Abstract
Anti-centromere autoantibodies (ACA) are commonly found in the serum of patients with a limited type of scleroderma and other systemic autoimmune diseases. CENP-A is one of the major antigens against ACA and a histone H3-like protein. To analyse the autoantigenic epitopes of CENP-A, a series of truncated peptides of human CENP-A were expressed in Escherichia coli and immunoblotting analysis was performed with 91 ACA+ sera. Eighty sera (88%) with the ACA reacted to the 52-amino acids N-terminal region which is not homologous to H3, while no sera reacted to the C-terminus which has a sequence similarity with H3. Moreover, ELISA was also employed in this study using two synthetic peptides corresponding to the amino acid sequences 3–17 (peptide A) and 25–38 (peptide B). Peptides A and B were reactive to 78 (86%) and 79 (87%) of ACA, respectively. Core antigens of hepatitis B virus (HBV) and hepatitis C virus (HCV) have similar sequences to peptide A and/or peptide B, but three sera containing HBV without ACA and five sera containing HCV without ACA were found to be reactive to neither peptide. Centromere localization of CENP-A is dependent on the H3-like C-terminal domain which is not autoantigenic, while the antigenic N-terminal domain, which might play unidentified functional roles, should be an important region for the induction of ACA.
Keywords: autoepitope, CENP-A, recombinant protein, synthetic peptide
Introduction
Sera from patients with various rheumatic diseases often contain circulating antinuclear autoantibodies [1]. Anti-centromere antibodies (ACA) have a close relationship with a limited form of systemic scleroderma (SSc) and they are also detected in patients with various rheumatic diseases [2,3]. The ACA reactivity is directed against multiple centromere proteins (CENPs) (reviewed in [4]), and among them, three major antigens, CENP-A (17 kD), CENP-B (80 kD), and CENP-C (140 kD), are mainly recognized by ACA [2–3]. Concerning these autoantigens, several epitope mapping studies have been reported. According to studies on CENP-B and -C, centromere autoantigens contain multiple epitopes, including both major and minor epitopes, and the epitope regions coincide with biologically functional sites [5–14]. These results are entirely consistent with basic concepts concerning autoantibodies in systemic autoimmune diseases [15]. Among the three antigens, the cDNA encoding CENP-A was last cloned [16] but a detailed mapping of CENP-A has not been published to date. In a previous study [17], we identified one antigenic epitope of CENP-A at the N-terminal charged region (amino acid sequence 3–17) using ELISA with a synthetic peptide. In this study we have made several deletion constructs derived from human CENP-A genes and investigated the reactivities of ACA against the fusion protein of CENP-A with glutathione S-transferase (GST) by immunoblotting. In our results, the histone H3-like C-terminal region was found to be not antigenic. Since other epitope(s) in addition to the previously identified linear epitope were assumed to exist at the N-terminus, we made two kinds of synthetic peptides (amino acid sequences 3–17 and 25–38) to determine more precisely the epitope regions. We identified two immunodominant regions at the N-terminus of CENP-A, which were not homologous with H3 in CENP-A.
MATERIALS and METHODS
Sera
Among 91 sera with ACA, 77 sera were used in our previous study [17]. The remaining 14 sera were confirmed to contain ACA by indirect immunofluorescence with HEp-2 cells and by immunoblotting with HeLa cell extract [2]. Among the patients with ACA were 57 with SSc, 13 with systemic lupus erythematosus (SLE), 10 with primary Sjögren’s syndrome (SS), three with Raynaud’s disease, two with dermatomyositis, two with discoid lupus erythematosus, one with rheumatoid arthritis (RA), one with overlapping SLE and RA, and two with miscellaneous conditions. Thirty sera were collected from healthy blood donors and used as normal controls. Twenty sera from atopic dermatitis patients and 20 sera from SSc patients without ACA were used as other controls.
