Abstract
Intravenous immunoglobulin preparations (IVIG) have shown positive effects in the treatment of immune defects and autoimmune diseases. It is not clear how IVIG interacts with the components of the immune system. To investigate this, we cloned previously a large number of phage displayed IgG Fab fragments derived from three patients with autoimmune thrombocytopenia (AITP) that were specifically bound by IVIG molecules. Many of these Fabs reacted with platelets. Sequencing revealed that the most frequently used germ-line gene segments of all IVIG-bound Fabs were identical to those observed for many other autoantibodies. Particularly, the loci 3–30 or 3–30/3–30.5, 3–23 and 3r, 3l, and 2a2 represented the most abundant genes used for the heavy (VH) and light chain V region (VL), respectively. This suggested a specific interaction of IVIG molecules with B cells that present B cell receptors derived from these germ-line genes. In the current study we determined the genetic origin of IVIG-reactive IgG and IgM cloned from a healthy person. A favoured selection of antibodies derived from the same germ-line origins as in AITP was observed. Because 3–30 and 3–23 are the most frequently rearranged VH germ-line gene segments among human B cells, our results suggest that this favoured anti-idiotypic interaction may have an important role for the development and control of the normal B cell repertoire.
Keywords: phage display, CDR, immunoglobulin gene, anti-idiotype, germ-line
INTRODUCTION
Commercial intravenous immunoglobulin preparations (IVIG) are prepared by purification of serum IgG from several thousand healthy donors. IVIG has shown positive effects in a variety of immune deficiencies as well as in autoimmune diseases such as autoimmune thrombocytopenia (AITP) [1] and Kawasaki disease [2]. However, the specific mechanisms of IVIG interactions with the immune system are not clear. Various studies suggested the inhibition of effector cells through blockade of Fc receptors [3], healing of persisting infections, induction of cytokines [4], inhibition of autoantibodies by anti-idiotypic antibodies [5], B cell modulation by anti-CD5 and anti-B cell antigen receptor antibodies, and the reconstruction of the ‘idiotypic’ or ‘V-region’ connected [6] network [7] by IVIG. However, the documented increase of serum IgM, decrease of certain autoantibodies and long-term therapeutic effects over 6 months duration far beyond the half-life of infused IVIG observed often in children treated for AITP [8] cannot be explained by non-specific mechanisms such as Fc receptor blockade alone. This effect requires the additional anti-idiotypic involvement of variable regions from IVIG molecules [5,6]. Anti-idiotypic IVIG molecules may fit the antigen-binding site, particularly the complementarity-determining regions (CDR) of their targets (Ab2 β, internal image). In addition, they may be directed against idiotypes distinct from the paratope, recognizing the framework regions (Ab2 α) [9].
To investigate the latter mechanisms, we analysed previously patient-derived monoclonal IgG Fab fragments that were bound by IVIG in an anti-idiotypic manner, applying the combinatorial antibody phage display system [10–13]. From three different patients with AITP, a large number of clones specifically bound by IVIG molecules were enriched. Sequencing revealed that the most frequently used germ-line gene loci of these IVIG-selected Fabs were identical to those reported for many other autoantibodies highly specific for the corresponding autoantigen such as dsDNA, acetylcholine receptor, thyroid peroxidase, blood group antigens and others. These loci were 3–23 and 3–30 or 3–30/3–30.5 in case of the heavy chains and 3l, 2a2, and 3r for the light chains [14]. Many of the IVIG-selected Fabs bound strongly to platelets, in contrast to IVIG-bound Fabs derived from a healthy individual [15]. In summary, the results suggested a specific interaction of IVIG molecules, particularly with autoantibodies and B cell receptors derived from those germ-line genes that are often used for the generation of autoantibodies.
Here, to investigate further the origin and nature of antibodies targeted by IVIG, we extended the comparative analysis of putative germ-line genes and mutation rates to IgG as well as IgM cloned from a healthy individual. As expected, there are clear differences in the mutation rates between the IgG and IgM sequences. Surprisingly, the data show that the most frequently used germ-line gene loci of those IVIG-selected immunoglobulins are the same as those observed in AITP, expanding our previous conclusions with regard to the normal B cell repertoire.
