Abstract
Atopic disorders are caused by disregulated activation of T helper 2 (Th2) cells that produce IL-4 and IL-5. Because the presence of IL-4 potently augments the differentiation of naive T cells into Th2 cells, it is important to seek the cell population which provides IL-4 for naive T cells. Recently, a unique subpopulation of T cells, natural killer (NK) T cells, has been shown to produce a large amount of IL-4 upon activation, suggesting their regulatory role in initiation of Th2 cell differentiation. To determine whether NK T cells play a regulatory role in human Th2 cell-mediated atopic diseases, we analysed the frequency of invariant Vα24Jα Q CD4−CD8− double-negative (DN) T cells, human NK T cells, in patients with atopic asthma and atopic dermatitis. We also studied cytokine production from Vα24+ Vβ11+ DN T cells, which comprise most of Vα24Jα Q DN T cells. We found that the invariant Vα24Jα Q DN T cells were greatly diminished in patients with asthma and atopic dermatitis. On the other hand, there was no significant difference in Vα24+ CD4+ T cells possessing invariant Vα24Jα Q TCR between healthy subjects and atopic patients. We also found that Vα24+ Vβ11+ DN T cells from healthy subjects predominantly produced interferon-gamma (IFN-γ) but not IL-4 upon activation. These results suggest that NK T cells may not be essential for human atopic disease and that the disappearance of NK T cells, most of which produce IFN-γ, may be involved in the pathogenesis of atopic diseases.
Keywords: Vα24JαQ TCR α-chain, natural killer T cells, atopic diseases, interferon-gamma
INTRODUCTION
A considerable amount of investigation has centred on the pathogenesis of atopic disorders, including asthma and atopic dermatitis (AD). The discovery of Th1 and Th2 subsets of CD4+ T cells has helped to explain the immunological basis for the diversity of T cell responses; Th1 cells produce IL-2, interferon-gamma (IFN-γ), and lymphotoxin, while Th2 cells produce IL-4, IL-5, IL-10 and IL-13 [1–4]. Thus, cytokine production in atopic disorders is known to exhibit a bias toward the Th2 cytokines, IL-4 and IL-5 [5,6], and the requirement of these cytokines has been confirmed using murine models of allergic inflammation [7–9]. However, the mechanisms that lead to Th2 effector cell differentiation in atopic disorders are poorly understood.
In mice, one mechanism underlying the Th2 rather than Th1 bias may be promoted by the activation of natural killer (NK)1+ T cells (reviewed in [10–12]). Murine NK1+ T cells are a specialized subset of CD4−CD8− double-negative (DN) T cell receptor (TCR) αβ T cells that express the NK1 antigen, a member of the family of NKR-P1 natural killer cell receptors [10–12]. Moreover, a subpopulation of CD4+ T cells also expresses the NK1 antigen on the cell surface in mice. These NK1+ T cells have unusual features in comparison with the mainstream T cells. First, NK1+ T cells possess an invariant TCR Vα14Jα281 that preferentially pairs with Vβ8, Vβ7, and Vβ2 [13,14]. This highly restricted TCR on NK1+ T cells presumably recognizes a monomorphic MHC class I-like molecule CD1d, rather than polymorphic MHC molecules [15–18]. NK1+ T cell development has recently been shown to be impaired in CD1-deficient mice [19–21]. Second, NK1+ T cells can promptly produce a large amount of IL-4 by in vivo administration of anti-CD3 antibody and promote IgE production [22,23]. These observations suggest that the NK1+ T cell is a unique T cell that is already programmed for the production of IL-4, and that NK1+ T cells may play a regulatory role in Th2 cell differentiation.
Invariant Vα24Jα Q DN T cells are thought to be a human counterpart of murine NK1+ T cells [24–27]. The TCR Vα24Jα Q chain has a high homology with murine Vα14Jα281 chain in both the amino acid and nucleotide sequences [24–27]. The Vβ chains pairing with the Vα24Jα Q are Vβ11 and Vβ13, which also have a high homology with murine Vβ8 and Vβ7 [26,27]. It has recently been shown that human DN T cell clones bearing the invariant Vα24Jα Q TCR also recognize CD1d molecule [28]. Moreover, Vα24Jα Q DN T cells express NKR-P1A molecule on the surface [28,29]. Furthermore, Vα24Jα Q DN T cell clones have been shown to produce both IL-4 and IFN-γ upon TCR stimulation [28,29]. However, the regulatory role of invariant Vα24Jα Q DN T cells (human NK T cells) in the pathogenesis of atopic diseases has not yet been clarified.
In order to determine whether NK T cells play the regulatory role in the pathogenesis of human Th2 cell-mediated atopic diseases, we analysed the frequency of invariant Vα24Jα Q DN T cells in patients with asthma and AD. We also studied cytokine production from Vα24+ Vβ11+ DN T cells, which comprise most of Vα24Jα Q DN T cells. We found that the NK T cells are drastically decreased in patients with asthma and AD and that the majority of human NK T cells produce IFN-γ but not IL-4 upon activation. These results suggest that the lack of IFN-γ-producing NK T cells may be involved in the pathogenesis of atopic diseases.
PATIENTS AND METHODS
Patients
Six patients diagnosed with atopic asthma [30] and three patients diagnosed with AD [31] were studied (Table 1). All were atopic patients who had > 300 U/ml of serum IgE determined with radioimmunosorbent test (RIST) and had positive results of radioallergosorbent test (RAST) against at least one of 30 allergens, including Dermatophagoides farinae, house dust, cat dander, grass pollens and five moulds. Asthmatic patients were taking inhaled β2-adrenergic agonists and/or oral theophyllines, some of them took inhaled beclomethasone, but none of them took oral corticosteroids. AD patients were treated with corticosteroid ointments, but no oral corticosteroids. All patients were in a stable condition of disease when analysed. Five healthy subjects with no history of atopic diseases and < 100 U/ml of serum IgE were also examined as controls.