CENP-A cDNA and deletion constructs
Two primer sets, an EcoRI universal primer (5′-GCGAATTCCGTTGCTGTC-3′) and a specific reverse primer (5′-AGAGTCCCCGGTATCATC-3′), and a forward primer (5′-TTCTCGAGCTCTGCGGCGTGTCATGG-3′) and the reverse primer (5′-TTCTCGAGCCGAGTCCCTCCTCAAGG-3′), were used to amplify the truncated N-terminus and the entire coding region of human CENP-A cDNA, respectively, as described [18]. Each polymerase chain reaction (PCR) product was introduced into a pGEM-T vector (Promega, Madison, WI), sequenced and cloned into a suitable pGEX4T vector (Pharmacia, Uppsala, Sweden), resulting in pGEX-AΔ N and pGEX-AF, respectively. Plasmids pGEX-A37-15 and pGEX-AΔ N-1 were constructed by introducing a BamHI stop codon linker (Nippon Gene, Toyama, Japan) into the HindIII site of pGEX-AF or pGEX-AΔ N, respectively. To obtain pGEX-AΔ EH, the EcoRI-HindIII fragment (amino acid 20–52) of pGEX-AΔ N was replaced with a SfiI ‘in-frame deletion cassette’ (5′-AATTTGGCCCTATTGGCCC-3′ and 5′-AGCTGGGCCAATAGGGCCA-3′), as described [12]. Plasmid pGEX-AΔ ES was constructed by removing the internal SfiI fragment (amino acid 53–90) of pGEX-AΔ EH.
Expression of recombinant CENP-A peptides and immunoblotting
Expression and purification of the GST fused proteins were performed as described [12–14]. Extracts were subjected to 12·5% SDS–PAGE, and immunoblotting assays were performed using a protocol described previously [2,14].
Overlap peptide synthesis
Overlap peptide synthesis was conducted on cellulose membranes using F-moc amino acids according to the manufacturer’s protocol (Auto spot robot ASP222; ABIMED Analysen-Technik GmbH, Langenfeld, Germany)[19]. The ASP222 software program was used to generate amino acid sequences of decapeptides and the spotting schedule for each cycle of amino acid addition. Peptides spanning the CENP-A autoantigen residues 11–58 were synthesized as a series of decapeptides with an overlapping of two amino acids. Immune reactions and colour developments were carried out according to the previous protocol [2].
Synthetic peptides
The published sequence of CENP-A [16] was used to construct two kinds of synthetic peptides. Peptide A, PRRRSRKPEAPRRRS, and peptide B, GPSRRGPSLGASSH, corresponding to residues 3–17 and residues 25–38, respectively, were synthesized with a C-terminal cysteine amide residue using a solid-phase method [20]. The peptide chain was elongated using an automated peptide synthesizer (Model 9050; Milligen/Biosearch, Burlington, MA) according to the standard operation programs. The peptides were purified using high performance liquid chromatography with a reverse phase column, Delta PakC18 on a Waters 600E Multisolvent Delivery System. The chromatography was performed with a linear gradient of 0·1% trifluoroacetic acid in acetonitrile. The molecular weight and amino acid sequence of the purified peptides were confirmed by a matrix-assisted laser desorption ionization time of flight mass spectrometer (KOMPACT MALDI II; Kratos-Shimadzu, Kyoto, Japan) and a protein sequencer (Applied Biosystems 477A; Foster City, CA). The synthetic peptides (2 mg) were conjugated to bovine serum albumin (BSA; 10 mg; Sigma, St Louis, MO) with the use of N-(4-maleimidobutyryloxy)-succinimide (GMBS; 2 mg; Dojin, Kumamoto, Japan) according to the manufacturer’s protocol.
ELISA
ELISA was performed basically according to the published protocol [17]. Nunc microtitre wells were coated with 50 ng of the synthetic peptides conjugated to BSA (0·5 μg/ml in PBS) at 4°C overnight. After incubation with PBS containing 0·5% Tween 20 (PBS–T) for 1 h at room temperature in order to block non-specific binding, serum samples (diluted 1:200 or serial two-fold in PBS–T containing 1% BSA) were added to the wells. After a 1-h incubation with shaking at room temperature, the wells were washed three times with PBS–T, and 100 μl of peroxidase-conjugated goat anti-human IgG (Dako, Glostrup, Denmark) diluted to 1:1000 in PBS–T were added. Following incubation for 1 h with shaking at room temperature, the wells were washed three times with PBS–T and twice with PBS. After a 1-h incubation with the substrate solution [17] at room temperature, absorbance (405 nm) was determined with a 340 ATTC (Tecan Co., Salzberg, Austria). The absorbance with the synthetic peptides following subtraction of the antibody reactivity to BSA was the value for each individual. Values >2 s.d. above the mean of 30 normal controls were set as the cut-off values. The cut-off values were 0·10 and 0·12 for peptides A and B, respectively.
