ConA and IL-2, and the CD4 expressions were compared in the proliferating cells. Figure 4(a) shows the CD3, CD4 and CD8 expressions in the cultured cells after 30 days of cultivation. Most proliferating cells shared CD3 and CD8 antigens but lacked detectable CD4 molecules in Agm1. The env gene fragment of provirus in cultured cells from Agm1 was confirmed by PCR and Southern blotting (Fig. 4b) and the expression of virus protein (gag) was detected in approximately 53% of cells at 30 days of cultivation (Fig. 4c). However, since almost complete loss of CD4+ cells was also observed in cultured cells from the SIVagm negative monkey, the lack of CD4+ cells in Agm1 was considered not to be due to SIVagm infection.
Fig. 4.
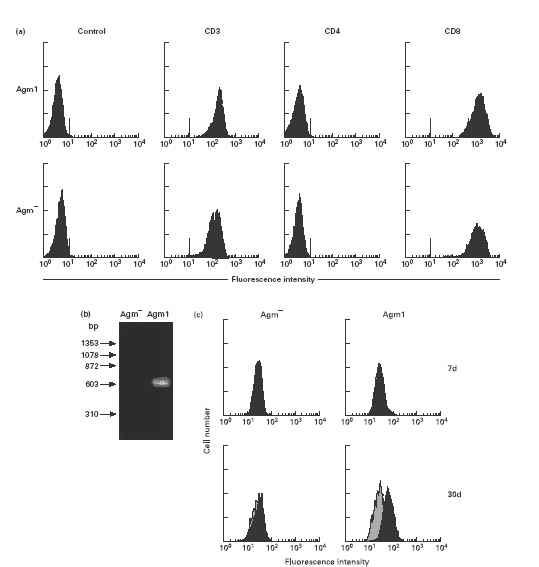
(a) CD3, CD4 and CD8 expressions in cultured lymphocytes. Peripheral blood mononuclear cells from Agm1 and Agm− were cultured with interleukin-2 and ConA for 30 days and examined by flow cytometry. Negative control cells were stained with control antibodies. (b) Detection of SIVagm provirus by polymerase chain reaction and Southern blotting in cultured lymphocytes. (c) Intracellular expression of gag protein in cultured lymphocytes stained with the anti-HIV gag monoclonal antibody, VAK-4. Negative control cells were incubated with mouse immunoglobulin G. The data for the control (grey histograms) and the sample stained with VAK-4 antibody (black histograms) can be seen to overlap on the figure. The VAK-4 positive percentages at 7 days (upper row) and 30 days (lower row) of cultivation in Agm1 were about 7% and 53%, respectively.
DISCUSSION
The present investigations revealed that the AGM retained helper T cell activity and showed no signs and symptoms of immunodeficiency, although CD4+ lymphocytes could barely be detected in the periphery. Great loss of peripheral CD4 lymphocytes and no detectable expression of CD4 mRNA were recognized in only one monkey presented here; however, both loss of CD4 cells and an asymptomatic state had been observed at every repeated examination performed for more than 1 year in this monkey. The disappearance of CD4 lymphocytes was not due to destruction of the CD4 gene, since we confirmed that 0.02% of lymphocytes were certainly CD4+ when very large numbers of lymphocytes (106) were counted by flow cytometric analysis. It is difficult to explain these immunological characteristics of the AGM on the basis of knowledge obtained for human and mouse immunology. Helper T cells, which are usually CD4+ in humans and mice, are required for most functional immune responses as well as PWM-induced antibody production. A dissociation between CD4 expression and helper activity was thus recognized in the AGM lymphocytes. Such a dissociation was suggested by us in a previous study [21], since helper activity could be detected in not only CD4 cells but also CD4− CD8low cells in the AGM. Thus, a low number of peripheral CD4 lymphocytes does not directly mean an immunodeficient state in the AGM.