Table 1.
Characteristics of atopic patients studied
| Patients | Age (years) | Sex | Diagnosis | Serum IgE (U/ml) | RAST | Blood eosinophils (/μl) |
|---|---|---|---|---|---|---|
| A-1 | 18 | M | Asthma | 680 | HDM* | 558 |
| A-2 | 32 | M | Asthma | 700 | HDM, C | 364 |
| A-3 | 41 | M | Asthma | 1500 | HDM, G, JC | 273 |
| A-4 | 23 | M | Asthma | 2000 | HDM, JC | 224 |
| A-5 | 59 | F | Asthma | 2400 | HDM | 233 |
| A-6 | 39 | M | Asthma | 3500 | HDM | 648 |
| A-7 | 25 | F | AD | 14 000 | HDM, C | 1460 |
| A-8 | 24 | F | AD | 720 | HDM, C, JC | 1008 |
| A-9 | 20 | F | AD | 1900 | HDM, G, JC | 376 |
M/F, Male/female; AD, atopic dermatitis.
Positive results of radioallergosorbent test (RAST) against house dust mite(HDM), cat dander (C), grass pollens (G), and Japanese cedar pollen (JC).
Flow cytometric analysis
Peripheral blood lymphocytes (PBL) were isolated from peripheral venous blood of nine atopic patients and five healthy subjects by Ficoll–Paque (Pharmacia Biotech Inc., Piscataway, NJ) density gradient centrifugation. For four-colour staining, cells were stained with FITC-, PE-, peridinin chlorophyll protein (PerCP)-, or allophycocyanin (APC)-conjugated MoAbs in PBS containing 1% fetal calf serum (FCS) for 30 min at 4°C. In some experiments, biotin-conjugated MoAbs were used and visualized by streptavidin-APC (Becton Dickinson, Mountain View, CA). The following MoAbs were used: CD4 (Leu-3a), CD8 (Leu-2a), TCRαβ (Becton Dickinson), TCR Vα24, and TCR Vβ11 (Cosmo Bio Co., Tokyo, Japan). Anti-human NKR-P1A MoAb (DX1) [32] was kindly provided by Dr L. Lanier (DNAX Research Institute, Palo Alto, CA). Stained cells were resuspended in PBS containing 1% FCS and analysed on FACScalibar (Becton Dickinson) using CELL Quest software.
Purification of CD4−CD8− DN and CD4+ T cells
CD4−CD8− DN T cells were sorted from PBL of atopic patients and healthy subjects by FACStar (Becton Dickinson) using anti-CD4 and anti-CD8 MoAbs. CD4+ T cells were also sorted from PBL by FACStar using anti-CD4 MoAb. The purity of the fractionated DN and CD4+ T cells was > 99%.
Cloning and sequencing of cDNAs encoding TCR Vα24 genes
Total RNA was isolated from sorted DN or CD4+ T cells using Isogen solution (Nippon Gene Co., Tokyo, Japan). The first strand complementary DNA (cDNA) was synthesized from total RNA using oligo-dT primer and avian myeloblastosis virus reverse transcriptase as described elsewhere [30]. cDNAs encoding TCR Vα24 were then amplified by polymerase chain reaction (PCR) using primers for Vα24 with an EcoRI restriction site (5′-CGAATTCCTCAGCGATTCAGCCTCCTAC-3′) and Cα (5′-CGAATTCGGTGAATAGGCAGACAGACTT-3′). PCR products were purified by phenol extraction, precipitated with ethanol, and digested with excess amounts of EcoRI. The DNA fragments with expected sizes of the cDNAs were enriched by preparative low melting point agarose gel electrophoresis. The recovered DNA fragments were ligated to M13mp19 plasmids at EcoRI site. Phages were grown on TG-1 Escherichia coli cells. After hybridization with a Cα probe [33], recombinant phage DNAs were purified for DNA sequence determination. Sequencing reactions were performed by the dye primer method using an automated sequencer (Applied Biosystems, Foster City, CA).
Intracellular staining for IFN-γ and IL-4
To analyse the cytokine production from Vα24+ Vβ11+ DN T cells, sorted DN T cells were stimulated with phorbol myristate acetate (PMA; 25 ng/ml) and ionomycin (1 μm) for 4 h in RPMI 1640 medium supplemented with 10% FCS. Simultaneously, monensin (2 μm; Sigma, St Louis, MO) was added to prevent cytokine release. Cells were harvested, washed twice with PBS, and resuspended in PBS containing 1% FCS. Cells were incubated with biotin-conjugated anti-TCR Vα24 (mouse IgG1) and unconjugated anti-TCR Vβ11 (mouse IgG2a) for 30 min at 4°C and these antibodies were visualized with streptavidin–APC and anti-mouse IgG2a–PerCP (Becton Dickinson), respectively. After stained cells were fixed with 4% paraformaldehyde and permeabilized with PBS containing 0·4% saponin, cells were incubated with anti-IL-4–PE and anti-IFN-γ–FITC (PharMingen, San Diego, CA) for 30 min at 4°C. Cells were washed, resuspended in PBS containing 1% FCS, and analysed on FACScalibar. To analyse cytokine production from CD3+ T cells, unsorted PBL were stimulated with PMA and ionomycin in the presence of monensin as described above. After cells were stained with anti-CD3–PerCP (Becton Dickinson), intracellular cytokines were analysed as described above.