The reactivity of each patient was calculated as follows:
As a result, for the antibodies to both peptides A and B, reactivities >1·00 were considered positive.
Results
Epitope mapping of CENP-A with recombinant protein in Escherichia coli
Various deletion constructs, named 37-15, ΔΝ-1, Δ EH and Δ ES, derived from human CENP-A genes were expressed as GST-fusion protein (Fig. 1). The quantities of each construct loaded on a gel were roughly adjusted to be equal by their reactivities against anti-GST antibodies (Fig. 2a). Non-fused GST recombinant protein was reactive to no ACA+ sera. Human CENP-A consists of 140 amino acids and shares significant homology with histone H3. This homology is restricted to the C-terminal portion ranging from amino acids 48 to 134 [16]. Hind III sites were suitable for creating deletion fragments spanning amino acids 1–52 and 52–140. Representative data on immunoblotting are shown in Fig. 2b–d. No sera had reactivities to C-terminal constructs Δ EH and Δ ES, while 80 and 73 ACA+ sera reacted to 37–15 and Δ N-1, respectively. Most sera (73 ACA) belonged to group I (Fig. 2b), which reacted to both 37–15 and Δ N-1. Seven sera belonged to group II (Fig. 2c), which reacted only to 37–15. Eleven sera belonged to group III (Fig. 2d), which reacted to neither 37–15 nor Δ N-1. Among these 11 ACA, nine sera were negative for anti-CENP-A in the immunoblotting analysis with HeLa cell extract. From these results, at least two immunodominant epitopes were considered to exist on amino acids 1–19 and 19–52 of CENP-A protein at the N-terminus.
Fig. 1.
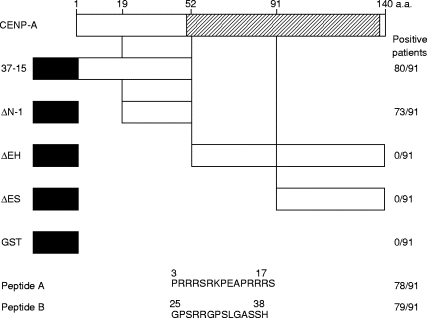
Deletion constructs and synthetic peptides of CENP-A for epitope mapping. A hatched bar shows the histone H3-like domain. Black bars show fusion partner glutathione S-transferase (GST). Numbers in the right column represent positive patients in 91 examined anti-centromere autoantibodies.
Fig. 2.
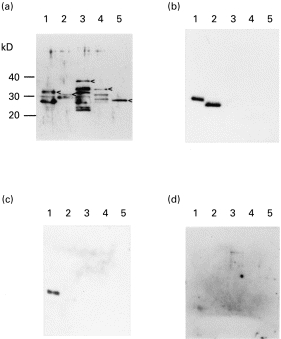
Immunoblotting of purified recombinant proteins. (a) Anti-glutathione S-transferase (GST) antibody. Arrowheads show purified recombinant proteins for each construct of 37–15 (lane 1), Δ N-1 (lane 2), Δ EH (lane 3), Δ ES (lane 4), and GST alone (lane 5) as depicted in Fig. 1. The smaller bands are considered to be degraded products. (b) Anti-centromere autoantibody (ACA) group I (n = 73). This group reacted to 37–15 and Δ N-1. (c) ACA group II (n = 7). This group reacted only to 37–15. (d) ACA group III (n = 11). This group reacted to no constructs.
Fine epitope mapping of the N-terminus of CENP-A
As we identified one linear autoepitope in the amino acid sequence 3–17 in the previous study [17], this sequence should represent an epitope in amino acids 1–19. To map the linear epitope finely in the amino acid sequence 19–52, eight ACA+ sera showing strong reactivity with the recombinant 37–15 were tested for reactivity against 20 consecutive cellulose-bound linear peptides of 10 amino acids in length and an eight amino acid overlap in the region of amino acids 11–58 of CENP-A. Seven of these eight sera showed significant reactions, predominantly against the region of amino acids 25–38, as shown in Fig. 3, which is a representative dot assay result. Sera of one normal donor and an anti-CENP-A−ACA in the immunoblotting analysis did not react with any of the cellulose-bound CENP-A peptides. According to the result, the amino acid 25–38 portion was considered to be a possible representative epitope in the 19–52 portion.