MATERIALS AND METHODS
Phage display library construction
The library was constructed from peripheral blood lymphocytes (PBL) of a healthy, adult female volunteer (control individual 4 in [15]). A detailed protocol has been published recently [15]. Briefly, total RNA was extracted using Trireagent (Bio Tech Trade + Service GmbH, St. Leon-Rot, Germany) from lymphocytes purified with Ficoll–Paque (Pharmacia, Freiburg, Germany) from PBL. Then 10 μl each of RNA were reverse-transcribed with constant region primers cG1z, cM1, cK1z and cL2, respectively, to synthesize the first-strand cDNA using a SuperScript RT kit (Gibco/BRL, Eggenstein, Germany). The L chains were amplified by polymerase chain reaction (PCR) with the cL2 3′ primer and eight different 5′ Vλ primers and the cK1d 3′ primer and five different 5′ Vκ primers, respectively [16]. Similarly, the H chain Fd regions were amplified with the cG1z and cM1 3′ primer, respectively, and eight different 5′ VH primers [16]. Of each amplified DNA sample, 5–10 μl were analysed on a 2% agarose gel.
Following the above procedure, the amplified L chain DNA was pooled, gel purified, digested with SacI and XbaI, again gel purified and cloned into the SacI/XbaI-linearized pComb3H vector [17]. The recombinant DNA was electroporated into Escherichia coli XL1-Blue. The size and the insert frequency of the resultant L chain library were determined, and phagemid DNA from the total library was prepared. Then, the amplified and gel-purified H chain Fd region DNA was digested with SpeI and XhoI, gel purified, ligated with the SpeI/XhoI-linearized phagemid DNA containing the L chain library and electroporated into XL1-Blue. Again, the size and the insert frequency of the resultant L chain–H chain Fd library were determined and Fab-phage production was initiated by addition of helper phage VCSM13. The recombinant phages were precipitated with 4% polyethylene glycol (PEG) and 3% NaCl (w/v), resuspended in Tris-buffered saline (TBS) containing 1% bovine serum albumin (BSA) and 0·02% NaN3, and stored frozen or at 4°C for immediate use. From the bacterial pellets, phagemid DNA was prepared to transform E. coli for a new phage rescue after longer storage. In addition, to optimize the libraries, the phagemid DNA was separated on an agarose gel. The gel-purified bands corresponding to pComb3H with both the H and L chain inserts were again electroporated into E. coli for production of Fab-phage.
Enrichment with IVIG
Fab-phages were selected by panning of approximately 1012 recombinant phages in Maxisorp immunotubes (Nunc, Wiesbaden, Germany) coated with IVIG at a concentration of 30 μg/ml PBS and blocked with 1% casein. Unbound phage was removed and the tubes were washed vigorously up to 10 times with 0·2% casein in PBS. Then the bound phages were eluted with 300 μl 0·1 m HCl/glycine pH 2·2, and neutralized with 60 μl 2 m Tris–HCl at pH 9. Eluted phages were used to infect 3 ml of a fresh E. coli XL1-Blue culture grown with tetracycline. After a 20-min incubation at 37°C on a shaker at 240 rev/min, the number of eluted phages was determined by plating dilutions on carbenicillin plates. The transformed bacteria were diluted in 10 ml super broth (SB) [17] supplemented with 10 mm MgCl2 and a low concentration of carbenicillin, grown for 1 h as above and then further diluted in 100 ml SB with a high concentration of carbenicillin. After 1 h incubation, 1011 VCSM13 helper phages were added. Following another 2 h incubation, kanamycin was added and the culture grown overnight. Phage preparation and further panning in IVIG-coated tubes were repeated three more times.