Statistical analysis
Data are summarized as mean ± s.d. The statistical analysis of results was performed by the unpaired t-test. P < 0·05 was considered significant.
RESULTS
Vα24+ DN T cells but not Vα24+ CD4+ T cells are decreased in patients with atopic diseases
To investigate the regulatory role of invariant Vα24Jα Q DN T cells in the pathogenesis of Th2 cell-mediated atopic diseases, we evaluated the frequency of Vα24Jα Q DN T cells in peripheral blood of patients with atopic asthma and AD (Table 1). We carried out a two-step frequency analysis of Vα24Jα Q DN T cells as described in Patients and Methods; the frequency of Vα24+ DN T cells was determined by three-colour FACS analysis and subsequently the frequency of Vα24Jα Q rearrangement among Vα24+ DN T cells was determined by sequencing. Results are summarized in Table 2 and representative FACS analyses of Vα24+ DN T cells are shown in Fig. 1. Interestingly, while Vα24+ CD4+ T cells were present in peripheral blood of asthma and AD patients at a similar frequency (0·17 ± 0·06% of PBL, n = 9) to that of healthy subjects (0·16 ± 0·04% of PBL, n = 5) (R1 region of panels i and ii in Fig. 1a), Vα24+ DN T cells were uniformly decreased in atopic patients (0·07 ± 0·04% of PBL, n = 9, P < 0·001) compared with those of healthy subjects (0·19 ± 0·05% of PBL, n = 5) (R2 region of panels iii and iv in Fig. 1a) (Table 2). In addition, there was no significant correlation between the decrease of Vα24+ DN T cells and the treatments for atopic diseases (data not shown).
Table 2.
TCR Vα24 CD4−CD8− double-negative (DN) T cell population in peripheral blood lymphocytes (PBL) from patients with atopic diseases
| Materials | Vα24+ DN T cells /PBL (%) | Vβ11+α24+ DN T cells (%) | NKR-P1A+α24+ DN T cells (%) | NKR-P1A+ Vβ11+α24+ DN T cells (%) |
|---|---|---|---|---|
| Cont-1 | 0·24 | 47·2 | 45·2 | 45·8 |
| Cont-2 | 0·13 | 44·9 | 51·2 | 43·7 |
| Cont-3 | 0·17 | 51·0 | 61·7 | 49·4 |
| Cont-4 | 0·19 | 60·8 | 62·3 | 59·5 |
| Cont-5 | 0·21 | 83·3 | 81·4 | 80·1 |
| Mean (s.d.) | 0·19 (0·04) | 57·4 (15·6) | 60·4 (13·8) | 55·7 (14·9) |
| A-1 | 0·06 | 9·9 | 15·3 | 7·5 |
| A-2 | 0·14 | 12·3 | 11·2 | 8·6 |
| A-3 | 0·07 | 8·2 | 14·4 | 7·6 |
| A-4 | 0·08 | 7·3 | 11·2 | 6·2 |
| A-5 | 0·06 | 11·4 | 12·6 | 8·8 |
| A-6 | 0·11 | 7·7 | 9·2 | 6·5 |
| A-7 | 0·03 | 5·2 | 21·2 | 4·2 |
| A-8 | 0·03 | 8·1 | 10·0 | 6·1 |
| A-9 | 0·06 | 9·3 | 9·9 | 6·3 |
| Mean (s.d.) | 0·07 (0·03)* | 8·8 (2·1)* | 12·8 (3·7)* | 6·9 (1·4)* |
The expression of TCR Vβ11 and NKR-P1A in peripheral blood Vα24 DN T cells from healthy subjects (cont-1 to -5) and patients with atopic diseases (A-1 to -9) was analysed by FACS using anti-CD4 peridinin chlorophyll protein (PerCP) plus anti-CD8 PerCP, anti-Vα24 biotin + streptavidin- allophycocyanin (APC), anti-Vβ11 FITC, and NKR-P1A + anti-mIgGl PE. PBL, Peripheral blood lymphocytes.
Significantly different from the mean value of healthy subjects, P <0·001.
Fig. 1.
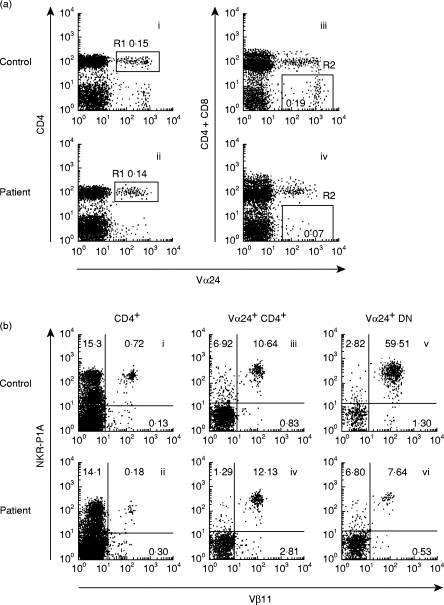
(a) Vα24+ double-negative (DN) T cells but not Vα24+ CD4+ T cells are decreased in patients with atopic diseases. TCR Vα24+ CD4+ T cells (left) and Vα24+ CD4−CD8− DN T cells (right) in peripheral blood lymphocytes (PBL) from healthy subjects (i, iii) and atopic patients (ii, iv) were analysed by FACS using anti-CD4 peridinin chlorophyll protein (PerCP) (plus anti-CD8 PerCP) and anti-TCR Vα24 biotin + streptavidin–allophycocyanin (APC). (b) Vα24+ Vβ11+ DN T cells are diminished in patients with atopic diseases. The expression of TCR Vβ11 chain and NKR-P1A was analysed on CD4+ T cells, Vα24+ CD4+ T cells, and Vα24+ DN T cells. i, ii: PBL were stained with anti-CD4 PerCP, anti-TCR Vβ11 FITC, and anti-NKR-P1A (mIgG1) + anti-mIgG1 PE. iii–vi: PBL were stained with anti-CD4 PerCP (plus anti-CD8 PerCP), anti-Vα24 biotin + streptavidin–APC, anti-Vβ11 FITC, and anti-NKR-P1A + anti-mIgG1 PE.