Fig. 3.
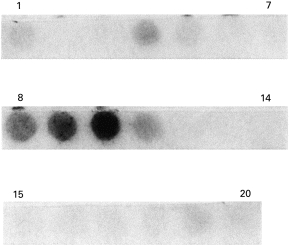
Reactivity of a representative anti-CENP-A+ serum with peptides spanning the N-terminal CENP-A protein (11–58 aa) by linear epitope mapping. Each spot represents a linear synthetic peptide of 10-aa length bound to a cellulose membrane. The numbers 1, 2, 3, ····, 20, indicate the individual peptides, 11–20 aa, 13–22 aa, 15–24 aa, ····, 49–58 aa. Marked positive reactions were detected in the spot numbers 8–10 (the region of 25–38 aa).
Immunoreactivities of ACA+ sera to both peptides in ELISA
In this study, the sequence of amino acids 25–38 named peptide B was synthesized in addition to amino acids 3–18 (peptide A) as a positive control. ELISA with each peptide conjugated to BSA was performed, and values >2 s.d. above the mean of 30 normal controls were determined as cut-off values. Twenty sera from atopic dermatitis patients without ACA and 10 sera from SSc patients without ACA were reactive to neither peptide A nor peptide B. Among 91 ACA+ sera, 78 sera were positive for peptide A, 79 sera for peptide B, and 73 sera were positive for both (Fig. 4). Of 13 anti-peptide A− ACA, eight were not reactive to 37–15 and the other four were reactive to peptide B and Δ N-1 in immunoblotting. The other one reacted to 37–15 but did not react to Δ N-1, which might imply the possible existence of a minor epitope around amino acid 19. None of 12 anti-peptide B− ACA was reactive to Δ N-1, which suggests that peptide B was the most immunodominant epitope in the region of amino acids 19–52. Judging from these results, the two sequences, peptides A and B, made up the major linear autoepitopes of CENP-A.
Fig. 4.
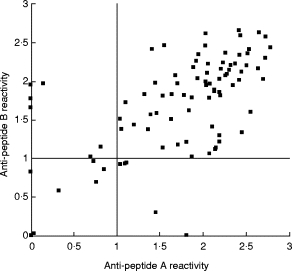
Binding assay in ELISA of peptide A and peptide B. Reactivities of anti-centromere autoantibody (ACA)-positive sera (n = 91) against each peptide A and B were plotted. x and y axes represent reactivities to peptides A and B, respectively. The cut-off line at 1·00 represents 2 s.d. above the mean binding levels of 30 normal control sera for both peptides.
There was no correlation between the antibody titres against peptide A and the clinical settings. However, the titres against peptide B in 17 patients with SSc and secondary SS (2·02 ± 0·52) were significantly higher (P < 0·05, Mann–Whitney U-test) than those in 13 patients with SLE (1·57 ± 0·58) or in 10 patients with primary SS (1·34 ± 0·60). They were also higher than those in 40 patients with SSc without secondary SS (1·69 ± 0·61), but not significantly.
Reactivities to both peptides A and B were correlated (y = 0·55x + 0·51, r = 0·579, P < 0·001), but there were no homologies in the sequences between them. In addition, cross-reactivities between the two peptides were not found by mutual inhibition ELISA with several sera (data not shown).
Sequence homology
The GenBank databank search showed that no other protein was found to contain the whole amino acid sequence of either peptide A or B. However, interestingly, hepatitis B and C virus (HBV and HCV) core proteins have similarities to the peptides (Fig. 5). In HBV core antigen, the amino acid 161–175(6) region is similar to peptide A. In HCV core antigen, PRRRS, which is repeated twice in peptide A, was found among amino acids 112–116, and the amino acid 38–48 portion is similar to peptide B. In our preliminary experiments with ELISA, all sera from three individuals with HBV and five individuals with HCV reacted to neither peptide A nor peptide B.
Fig. 5.