After the last panning step, single colonies were used to grow recombinant Fab-phages in 48-well plates in 300 μl SB containing carbenicillin and 10 mm MgCl2. Diluted VCSM helper phages were added after 5·5 h at 37°C on a shaker. After 1 h further incubation, kanamycin was added and the cultures grown overnight. Phage supernatants were collected after 30 min centrifugation at 490 g using a plate rotor. The resulting pellets were used for analytical DNA preparations as described [18].
IVIG-ELISA of recombinant Fab-phages
Microtitre plates (Maxisorp, Nunc) were coated with equal concentrations of IVIG (Sandoglobin; Sandoz, Basel, Switzerland), human Fc-fragments (Dianova, Hamburg, Germany) and other tested antigens at 600 ng/well each or PBS at 4°C overnight. After washing with PBS containing 0·2% casein, the plates were blocked with 1% casein for 1 h at 37°C. Then, 48-well phage supernatants were added and incubated at 37°C for 1 h. After washing with PBS, the bound Fab-phages were detected with peroxidase-labelled goat anti-M13 phage antibody (Pharmacia).
To control for the expression of comparable Fab-phage titres, additional wells were coated with unlabelled anti-M13 antibodies, incubated with 1:10 dilutions of individual investigated Fab-phages and developed as above with peroxidase-labelled anti-M13 antibody.
Determination and analysis of nucleotide and amino acid sequences
The V regions of selected Fab fragments were determined from phage or double-stranded DNA purified with either Midiprep (Qiagen, Hilden, Germany) or Wizard columns (Promega, Mannheim, Germany). Sequencing was done in an automatic sequencer (Li-Cor, MWG Biotech, Ebersberg, Germany) using the cycle sequencing kit (Amersham, Braunschweig, Germany) and infrared fluorophore (IRD41) labelled primers [15]. The IRD labelled sequencing primers were PelB and SeqGb or SeqMb for the H chains and OmpA and SeqLb or SeqKb for the L chains [17,19]. Automatic sequence analysis was controlled by comparing sequences from both directions and by manually analysing the scf-files and repeating the DNA preparation and sequencing in case of unclear sequences. The computer program MacDNAsis was used to align all sequences.
V-BASE (version May 31, 1999) [20] was searched via Internet (http://www.mrc-cpe.cam.ac.uk/imt-doc/public/INTRO.html) using the program DNA-Plot (developed by W. Müller and H.-H. Althaus, University of Köln, Germany) for determination of germ-line segments and mutations. To enable easy comparison with our previous and other studies in this field, we applied the widely used Kabat definition of CDR and framework regions (FR) [21] as well as the V-BASE nomenclature. In agreement with previous studies [14] we used the full length of the germ-line V-genes in these calculations, although the last bases may have been replaced by D-segments in some recombinations. For calculating the mutation rates of individual V-genes, the regions originating from pComb3H or the PCR primers were not included. In cases of unknown D-genes we also consulted the IMGT database (http://imgt.cnusc.fr:8104) [22]. However, sometimes the homology to D-genes was too low for exact determinations, and this is indicated by a ‘?’ in Table 2. While the VH/l loci identified by IMGT were identical to V-BASE, IMGT listed various allelic variations and a corresponding nomenclature of the genes (not shown).
Table 2.