Since it has been shown that the majority of Vα24+ DN T cells in normal subjects co-express TCR Vβ11 chain and a NK marker NKR-P1A [26–29], we then determined whether the decreased Vα24+ DN T cells in atopic patients still co-expressed TCR Vβ11 chain and NKR-P1A. As controls, the expression of TCR Vβ11 chain and NKR-P1A was also analysed on CD4+ T cells and Vα24+ CD4+ T cells. As shown in Fig. 1b, NKR-P1A molecule was present on approximately 15% of peripheral blood CD4+ T cells in atopic patients as well as in healthy subjects, which was in agreement with the results of normal subjects reported originally by Lanier et al. [32]. Consistent with a similar frequency of Vα24+ CD4+ T cells between healthy subjects and atopic patients (Fig. 1a), the NKR-P1A+ Vβ11+ population among Vα24+ CD4+ T cells was found at a similar frequency between healthy subjects (11·0 ± 3·7%, n = 5) and atopic patients (10·3 ± 2·6%, n = 9) (Fig. 1b). However, the majority of Vα24+ DN T cells in healthy subjects co-expressed TCR Vβ11 and NKR-P1A (55·7 ± 14·9%, n = 5), whereas the frequency of Vα24+ DN T cells co-expressing NKR-P1A and TCR Vβ11 in atopic patients was significantly decreased (6·9 ± 1·4%, n = 9, P < 0·001) (Fig. 1b and Table 2).
Invariant Vα24JαQ DN T cells are diminished in patients with atopic diseases
Because Vα24Jα Q DN T cells have been shown to co-express TCR Vβ11 and NKR-P1A [28,29], the low frequency of Vβ11+ NKR-P1A+ cells in Vα24+ DN T cells in atopic patients suggested that the frequency of Vα24Jα Q rearrangement among Vα24+ DN T cells might be decreased. To investigate this possibility, we analysed nucleotide sequences of complementarity determining region 3 (CDR3) of TCR Vα24 genes in sorted DN T cells. In agreement with previous reports [24–27,33], the majority (86·7%) of Vα24+ DN T cells exhibited invariant Vα24Jα Q rearrangement in healthy subjects (Table 3). In contrast, the nucleotide sequences of CDR3 in Vα24+ DN T cells were random in atopic patients and Vα24Jα Q rearrangement was hardly detectable in the patients (Table 3). Thus, the actual number of Vα24Jα Q DN T cells in peripheral blood was severely decreased in atopic patients (0·04 ± 0·06/mm3, n = 9, P < 0·001) compared with that of healthy subjects (2·7 ± 0·5/mm3, n = 5) (Table 3). On the other hand, very few Vα24+ CD4+ T cells possessed invariant Vα24Jα Q TCR in both healthy subjects (2·5 ± 3·5%, n = 5) and atopic patients (3·5 ± 4·5%, n = 9).
Table 3.
Frequencies of invariant Vα24Jα Q double-negative (DN) T cells in patients with atopic diseases
| Source | Vα24Jα Q/Vα24† | Vα24+ DN T cells‡ (/mm3) | Vα24Jα Q DN T cells (/mm3) |
|---|---|---|---|
| Cont-1 | 15/20 | 3·9 | 2·9 |
| Cont-2 | 16/20 | 2·3 | 1·8 |
| Cont-3 | 16/18 | 2·9 | 2·6 |
| Cont-4 | 17/19 | 3·4 | 3·0 |
| Cont-5 | 18/18 | 3·2 | 3·2 |
| Mean (s.d.) | 86·7% (9·6%) | 3·1 (0·6) | 2·7 (0·5) |
| A-1 | 2/17 | 1·0 | 0·11 |
| A-2 | 1/16 | 2·9 | 0·18 |
| A-3 | 0/16 | 1·2 | 0 |
| A-4 | 0/16 | 1·6 | 0 |
| A-5 | 0/16 | 1·4 | 0 |
| A-6 | 0/16 | 2·5 | 0 |
| A-7 | 3/16 | 0·4 | 0·05 |
| A-8 | 0/16 | 0·5 | 0 |
| A-9 | 0/16 | 0·8 | 0 |
| Mean (s.d.) | 4·1% (6·3%)* | 1·4 (0·8)* | 0·04 (0·06)* |
TCR Vα24 cDNA clones were randomly isolated from the polymerase chain reaction (PCR)-amplified libraries of DN T cells from healthy subjects (cont-1 to -5) and patients with atopic diseases (A-1 to -9), and the frequency of Vα24J αQ rearrangement among Vα24 cDNA clones was determined by sequencing of complementarity determining region 3 (CDR3) of TCR Vα24 genes.
TCR Vα24 DN T cells in peripheral blood lymphocytes from healthy subjects and patients with atopic diseases were analysed by FACS.
Significantly different from the mean value of healthy subjects, P <0·001.