Alignments of hepatitis B virus (HBV) core antigen with peptide A (a) and of hepatitis C virus (HCV) core antigen with peptide A and peptide B (b) in amino acid sequences. Numbers show amino acid positions. Identical and similar amino acids, respectively.
Discussion
Among the major centromere autoantigens, CENP-A was first characterized in a number of ACA+ sera by Guldner et al. [21]. Although there has been no published report on successful cDNA cloning of the CENP-A gene by immunoscreening since then, Palmer et al. purified bovine CENP-A and determined the partial amino acid sequences [22]. Sullivan’s group reported a full-length human cDNA clone of CENP-A [17] and Hoch’s group established the ELISA system for anti-CENP-A antibodies with recombinant CENP-A using a baculovirus system [23]. However, as for epitope mapping studies on CENP-A, there has been no systematic report to date.
Brendel et al. suggested that autoantigens as a group contain more long charge runs or clusters of charged groups, which are suggested to be exposed on the surface of the proteins and to be strongly immunogenic [24]. They also suggested that a possibly more generally conserved feature of the autoantigens consists of extended surface structures that are charge-, proline-, or glycine-rich. In our previous study [17] we focused on the characteristic amino acid sequences in CENP-A and synthesized peptides composed of amino acid residues 3–17 (named peptide A in this study). That region contains 7 Arg (R), 1 Lys, and 3 Pro (P), and two repeats of PRRRS. Also in this study, more than 90% of anti-CENP-A+ sera showed reactivity to peptide A. Moreover, Valdivia et al. succeeded in producing powerful experimental antibodies in rabbits using the peptide A sequence [25], which suggests the strong antigenicity of peptide A. However, the existence of other autoepitope(s) in addition to this region was considered [17,25].
After obtaining the full-length clone of CENP-A, we employed recombinant CENP-A produced in Escherichia coli in an epitope mapping study. A 2/3 part of the C-terminal domain, which shared 62% identity with histone H3, was confirmed to show no reactivity to ACA. We identified another major autoepitope, amino acid sequence 25–38 (peptide B). In contrast to peptide A, peptide B had a few charged residues (2 Arg and 1 His). However, it contained two dipeptide Pro-Ser or one Ser-Ser, leaving each of the three residues (PPXXXPSXXXSS) and two repeats of Gly-Pro-Ser, although it was not clear whether these sequences are critical. According to the study with 10-mer continuous primary structure peptides covering the CENP-A sequence from residue 11–58, additional determinants except for the regions of peptides A and B were considered on CENP-A, i.e. amino acids 39–52 (data not shown), but they reacted to minor populations of ACA and the reactivities were very low. These results fitted well with the fact that there was only one of seven ACA without both anti-peptides A and B antibodies that reacted to the recombinant N-terminal part. We concluded that the major autoepitopes on CENP-A are two regions, amino acid sequences 3–17 and 25–38.
In the core antigens of HBV and HCV, sequences similar to peptides A and B were found. Especially, the HCV core antigen contains a sequence similar to peptide B and the PRRRS sequence which repeats in peptide A. However, the regions of viruses described in Fig. 5 were not generally recognized by HBV- or HCV-infected sera according to the data from the previous epitope mapping studies [26–28]. Although our preliminary study has not confirmed the cross-reactivity, further studies on the reactivities of ACA to those sequences are required.
In this study we used an E. coli system for producing the recombinants. However, we did not apply the full-length peptide due to unacceptable proteolysis (data not shown). The ELISA system developed by Hoch’s group was very sensitive for ACA [23]. It might be due to the usage of the baculovirus system, which can induce modifications in the protein [23,29]. In the N-terminal part of CENP-A, four copies of the peptide Ser/Thr-Pro, which are good candidates for phosphorylation, are present [16]. Some ACA for CENP-A might show a striking preference for conformational epitopes by protein modifications such as phosphorylation.
Since autoantibody-defined epitopes on nuclear antigens are often biologically functional regions [15], this theory may be applicable to ACA [5–14]. Although the C-terminus of CENP-A is necessary for targeting to the centromere [16], the functional role of the N-terminus has not been clarified yet. It might be worthwhile focusing on the antigenic N-terminal part, which could play role(s) in other functions, e.g. DNA binding, oligomerization, etc., as previously suggested for CENP-A [30,31]. From our data, the representative autoepitopes on major antigens recognized by ACA have been characterized. Since the N-terminal part of CENP-A as well as the C-terminal part of CENP-B [6,9,10] is the most reactive epitope, further analysis of immune responses concerning this area is important for an understanding of the induction of specific autoantibodies against centromere antigens.