Putative germ-line sequences of intravenous immunoglobulin preparation (IVIG)-selected heavy and light chain variable regions of the MM and MG libraries*
| Clone | Frequency | VH/L family | VH/L | VH/L locus | Percent homology | DH | JH/L |
|---|---|---|---|---|---|---|---|
| Heavy chains of MM | |||||||
| 03 | 3 | DP-47/V3–23… + (Z12347) | 3–23 | 99·6 | D1–26? (X97051) | JH3b (X86355) | |
| 05 | 2× | 3 | COS-6/DA-8… + (Z17392) orDP-53/hmlv148…. + (Z12353) | 3–74 or 3–74 | 96·3 | D6–19 (X97051) | JH6b (X86355) |
| 20 | 4× | 3 | DP-38/9–1… + (Z12338) | 3–15 | 99·3 | Not found | JH4b(X86355) |
| 29 | 3 | DP77/WHG16 + (Z14073)or V3–21 + (M99658)or HHG4 (X62129) | 3–21or 3–21or unkn. | 99·6 | D6–13/DN1(X97051) | JH5b(X86355) | |
| 53 | 3 | DP-47/V3–23… + (Z12347) | 3–23 | 95·2 | DXP4? (= IGHD 3–3,X93618)† | JH3b (X86355) orJH3a (J00256) | |
| 66 | 4 | DP-79/4d154… + (Z14075) | 4–39 | 99·6 | D3–3/DXP4(X97051) | JH3b (X86355) | |
| 74 | 3 | DP-47/V3–23… + (Z12347) | 3–23 | 100 | D3–16 (X97051) orD1–26 (X97051)? | JH4b (X86355) | |
| 117 | 3 | DP-47/V3–23… + (Z12347) | 3–23 | 99·6 | D6–13/DN1(X97051) | JH6b (X86356) | |
| Heavy chains of MG | |||||||
| 13b | 5× | 3 | V3–30 + (M99663)‡or b28e (M77334) | 3–30or unkn. | 93·7 | D6–13/DN1 orD6–19 (X97051) | JH4b (X86355) |
| 14b | 4 | DP-70/4d68… + (Z12370) | 4–04 | 99·3 | D1–14/DM2?(X97051) | JH3b (X86355) | |
| 16b | 1 | DP-75/VI-2… + (Z14071) | 1–02 | 97 | D4–17 (X97051) | JH6b (X86355) | |
| 18a | 3 | V3–30 + (M99663) | 3–30 | 95·9 | DM2? (= IGHD1–14*01, X13972)† | JH6b (X86355) | |
| 20a | 4 | DP-79/4d154… + (Z14075) | 4–39 | 94·5 | D1–7/DM1 (X97051) | JH3a (J00256) | |
| 24a | 3× | 3 | V3–30 + (M99663) orDP-49/1.9III… + (Z12349) | 3–30 or3–30.5 | 95·2 | D1–14/DM2 (X97051) | JH6b (X86355) |
| 26a | 4 | DP-79/4d154… + (Z14075) | 4–39 | 92·3 | D1–26 (X97051) | JH4b (X86355) | |
| 29a | 3 | V3–30 + (M99663) orDP-49/1.9III… + (Z12349) | 3–30 or3–30.5 | 96·3 | DM2? (= IGHD1–14*01, X13972)† | JH6b (X86355) | |
| 30b | 1 | 4M28 orhv1263/3M28 (M83132) | Unkn. | 99·6 | D6–6/DN4?(X97051) | JH3b (X86355) | |
| Light chains of MG§ | |||||||
| 14b | 3 | DPL16/VL3.1 (Z22202) | 3l | 100 | – | JL2/3a (M15614) | |
| 20a | 3 | DPL16/VL3.1 (Z22202) | 3l | 91·6 | – | JL3b (D87017) | |
| 24a | 3× | 3 | 3r.9 Cs/DPL23… + (Z73647) | 3r | 93·3 | – | JL2/3a (M15614) |
| 29a | 2 | 2b2.400B5 + (Z73665) | 2b2 | 98·1 | – | JL2/3a (M15614) | |
| 30b | 2 | 2a2.272A12/DPL11… | 2a2 | 89·8 | – | JL2/3a (M15614) | |
Numbers in parentheses indicate EMBL accession numbers. Sequence names are according to the V-BASE database from May 1999; dots indicate abbreviated names and/or the existence of synonyms; +, a mapped chromosomal location; unkn., an unknown gene locus; frequency indicates the number of identical clones selected.
Derived from the IMGT database.
IMGT listed the V3–33*05 allele from locus 3–33 with the same homology as V3–30.*03.
IMGT synonyms for 3l, 3r, and 2a2 are IGLV3–19, IGLV3–1, and IGLV2–14, respectively.