The present CD4− AGM was infected with SIVagm, and infectious virus could be isolated from the CD4+ and CCR5− CemX174 cells [36] cocultured with peripheral CD8 cells. Sequence analysis indicated that the env region including the CD4 binding site of verCD8 was highly homologous with that of verCD4 derived from another AGM kept in the same colony. It should be noted that such PCR-derived sequences do not represent the entire virus population in the culture. The present data determined only the major genotype present in the culture. In addition, SIVagm might accumulate mutations after 1 month of cultivation. Further studies are required to elucidate the divergence of these viruses.
Several possible explanations can be put forward for the origin of peripheral CD4−CD8+SIVagm+ cells. Virus-infected CD4 cells might be eliminated by cytotoxic lymphocytes (CTL), and CD8+ CTL might become infected with virus in the process of killing SIV+ target cells [37]. Virus infection could occur in activated lymphocytes, which expressed both CD4 and CD8 antigens [32,38]. A CD4-independent pathway might be included in the virus infection of CD4− cells [39–41]. We cloned and sequenced the AGM CCR5 cDNA (DDBJ accession number, AB015944). Some CD4− cell lines were transfected with AGM CCR5 cDNA containing expression vector and the susceptibility to SIVagm infection was examined in AGM CCR5 transfectant cells. No apparent infection was noted in these transfectant cells throughout the culture and therefore, there exists no direct evidence at present that the SIVagm obtained from Agm3 can be propagated in a CD4-independent manner.
CD4−CD8+SIVagm+ cells were also obtained by the activation and cultivation of SIVagm+ PBMC including CD4 lymphocytes. Similar possibilities to those outlined above might be applied to the in vitro induction of CD4−CD8+SIVagm+ cells. However, most of the proliferating cells derived from SIVagm− PBMC were also CD4−, suggesting that SIVagm infection is not necessary for loss of CD4+ cells. We previously reported that the cell surface CD4 expression decreased after stimulation and most proliferating cells lacked detectable CD4 molecules within 2 weeks when peripheral CD4+CD8low cells were highly purified by sorting and cultured with ConA and IL-2 [21]. The down regulation of CD4 was not accompanied by internalization or modulation of surface CD4 molecules, whereas the expression of CD4 mRNA in the cultured cells decreased gradually [21]. These findings demonstrated that lymphocyte activation induced the down regulation of CD4 mRNA expression, resulting in the loss of surface expression of CD4 molecule. Taking into account the peculiar differentiation of AGM lymphocytes, the possibility cannot be excluded that CD4 expression was decreased in virus-infected CD4 cells after lymphocyte activation and that these cells then developed into CD4− cells.
It has been reported that IL-16 derived from the AGM, which is a soluble factor produced by CD8 cells, has the effect of suppressing HIV replication in activated human CD4 cells [42]. Human IL-16 also suppresses HIV replication [43,44] and acts as a growth factor specific for CD4 cells in cooperation with IL-2 [45,46]. A new field of AIDS therapy has clearly been opened up by IL-16. However, an effect of IL-16 on SIVagm replication in AGM CD4 lymphocytes had not been demonstrated, and it is still unclear whether apathogenicity in the AGM is dependent on the effect of IL-16 or not.
The regulation of CD4 gene expression in the AGM is distinct from that in humans. The immunological characteristics of the AGM must not be disregarded when attempting to understand resistance to SIV infection. Our present and previous data indicate the existence of CD4− helper T cells in the AGM. It can be inferred therefore that the infection efficiency in AGM helper T cells might be lower than that in humans, since masking of CD4 molecules could block chain-reacting infection among helper T lymphocytes. In view of the fact that the half-life of HIV-producing cells has been estimated to be approximately 2 days [47], continuous and repeated infection of noninfected CD4 cells would be required to maintain a high viral load for the development of AIDS. Since the lifetime of helper T cells is longer than that of virus-producing cells, the immune system in the AGM can maintain its function even if CD4 cells disappear in the periphery. Such a hypothetical strategy in which an immune system equipped with CD4− helper T cells does not break down with virus growth could explain the coexistence of SIVagm and AGM.