Acknowledgments
This work was supported in part by Grants C:06660120 (K.S.) and 08770652 and 09770628 (Y.M.) from the Ministry of Education of Japan.
REFERENCES
- 1.von Muhlen CA, Tan EM. Autoantibodies in the diagnosis of systemic rheumatic diseases. Semin Arthritis Rheum. 1995;24:323–58. doi: 10.1016/s0049-0172(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 2.Muro Y, Sugimoto K, Okazaki T, Ohashi M. The heterogeneity of anticentromere antibodies in immunoblotting analysis. J Rheumatol. 1990;17:1042–7. [PubMed] [Google Scholar]
- 3.Earnshaw W, Bordwell B, Marino C, Rothfield N. Three human chromosomal autoantigens are recognized by sera from patients with anti-centromere antibodies. J Clin Invest. 1986;77:426–30. doi: 10.1172/JCI112320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueroa J, Saffrich R, Ansorge W, Valdivia M. Microinjection of antibodies to centromere protein CENP-A arrests cells in interphase but does not prevent mitosis. Chromosoma. 1998;107:397–405. doi: 10.1007/s004120050323. [DOI] [PubMed] [Google Scholar]
- 5.Earnshaw WC, Sullivan KF, Machlin PS, et al. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol. 1987;104:817–29. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earnshaw WC, Machlin PS, Bordwell BJ, Rothfield NF, Cleveland DW. Analysis of anticentromere autoantibodies using cloned autoantigen CENP-B. Proc Natl Acad Sci USA. 1987;84:4979–83. doi: 10.1073/pnas.84.14.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugimoto K, Muro Y, Himeno M. Anti-helix-loop-helix domain antibodies: discovery of autoantibodies that inhibit DNA binding activity of human centromere protein B (CENP-B) J Biochem. 1992;111:478–83. doi: 10.1093/oxfordjournals.jbchem.a123783. [DOI] [PubMed] [Google Scholar]
- 8.Yoda K, Kitagawa K, Masumoto H, Muro Y, Okazaki T. A human centromere protein, CENP-B, has a DNA binding domain containing four potential α helices at the NH2 terminus, which is separable from dimerizing activity. J Cell Biol. 1992;119:1413–27. doi: 10.1083/jcb.119.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugimoto K, Migita H, Hagishita Y, Yata H, Himeno M. An antigenic determinant on human centromere protein B (CENP-B) available for production of human-specific anticentromere antibodies in mouse. Cell Struct Funct. 1992;17:129–38. doi: 10.1247/csf.17.129. [DOI] [PubMed] [Google Scholar]
- 10.Muro Y, Sugimoto K, Himeno M, Ohashi M. The clinical expression in anticentromere antibody-positive patients is not specified by the epitope recognition of CENP-B antigen. J Dermatol. 1992;19:584–91. doi: 10.1111/j.1346-8138.1992.tb03734.x. [DOI] [PubMed] [Google Scholar]
- 11.Verheijen R, de Jong BAW, Oberye EHH, van Venrooij WJ. Molecular cloning of a major CENP-B epitope and its use for the detection of anticentromere autoantibodies. Mol Biol Rep. 1992;16:49–59. doi: 10.1007/BF00788753. [DOI] [PubMed] [Google Scholar]
- 12.Sugimoto K, Yata H, Muro Y, Himeno M. Human centromere protein C (CENP-C) is a DNA-binding protein which possesses a novel DNA-binding motif. J Biochem. 1994;116:877–81. doi: 10.1093/oxfordjournals.jbchem.a124610. [DOI] [PubMed] [Google Scholar]
- 13.Sugimoto K, Kuriyama K, Shibata A, Himeno M. Characterization of internal DNA-binding and C-terminal dimerization domains of human centromere/kinetochore autoantigen CENP-C in vitro: role of DNA-binding and self-associating activities in kinetochore organization. Chromosome Res. 1997;5:132–41. doi: 10.1023/a:1018422325569. [DOI] [PubMed] [Google Scholar]
- 14.Sugimoto K, Kuriyama K, Himeno M, Muro Y. Epitope mapping of human centromere autoantigen centromere protein C (CENP-C); heterogeneity of anti-CENP-C response in rheumatic diseases. J Rheumatol. 1998;25:474–81. [PubMed] [Google Scholar]