RESULTS
Library optimization
From a female adult volunteer without history of autoimmune disease, combinatorial phage display antibody libraries of IgM and IgG type were established. The complete H and L chain containing IgM library (MM) had about 8 × 106 members. Eight out of 10 (80%) clones contained both H and L chain inserts, as estimated by analysing restricted DNA from single colonies and the total phagemid DNA. The IgG library MG consisted of about 4 × 106 members. Eight out of 10 (80%) clones contained both H and L chain inserts. To optimize the libraries, the recombinant plasmids were purified and separated on a gel. After retransformation of E. coli with the purified band corresponding to pComb3H with both the H and L chain inserts, the MM library had 7 × 106 and the MG library 1·1 × 107 independent clones. All analysed clones had both inserts of the right size.
IVIG-selected Fab-phages
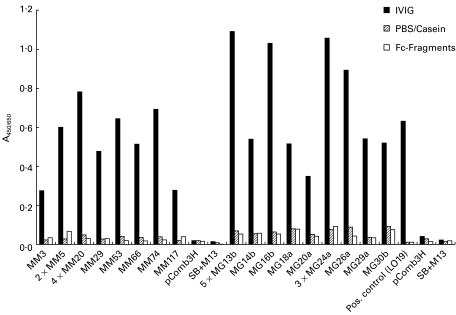
After the final panning step, most Fab-phage supernatants from individual colonies from both libraries grown in a 48-well plate were bound to IVIG-coated wells with an A450 above 0·2 (Table 1,Fig. 01). Controls consisting of SB medium with helper phage or pComb3H-transfected E. coli phage supernatant were negative. Most selected clones showed a medium to high A450 for IVIG, but no binding to Fc fragments or PBS/casein-coated wells was seen. A450 of IgG Fab-expressing phage appeared somewhat higher than that of IgM Fab (Table 1,Fig. 1). In contrast, Fab-phage supernatants from the original, unpanned libraries were negative (A450 < 0·2) or showed only a weak reaction with IVIG (Table 1).
Table 1.
Summary of ELISA data of individual Fab phage supernatants on intravenous immunoglobulin preparations (IVIG) before and after panning with IVIG*
| A450/650 | < 0·1 | > 0·1 | > 0·2 | > 0·3 | > 0·4 | > 0·5 | > 0·6 | > 0·7 | > 0·8 | > 0·9 | > 1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM unpanned | n = 82 | 39 | 38 | 4 | 1 | |||||||
| MM after panning | n = 75 | 9 | 16 | 9 | 12 | 9 | 12 | 3 | 4 | 1 | ||
| MG unpanned | n = 86 | 42 | 39 | 3 | 2 | |||||||
| MG after panning | n = 41 | 2 | 3 | 4 | 4 | 4 | 9 | 5 | 2 | 2 | 2 | 4 |
The positive control autoimmune thrombocytopenia (AITP) clone LO19 had an A450 of > 0·6; the negative controls of pComb3H- transformed phage supernatants and VCSM13-containing SB medium were < 0·1.
Fig. 1.
Summary of two independent ELISAs with representative Fab-phage supernatants from the MM and MG libraries after the 4th round of panning on intravenous immunoglobulin preparations (IVIG). Coated proteins consisted of IVIG or Fc fragments in equal concentrations as well as PBS/casein blocking buffer to also detect non-specific binding. Negative controls for each ELISA were pComb3H-transfected Escherichia coli phage supernatant (pComb3H) and SB medium containing VCSM13 helper phage (SB + M13). Bound Fab-phages were detected with horseradish peroxidase-labelled anti-M13 antibodies.