The majority of Vα24+ Vβ11+ DN T cells produce IFN-γ but not IL-4
Murine NK T cells have been shown to produce large amounts of IL-4 upon activation [22,23]. Human Vα24Jα Q DN T cell clones have been shown to produce both IL-4 and IFN-γ upon TCR stimulation, and hence their cytokine profiles have been described as Th0 type [28,29]. Thus, in contrast to murine NK T cells, the production of IL-4 from human Vα24+ Vβ11+ T cells might be limited. Therefore, we decided to analyse cytokine production from Vα24+ Vβ11+ DN T cells, which are selectively decreased in atopic patients (Table 2). In these experiments, purified DN T cells from healthy subjects were stimulated with PMA and ionomycin for 4 h and subjected to four-colour FACS analysis with surface staining for Vα24+ and Vβ11+ and intracellular staining for IL-4 and IFN-γ. The majority (78·6%) of human Vα24+ Vβ11+ DN T cells produced IFN-γ but not IL-4 (Th1 type) and only a small portion (7·1%) of them produced both IL-4 and IFN-γ (Th0 type) (n = 5) (Fig. 2 and Table 4). Furthermore, the frequency of Th1 cells among Vα24+ Vβ11+ DN T cells was significantly higher than that among CD3+ T cells (16·5 ± 2·7%, n = 5, P < 0·001) (Fig. 2). These results indicate that, in contrast to murine NK T cells, human Vα24+ Vβ11+ DN T cells predominantly produce IFN-γ.
Fig. 2.
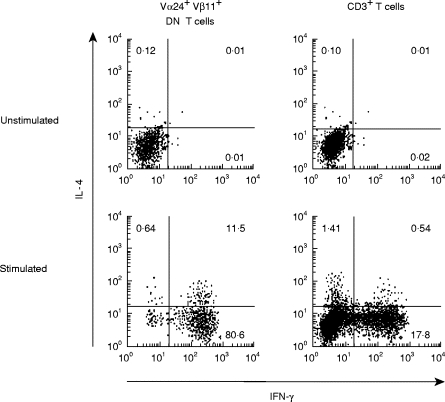
IFN-γ production from Vα24+ Vβ11+ double-negative (DN) T cells upon activation. Left: sorted DN T cells from healthy subjects were unstimulated or stimulated with phorbol myristate acetate (PMA; 25 ng/ml) plus ionomycin (1 µm) for 4 h, and cytokine production was evaluated by intracellular staining for IL-4 and IFN-γ on Vα24+ Vβ11+ cells. Right: unsorted peripheral blood lymphocytes (PBL) were unstimulated or stimulated with PMA plus ionomycin and cytokine production was evaluated by intracellular staining for IL-4 and IFN-γ on the CD3+ population.
Table 4.
Vα24+ Vβ11+ double negative (DN) T cells from healthy subjects predominantly produce IFN-γ
| Th0 (%) | Th1 (%) | Th2 (%) | |
|---|---|---|---|
| Cont-1 | 8·51 | 75·20 | 1·39 |
| Cont-2 | 5·80 | 73·41 | 1·10 |
| Cont-3 | 11·45 | 80·60 | 0·64 |
| Cont-4 | 6·30 | 81·20 | 0·54 |
| Cont-5 | 3·52 | 82·59 | 0·82 |
| Mean (s.d.) | 7·11 (3·00) | 78·60 (4·72) | 0·90 (0·34) |
Sorted DN T cells from healthy subjects were stimulated with phorbol myristate acetate (PMA; 25 ng/ml) plus ionomycin (1 μm) for 4 h, and cytokine production was evaluated by intracellular staining for IL-4 and IFN-γ on Vα24 Vβ11 cells. Th0, IL-4+; IFN-γ+; Th1, IL-γ+ Th2, IL-4+ IFN-γ−.
DISCUSSION
Cytokine production in atopic disorders is known to exhibit a bias toward the Th2 cytokines, IL-4 and IL-5. It has been shown that cytokines present during the initiation of an immune response determine the outcome of a particular Th subset [3,4]. Although IL-4 signal may not be essential for Th2 cell differentiation [34], the presence of IL-4 potently augments the differentiation of naive T cells into Th2 cells [35,36]. Most compelling evidence for this is obtained by studies showing that mice lacking either IL-4 [37,38] or Stat6 [39–41], a transcription factor activated by IL-4, fail to generate Th2 cells and the IgE responses. Despite the importance of IL-4, however, it is not clear how IL-4 synthesis is ever initiated. Several candidates for the initial source of IL-4 have been suggested. In addition to Th2 cells themselves, mast cells [42], γδ+ T cells [43], and NK T cells [44] are all capable of producing IL-4. In this regard, it is striking that NK T cells can produce IL-4 without cell division, while naive αβ T cells can not produce IL-4 until they proceed through at least three cell divisions even in the presence of IL-4 [45]. Nevertheless, it has become clear that murine NK T cells are not essential for in vivo Th2 responses, because mice lacking detectable NK T cells can still mount Th2 responses [19–21,46,47]. In addition, the Th2 allergic pulmonary responses induced by exogenous antigens have been shown to be independent of NK T cells in mice [47], although these responses are apparently IL-4-dependent [9].
In this study, we found that the invariant Vα24Jα Q DN T cells, which are a human counterpart of murine NK T cells [24–27], were greatly diminished in patients with asthma and AD (Tables 2 and 3) suggesting that Th2 cytokines from NK T cells may not be essential for human Th2 cell-mediated atopic diseases. On the other hand, there was no significant difference in Vα24+ CD4+ T cells possessing invariant Vα24Jα Q TCR between healthy subjects and atopic patients. Second, we also found that human NK T cells predominantly produced IFN-γ but not IL-4 upon activation (Fig. 2 and Table 4). Given that IFN-γ is known to inhibit Th2 responses in allergic airway inflammation [48], these results suggest that the disappearance of NK T cells, most of which produce IFN-γ, may be one of the mechanisms underling the pathogenesis of atopic diseases.