We have described a peculiar example of coexistence of SIVagm and its host. This may provide a clue to interpreting the resistance to simian AIDS observed in the AGM. Since the SIV growth was very low in cells converted from being CD4+ to CD4− cells following cultivation prior to SIV infection [21], we believe that distinct regulation of CD4 expression is closely associated with apathogenicity in the AGM. We will now proceed to a functional analysis of the regulatory regions of the AGM for CD4 gene expression and examine the possibilities for a new AIDS therapy based on the induction of helper T cells with resistance to HIV infection.
REFERENCES
- 1.Fukasawa M, Miura T, Hasegawa A, et al. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature. 1988;333:457–61. doi: 10.1038/333457a0. [DOI] [PubMed] [Google Scholar]
- 2.Tsujimoto H, Cooper RW, Kodama T, et al. Isolation and characterization of simian immunodeficiency virus from mandrills in Africa and its relationship to other human and simian immunodeficiency viruses. J Virol. 1988;62:4044–50. doi: 10.1128/jvi.62.11.4044-4050.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsujimoto H, Hasegawa A, Maki N, et al. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature. 1989;341:539–41. doi: 10.1038/341539a0. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch VM, Dapolito GA, Goldstein S. A distinct African lentivirus from Sykes' monkeys. J Virol. 1993;67:1517–28. doi: 10.1128/jvi.67.3.1517-1528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fultz PN, McClure HM, Anderson DC, Swenson RB, Anand R, Srinivasan A. Isolation of a T-lymphotropic retrovirus from naturally infected sooty mangabey monkeys (Cercocebus atys) Proc Natl Acad Sci USA. 1986;83:5286–90. doi: 10.1073/pnas.83.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marx PA, Li Y, Lerche NW, et al. Isolation of a simian immunodeficiency virus related to human immunodeficiency virus type-2 from a West African pet sooty mangabey. J Virol. 1991;65:4480–5. doi: 10.1128/jvi.65.8.4480-4485.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–9. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- 8.Peeters M, Fransen K, Delaporte E, et al. Further characterization of 2 immunodeficiency virus isolates from wild captured chimpanzees. AIDS Res Hum Retrovir. 1991;7:161. [Google Scholar]
- 9.Hirsch VM, Dapolito G, Goeken R, Campbell BJ. Phylogeny and natural history of the primate lentiviruses, SIV and HIV. Curr Opin Genet Dev. 1995;5:798–806. doi: 10.1016/0959-437x(95)80014-v. [DOI] [PubMed] [Google Scholar]
- 10.Lernould JM. Classification and geographical distribution of guenons: a review. In: Gautier-Hion A, Bourliere F, Gautier JP, editors. A primate radiation: evolutionary biology of the African guenons. Cambridge: Cambridge University Press; 1988. pp. 54–78. [Google Scholar]
- 11.Hendry RM, Wells MA, Phelan MA, Schneider AL, Epstein JS, Quinnan GV. Antibodies to simian immunodeficiency virus in African-green monkeys in Africa in 1957–62. Lancet. 1986;2:455. doi: 10.1016/s0140-6736(86)92156-2. [DOI] [PubMed] [Google Scholar]
- 12.Lowenstine LJ, Pederson NC, Higgins J. Seroepidemiologic survey of captive Old World primates for antibodies to human and simian retroviruses, and isolation of a lentivirus from sooty mangabeys (Cercocebus atys) Int J Cancer. 1986;38:563–73. doi: 10.1002/ijc.2910380417. [DOI] [PubMed] [Google Scholar]
- 13.Ohta Y, Masuda T, Tsujimoto H, et al. Isolation of simian immunodeficiency virus from African-green monkeys and seroepidemiologic survey of the virus in various non-human primates. Int J Cancer. 1988;41:115–22. doi: 10.1002/ijc.2910410121. [DOI] [PubMed] [Google Scholar]