- 15.Tan EM, Muro Y, Pollard KM. Autoantibody-defined epitopes on nuclear antigens are conserved, conformation-dependent and active site regions. Clin Exp Rheumatol. 1994;12(Suppl. 11):S27–31. [PubMed] [Google Scholar]
- 16.Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targetting to the centromere. J Cell Biol. 1994;127:581–92. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muro Y, Iwai T, Ohashi M. A charged segment mainly composed of basic amino acids forms an autoepitope of CENP-A. Clin Immunol Immunopathol. 1996;78:86–89. doi: 10.1006/clin.1996.0013. [DOI] [PubMed] [Google Scholar]
- 18.Sugimoto K, Himeno M. A rapid isolation of the unknown 5′-flanking sequence of human CENP-B cDNA with polymerase chain reactions. Agric Biol Chem. 1991;55:2687–92. [PubMed] [Google Scholar]
- 19.Frank R, Overwin H. SPOT synthesis. Epitope analysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol Biol. 1996;66:49–169. doi: 10.1385/0-89603-375-9:149. [DOI] [PubMed] [Google Scholar]
- 20.Appel JR, Pinilla C, Niman H, Houghten RA. Elucidation of discontinuous linear determinants in peptides. J Immunol. 1990;144:976–83. [PubMed] [Google Scholar]
- 21.Guldner HH, Lakomek HJ, Bautz FA. Human anti-centromere sera recognise a 19.5 kD non-histone chromosomal protein from HeLa cells. Clin Exp Immunol. 1984;58:13–20. [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer DK, O’Day K, Trong HL, Charbonneau H, Margolis RL. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci USA. 1991;88:3734–8. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun D, Martinez A, Sullivan KF, Sharp GC, Hoch SO. Detection of anticentromere antibodies using recombinant human CENP-A protein. Arthritis Rheum. 1996;39:863–7. doi: 10.1002/art.1780390520. [DOI] [PubMed] [Google Scholar]
- 24.Brendel V, Dohlman J, Blaisdell BE, Karlin S. Very long charge runs in systemic lupus erythematosus-associated autoantigens. Proc Natl Acad Sci USA. 1991;88:1536–40. doi: 10.1073/pnas.88.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valdivia MM, Figueroa J, Iglesias C, Ortiz M. A novel centromere monospecific serum to a human autoepitope on the histone H3-like protein CENP-A. FEBS Letters. 1998;422:5–9. doi: 10.1016/s0014-5793(97)01583-4. [DOI] [PubMed] [Google Scholar]
- 26.Sallberg M, Ruden U, Wahren B, Magnius LO. Immune recognition of linear antigenic regions within the hepatitis B pre-C and C-gene translation products using synthetic peptides. J Med Virol. 1994;42:7–15. doi: 10.1002/jmv.1890420103. [DOI] [PubMed] [Google Scholar]
- 27.Ching W-M, Wychowski C, Beach MJ, et al. Interaction of immune sera with synthetic peptides corresponding to the structural protein region of hepatitis C virus. Proc Natl Acad Sci USA. 1992;89:3190–4. doi: 10.1073/pnas.89.8.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akatsuka T, Donets M, Scaglione L, et al. B-cell epitopes on the hepatitis C virus nucleocapsid protein determined by human monospecific antibodies. Hepatology. 1993;18:503–10. [PubMed] [Google Scholar]
- 29.Martinez A, Sun D, Billings PB, Swiderek KM, Sullivan KF, Hoch SO. Isolation and comparison of natural and recombinant human CENP-A autoantigen. J Autoimmun. 1998;11:611–9. doi: 10.1006/jaut.1998.0249. [DOI] [PubMed] [Google Scholar]
- 30.Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–13. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vafa O, Sullivan KF. Chromatin containing CENP-A and α-satellite DNA is a major component of the inner kinetochore plate. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]