The number of NK T cells is regulated by the balance between their generation and disappearance. Thus, the diminished NK T cells in atopic patients should result from decreased generation and/or increased disappearance. Although there is as yet no information on the regulation of NK T cell development in humans, recent findings using gene-targeting mice revealed the mechanisms of NK T cell development in mice. Mice lacking rearrangement for Vα14Jα281 were found to lack NK T cells, suggesting that Vα14Jα281 rearrangement is essential for NK T cell development [49]. However, it is unlikely that atopic patients are defective in TCR Vα24Jα Q rearrangement because all atopic patients exhibited Vα24Jα Q rearrangement in CD4+ T cells at a similar frequency to that of healthy subjects. Beside TCR Vα rearrangement, cytokine signalling from common cytokine receptor γ-chain (γc), which is a shared component of the receptors for IL-2, IL-4, IL-7, IL-9 and IL-15 [50], is known to be essential for NK T cell development [51]. Because NK T cells can develop in mice lacking IL-2 or IL-7Rα[50] but not in mice lacking IL-2Rβ[52], which is a shared component of the receptors for IL-2 and IL-15, it is likely that IL-15 is important for NK T cell development. In fact, it has recently been reported that NK T cell development is impaired in IL-15R-deficient mice [53]. Therefore, it is possible that IL-15 itself or IL-15-related signalling molecules, such as Stat5, may be involved in the diminished NK T cells in atopic patients.
Recently, Eberl et al. [54] have shown that the activation of murine NK T cells results in rapid disappearance of these cells. In contrast, unstimulated NK T cells exhibit a slow turnover rate [12]. Therefore, it is possible that NK T cells are continuously activated in patients with atopic diseases. Although it is difficult to analyse the turnover rate of NK T cells in human diseases, these analyses may give us a new insight into the role of NK T cells in human disease. In this regard, it remains unclear how NK T cells regulate the immune responses to exogenous antigens in an antigen-specific fashion. Because NK T cells express the limited range of TCR and because these TCR have been shown to recognize lipid ligands on CD1 molecules [16,17], the activation of NK T cells may be regulated by a distinct mechanism from that of mainstream αβ T cells. Further studies including the mechanisms of NK T cell activation and the turnover of NK T cells in the site of allergic inflammation may reveal the role of NK T cells in atopic diseases.
We showed that the majority (79%) of human Vα24+ Vβ11+ DN T cells produced IFN-γ but not IL-4 (Th1 type) and only a small proportion (7%) of them produced both IL-4 and IFN-γ (Th0 type) (Fig. 2 and Table 4). In contrast, previous studies reported that most Vα24+ Vβ11+ DN T cell clones derived from peripheral blood T cells of healthy donors produced substantial amounts of both IL-4 and IFN-γ (Th0 type) upon stimulation [28,29]. Exley et al. [28] also described that, compared with CD4+ T cells, there was a trend towards higher levels of IL-4 production by the Vα24+ Vβ11+ DN T cell clones, assessed by IL-4 production and IL-4/IFN-γ ratios. The difference of Th1 versus Th0 cytokine profiles may be due to the cells studied, i.e. uncultured peripheral blood Vα24+ Vβ11+ DN T cells in our study versus Vα24+ Vβ11+ DN T cell clones expanded by phytohaemagglutinin (PHA) and IL-2, which may have differentiated towards IL-4-producing cells (Th0 type). Prussin et al. [55] also found that using mixed CD4+ and DN populations of human Vα24+ Vβ11+ T cells, cytokine profiles of those cells were either Th0 (IL-4+ IFN-γ+) or Th1 (IL-4− IFN-γ+) type.
At present there is no report on cytokine profiles of NK T cells in atopic patients. Because Vα24Jα Q DN T cells were severely decreased in atopic patients compared with those of healthy subjects, and the majority of Vα24+ Vβ11+ DN T cells do not represent Vα24Jα Q DN T cells in atopic patients (Table 3), it was extremely difficult to analyse cytokine profiles of Vα24Jα Q DN T cells in atopic patients by our FACS analysis of Vα24+ Vβ11+ DN T cells. Therefore, this issue remains to be elucidated until cytokine profiles of Vα24Jα Q DN T cells can be analysed by single-cell reverse transcriptase (RT)-PCR methods [56] in the future.
In conclusion, we have shown that the invariant Vα24Jα Q DN T cells, human NK T cells, are greatly diminished in patients with atopic diseases and predominantly produce IFN-γ but not IL-4 upon activation. These results suggest that NK T cells may not be essential for human Th2 cell-mediated atopic diseases, and that the disappearance of NK T cells, most of which produce IFN-γ, may be involved in the pathogenesis of atopic diseases.
Acknowledgments
We thank Dr L. Lanier for providing anti-NKR-P1A MoAb and Dr M. Taniguchi for helpful discussion and critical comments. This work was supported in part by grants from the Ministry of Education, Science and Culture, Japan.