- 14.Allan JS, Kanda P, Kennedy RC, Cobb EK, Anthony M, Eichberg JW. Isolation and characterization of simian immunodeficiency viruses from two subspecies of African green monkeys. AIDS Res Hum Retrovir. 1990;6:275–85. doi: 10.1089/aid.1990.6.275. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch VM, McGann C, Dapolito G, et al. Identification of a new subgroup of SIVagm in tantalus monkeys. Virology. 1993;197:426–30. doi: 10.1006/viro.1993.1606. [DOI] [PubMed] [Google Scholar]
- 16.Müller MC, Saksena NK, Nerrienet E, et al. Simian immunodeficiency viruses from Central and Western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J Virol. 1993;67:1227–35. doi: 10.1128/jvi.67.3.1227-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norley SG, Kraus G, Ennen J, Bonilla J, Konig H, Kurth R. Immunological studies of the basis for the apathogenicity of simian immunodeficiency virus from African green monkeys. Proc Natl Acad Sci USA. 1990;87:9067–71. doi: 10.1073/pnas.87.22.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norley SG. SIVagm infection of its natural African green monkey host. Immunol Lett. 1996;51:53–8. doi: 10.1016/0165-2478(96)02555-2. [DOI] [PubMed] [Google Scholar]
- 19.Beer B, Scherer J, zur Megede J, Norley S, Baier M, Kurth R. Lack of dichotomy between virus load of peripheral blood and lymph nodes during long-term simian immunodeficiency virus infection of African green monkeys. Virology. 1996;219:367–75. doi: 10.1006/viro.1996.0262. [DOI] [PubMed] [Google Scholar]
- 20.Beer B, Denner J, Brown CR, et al. Simian immunodeficiency virus of African green monkeys is apathogenic in the newborn natural host. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;18:210–20. doi: 10.1097/00042560-199807010-00003. [DOI] [PubMed] [Google Scholar]
- 21.Murayama Y, Amano A, Mukai R, et al. CD4 and CD8 expressions in African green monkey helper T lymphocytes: implication for resistance to SIV infection. Int Immunol. 1997;9:843–51. doi: 10.1093/intimm/9.6.843. [DOI] [PubMed] [Google Scholar]
- 22.Hattori T, Sagawa K, Matsushita S, et al. Characterization of three monoclonal antibodies (VAK3–5) that identify p24, core protein of human immunodeficiency virus, and its precursors. Jpn J Cancer Res. 1987;78:235–41. [PubMed] [Google Scholar]
- 23.Wang J, Yan T, Simmer B, Emmrich F. The effect of anti-CD4 on helper function of CD4, 45RA+ versus CD4, 45RO+ T cells. Clin Exp Immunol. 1994;95:128–34. doi: 10.1111/j.1365-2249.1994.tb06026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Rensburg EJ, Engelbrecht S, Mwenda J, et al. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: detection of a SIVagm variant from a chacma baboon. J Gen Virol. 1998;79:1809–14. doi: 10.1099/0022-1317-79-7-1809. [DOI] [PubMed] [Google Scholar]
- 26.Jin MJ, Hui H, Robertson DL, et al. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 1994;13:2935–47. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson PR, Fomsgaard A, Allan J, et al. Simian immunodeficiency viruses from African green monkeys display unusual genetic diversity. J Virol. 1990;64:1086–92. doi: 10.1128/jvi.64.3.1086-1092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin MJ, Rogers J, Phillips-Conroy JE, et al. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J Virol. 1994;68:8454–60. doi: 10.1128/jvi.68.12.8454-8460.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirsch VM, Dapolito G, Johnson PR, et al. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J Virol. 1995;69:955–67. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baier M, Garber C, Muller C, Cichutek K, Kurth R. Complete nucleotide sequence of a simian immunodeficiency virus from African green monkeys: a novel type of intragroup divergence. Virology. 1990;176:216–21. doi: 10.1016/0042-6822(90)90246-n. [DOI] [PubMed] [Google Scholar]