References
- 1.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–57. [PubMed] [Google Scholar]
- 2.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–73. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 3.Paul WE, Seder RA. Lymphocyte responses and cytokines. Cell. 1994;76:241–51. doi: 10.1016/0092-8674(94)90332-8. [DOI] [PubMed] [Google Scholar]
- 4.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–93. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 5.Kay AB, Ying S, Varney V, Gaga M, Durham SR, Moqbel R, Wardlaw AJ, Hamid Q. Messenger RNA expression of the cytokine gene cluster, IL-3, IL-4, IL-5, and GM-CSF in allergen-induced late-phase cutaneous reactions in atopic subjects. J Exp Med. 1991;173:775–8. doi: 10.1084/jem.173.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamid Q, Azzawi M, Ying S, et al. Expression of mRNA for interleukin-5 in mucosal bronchial biopsies from asthma. J Clin Invest. 1991;87:1541–6. doi: 10.1172/JCI115166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakajima H, Iwamoto I, Tomoe S, Matsumura R, Tomioka H, Takatsu K, Yoshida S. CD4+ T lymphocytes and interleukin-5 mediate antigen-induced eosinophil infiltration into the mouse trachea. Am Rev Resp Dis. 1992;146:374–7. doi: 10.1164/ajrccm/146.2.374. [DOI] [PubMed] [Google Scholar]
- 8.Foster PS, Hogan SP, Ramsay AJ, Matthaei KI, Young IG. Interleukin 5 deficiency abolishes eosinophilia, airways hyperreactivity, and lung damage in a mouse asthma model. J Exp Med. 1996;183:195–201. doi: 10.1084/jem.183.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brusselle GG, Kips JC, Tavernier JH, van der Heyden JG, Cuvelier CA, Pauwels RA, Bluethmann H. Attenuation of allergic airway inflammation in IL-4 deficient mice. Clin Exp Allergy. 1994;24:73–80. doi: 10.1111/j.1365-2222.1994.tb00920.x. [DOI] [PubMed] [Google Scholar]
- 10.Bendelac A. Mouse NK1+ T cells. Curr Opin Immunol. 1995;7:367–74. doi: 10.1016/0952-7915(95)80112-x. [DOI] [PubMed] [Google Scholar]
- 11.MacDonald HR. NK1.1+ T cell receptor α/β cells: new clues to their origin, specificity and function. J Exp Med. 1995;182:633–8. doi: 10.1084/jem.182.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bendelac A, Rivera MN, Park S-H, Roark JH. Mouse CD1-specific NK1 cells: development, specificity and function. Annu Rev Immunol. 1997;15:535–62. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 13.Lantz O, Bendelac A. An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4–CD8− T cells in mice and humans. J Exp Med. 1994;180:1097–106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makino Y, Kanno R, Ito T, Higashino K, Taniguchi M. Predominant expression of invariant Vα14+ TCRα chain in NK1.1+ T cell populations. Int Immunol. 1995;7:1157–61. doi: 10.1093/intimm/7.7.1157. [DOI] [PubMed] [Google Scholar]
- 15.Bendelac A, Lantz O, Quimby ME, Yewdell JW, Bennink JR, Brutkiewicz RR. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–5. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 16.Bendelac A. CD1: presenting unusual antigens to unusual T lymphocytes. Science. 1995;269:185–6. doi: 10.1126/science.7542402. [DOI] [PubMed] [Google Scholar]
- 17.Kawano T, Cui J, Koezula Y, et al. CD1d-restricted and TCR-mediated activation of Vα14 NK T cells by glycosylceramides. Science. 1997;278:1626–9. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 18.Joyce S, Wood AS, Yewdell JW, et al. Natural ligand of mouse CD1d1: cellular glycosylphosphatidylinositol. Science. 1998;279:1541–4. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 19.Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E production in the absence of interleukin-4-secreting CD1-dependent cells. Science. 1997;275:977–9. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 20.Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK1.1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity. 1997;6:459–7. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 21.Mendiratta SK, Martin WD, Hong S, Boesteanu A, Joyce S, Van Kaer L. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity. 1997;6:469–77. doi: 10.1016/s1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimoto T, Paul WE. CD4pos, NK1.1pos T cells promptly produce interleukin 4 in response to in vivo challenge with anti-CD3. J Exp Med. 1994;179:1285–95. doi: 10.1084/jem.179.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshimoto T, Bendelac A, Watson C, Hu-Li J, Paul WE. Role of NK1.1+ T cells in a TH2 response and in immunoglobulin E production. Science. 1995;270:1845–7. doi: 10.1126/science.270.5243.1845. [DOI] [PubMed] [Google Scholar]
- 24.Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4–CD8−α/β T cells demonstrates preferential use of several Vβ genes and an invariant TCR α chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dellabona P, Casorati G, Friedli B, Angman L, Sallusto F, Tunnacliffe A, Roosneek E, Lanzavecchia A. In vivo persistence of expanded clones specific for bacterial antigens within the human T cell receptor α/β CD4–8− subset. J Exp Med. 1993;177:1763–71. doi: 10.1084/jem.177.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. An invariant Vα24-JαQ/Vβ11 T cell receptor is expressed in all individuals by clonally expanded CD4–CD8− T cells. J Exp Med. 1994;180:1171–6. doi: 10.1084/jem.180.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porcelli S, Gerdes D, Fertig AM, Balk SP. Human T cells expressing an invariant Vα24-JαQ TCRα are CD4− and heterogeneous with respect to TCRβ expression. Hum Immunol. 1996;48:63–67. doi: 10.1016/0198-8859(96)00090-0. [DOI] [PubMed] [Google Scholar]
- 28.Exley M, Garcia J, Balk SP, Porcelli S. Requirements for CD1d recognition by human invariant Vα24+ CD4–CD8− T cells. J Exp Med. 1997;186:109–20. doi: 10.1084/jem.186.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davodeau F, Peyrat M-A, Necker A, et al. Close phenotypic and functional similarities between human and murine αβ T cells expressing invariant TCR α-chain. J Immunol. 1997;158:5603–11. [PubMed] [Google Scholar]
- 30.American Thoracic Society. Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease (COPD) and asthma. Am Rev Respir Dis. 1987;136:225–44. doi: 10.1164/ajrccm/136.1.225. [DOI] [PubMed] [Google Scholar]
- 31.Hanifin JM, Rajka G. Diagnostic features of atopic dermatitis. Acta Derm Venereol. 1980;92:44–47. [Google Scholar]
- 32.Lanier LL, Chang C, Phillips JH. Human NKR-P1A: a disulfide-linked homodimer of the c-type lectin superfamily expressed by a subset of NK and T lymphocytes. J Immunol. 1994;153:2417–28. [PubMed] [Google Scholar]
- 33.Sumida T, Sakamoto A, Murata H, Makino Y, Takahashi H, Yoshida S, Iwamoto I, Taniguchi M. Selective reduction of T cells bearing invariant Vα24JαQ antigen receptor in patient with systemic sclerosis. J Exp Med. 1995;182:1163–8. doi: 10.1084/jem.182.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noben-Trauth N, Shultz LD, Brombacher F, Urban Jf, Jr, Gu H, Paul WE. An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc Natl Acad Sci USA. 1997;94:10838–43. doi: 10.1073/pnas.94.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swain SL, Weinberg AD, English M, Huston G. IL-4 directs the development of Th-2 like helper effectors. J Immunol. 1990;145:3796–806. [PubMed] [Google Scholar]
- 36.Seder RA, Paul WE, Davis MM, Fazekas De St Groth B. The presence of IL-4 during in vivo priming determines the lymphokine producing potential of CD4+ T cells from T cell receptor transgenic mice. J Exp Med. 1992;176:1091–8. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhn R, Rajewsky K, Muller W. Generation and analysis of interleukin-4 deficient mice. Science. 1991;245:707–10. doi: 10.1126/science.1948049. [DOI] [PubMed] [Google Scholar]
- 38.Kopf M, Le Gros G, Bachmann M, Lamers MC, Bluethmann H, Kohler G. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 1993;362:245–8. doi: 10.1038/362245a0. [DOI] [PubMed] [Google Scholar]
- 39.Shimoda K, van Deursen J, Sangster MY, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–3. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 40.Takeda K, Tanaka T, Shi W, et al. Essential role of Stat6 in IL-4 signaling. Nature. 1996;380:627–30. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 41.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–9. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 42.Brown MA, Pierce JH, Watson CJ, Falco J, Ihle JN, Paul WE. B cell stimulatory factor-1/interleukin-4 mRNA is expressed by normal and transformed mast cells. Cell. 1987;50:809–18. doi: 10.1016/0092-8674(87)90339-4. [DOI] [PubMed] [Google Scholar]
- 43.Ferrick DA, Schrenzel MD, Mulvania T, Hsieh B, Ferlin WG, Lepper H. Differential production of interferon-γ and interleukin-4 in response to Th1- and Th2-stimulating pathogens by γδ T cells in vivo. Nature. 1995;373:255–7. doi: 10.1038/373255a0. [DOI] [PubMed] [Google Scholar]
- 44.Arase H, Arase N, Nahagawa K, Good RA, Onoe K. NK1.1+ CD4+ CD8− thymocytes with specific lymphokine secretion. Eur J Immunol. 1993;23:307–10. doi: 10.1002/eji.1830230151. [DOI] [PubMed] [Google Scholar]
- 45.Bird JJ, Brown DR, Mullen AC, et al. Helper T cell differentiation is controlled by cell cycle. Immunity. 1998;9:229–37. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 46.Brown DR, Fowell DJ, Corry DB, Wynn TA, Moskowitz NH, Cheever AW, Locksley RM, Reiner SL. β2-microglobulin-dependent NK1.1+ T cells are not essential for T helper cell 2 immune responses. J Exp Med. 1996;184:1295–304. doi: 10.1084/jem.184.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Rogers KH, Lewis DB. β2-microglobulin-independent T cells are dispensable for allergen-induced T helper 2 responses. J Exp Med. 1996;184:1507–12. doi: 10.1084/jem.184.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwamoto I, Nakajima H, Endo H, Yoshida S. Interferon-γ regulates antigen-induced eosinophil recruitment into the mouse airways by inhibiting the infiltration of CD4+ T cells. J Exp Med. 1993;177:573–6. doi: 10.1084/jem.177.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui J, Shin T, Kawano T, et al. Requirement for Vα14 NK T cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–6. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 50.Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 51.Boesteanu A, De Silva AD, Nakajima H, Leonard WJ, Peschon JJ, Joyce S. Distinct role for signals relayed through the common cytokine receptor γ chain and interleukin 7 receptor α chain in natural T cell development. J Exp Med. 1997;186:331–6. doi: 10.1084/jem.186.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohteki T, Ho S, Suzuki H, Mak TW, Ohashi PS. Role for IL-15/IL-15 receptor β-chain in natural killer 1.1+ T cell receptor-αβ+ cell development. J Immunol. 1997;159:5931–5. [PubMed] [Google Scholar]
- 53.Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, Ma A. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 1998;9:669–76. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 54.Eberl G, Robson MacDonald H. Rapid death and regeneration of NK T cells in anti-CD3ε- or IL-12-treated mice: a major role for bone marrow in NK T cell homeostasis. Immunity. 1998;9:345–53. doi: 10.1016/s1074-7613(00)80617-2. [DOI] [PubMed] [Google Scholar]
- 55.Prussin C, Foster B. TCR Vα24 and Vβ11 coexpression defines a human NK1 T cell analog containing a unique Th0 subpopulation. J Immunol. 1997;159:5862–70. [PubMed] [Google Scholar]
- 56.Kelso A, Groves P, Ramm L, Doyle AG. Single cell analysis by RT-PCR reveals differential expression of multiple type 1 and 2 cytokine genes among cells within polarized CD4 + T cell populations. Int Immunol. 1999;11:617–21. doi: 10.1093/intimm/11.4.617. [DOI] [PubMed] [Google Scholar]