- 31.Tsubota H, Ringler DJ, Kannagi M, et al. CD8+CD4− lymphocyte lines can harbor the AIDS virus in vitro. J Immunol. 1989;143:858–63. [PubMed] [Google Scholar]
- 32.Dean GA, Reubel GH, Pedersen NC. Simian immunodeficiency virus infection of CD8+ lymphocytes in vivo. J Virol. 1996;70:5646–50. doi: 10.1128/jvi.70.8.5646-5650.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livingstone WJ, Moore M, Innes D, Bell JE, Simmonds P. Frequent infection of peripheral blood CD8-positive T-lymphocytes with HIV-1. Edinburgh Heterosexual Transmission Study Group. Lancet. 1996;348:649–54. doi: 10.1016/s0140-6736(96)02091-0. [DOI] [PubMed] [Google Scholar]
- 34.Semenzato G, Agostini C, Ometto L, et al. CD8+ T lymphocytes in the lung of acquired immunodeficiency syndrome patients harbor human immunodeficiency virus type 1. Blood. 1995;85:2308–14. [PubMed] [Google Scholar]
- 35.De Maria A, Pantaleo G, Schnittman SM, et al. Infection of CD8+ T lymphocytes with HIV. Requirement for interaction with infected CD4+ cells and induction of infectious virus from chronically infected CD8+ cells. J Immunol. 1991;146:2220–6. [PubMed] [Google Scholar]
- 36.Kirchhoff F, Pöhlmann S, Hamacher M, et al. Simian immunodeficiency virus variants with differential T-cell and macrophage tropism use CCR5 and an unidentified cofactor expressed in CEMx174 cells for efficient entry. J Virol. 1997;9:6509–16. doi: 10.1128/jvi.71.9.6509-6516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Maria A, Colombini S, Schnittman SM, Moretta L. CD8+ cytolytic T lymphocytes become infected in vitro in the process of killing HIV-1-infected target cells. Eur J Immunol. 1994;24:531–6. doi: 10.1002/eji.1830240306. [DOI] [PubMed] [Google Scholar]
- 38.Yang LP, Riley JL, Carroll RG, et al. Productive infection of neonatal CD8+ T lymphocytes by HIV-1. J Exp Med. 1998;187:1139–44. doi: 10.1084/jem.187.7.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Endres MJ, Clapham PR, Marsh M, et al. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–56. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 40.Edinger AL, Mankowski JL, Doranz BJ, et al. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci USA. 1997;94:14742–7. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin KA, Wyatt R, Farzan M, et al. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–3. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 42.Baier M, Werner A, Bannert N, Metzner K, Kurth R. HIV suppression by interleukin-16. Nature. 1995;378:563. doi: 10.1038/378563a0. [DOI] [PubMed] [Google Scholar]
- 43.Maciaszek JW, Parada NA, Cruikshank WW, Center DM, Kornfeld H, Viglianti GA. IL-16 represses HIV-1 promoter activity. J Immunol. 1997;158:5–8. [PubMed] [Google Scholar]
- 44.Zhou P, Goldstein S, Devadas K, Tewari D, Notkins AL. Human CD4+ cells transfected with IL-16 cDNA are resistant to HIV-1 infection: inhibition of mRNA expression. Nat Med. 1997;3:659–64. doi: 10.1038/nm0697-659. [DOI] [PubMed] [Google Scholar]
- 45.Viglianti GA, Parada NA, Maciaszek JW, Kornfeld H, Center DM, Cruikshank WW. IL-16 anti-HIV-1 therapy. Nat Med. 1997;3:938. doi: 10.1038/nm0997-938. [DOI] [PubMed] [Google Scholar]
- 46.Parada NA, Center DM, Kornfeld H, et al. Synergistic activation of CD4+ T cells by IL-16 and IL-2. J Immunol. 1998;160:2115–20. [PubMed] [Google Scholar]
- 47.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–6. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]