Genetic origin and mutation rates of IVIG-bound Fab
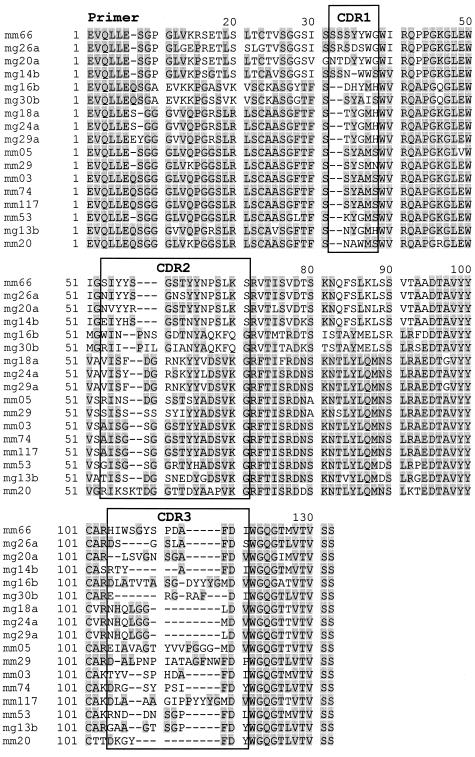
The variable regions of a panel of IVIG-selected Fab from both libraries were sequenced (Figs 2 and 3). EMBL accession numbers are AJ132847–AJ132856 for MG-20a, 24a, 29a, 14b, 30b, and AJ271547–AJ271557 and AJ272334 for the rest. All functional heavy chains were further investigated here, although none of the IVIG-binding IgM clones was associated with a functional light chain and four IgG lacked functional light chains. The sequences belonged to various VH families, with the heavy chains belonging to families 1, 3 and 4, and the light chains to λ 2 and 3 (Table 2). The VHCDR3 regions displayed an overall high degree of variation in length and sequence, ranging between eight (MG-14b) and 18 (MG-16b) amino acids (Fig. 2). From both libraries some individual clones were selected repetitively (Table 2). Interestingly, the three clones MG-18a, 24a, and 29a had the same VHCDR3 except one amino acid exchange from leucine to methionine for clone MG-24a. However, they displayed differences elsewhere in the CDR2 and framework regions (Fig. 2) and were paired with different light chains, arguing against—but not excluding—PCR artefacts.
Fig. 2.
Alignment of the VH amino acids from intravenous immunoglobulin preparation (IVIG)-selected Fabs of both libraries according to their highest homology. CDR regions according to the Kabat definition [21] are indicated. The start of the sequences (approximately eight amino acids) is determined by the polymerase chain reaction primers.
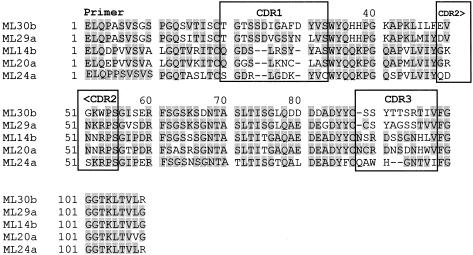
Fig. 3.
Alignment of the functional VL amino acids from intravenous immunoglobulin preparation (IVIG)-bound IgG according to their highest homology.
The identity of the VH regions, listed according to the Kabat numbering scheme [21], to putative germ-line genes was between 95·2% and 100% and 92·3% and 99·6% for the IgM and IgG clones, respectively. The homology for light chains to germ-line genes was between 89·8% and 100% (Table 2). While 5/9 of the different IgG heavy chains had a ratio of replacement mutations above 2·9 in the CDRs, this was observed for only 1/8 IgM clones (Table 3). In contrast, 6/8 IgM VH had no replacement mutation at all in the CDRs compared with 2/9 IgG VH (Table 3). Four of five light chains showed a high mutation rate, suggesting that they were derived from antigen-selected B cells, while the L chain of MG14b was in germ-line configuration.
Table 3.
Mutation rates in intravenous immunoglobulin preparation (IVIG)-selected VH and VL of MM and MG*
| CDRs | FRs | |||||
|---|---|---|---|---|---|---|
| Entire V-region | ||||||
| No./total | % | No./total | % | |||
| Clone | R/S | Ratio | R/S | Ratio | No./total | % |
| Heavy chains of MM | ||||||
| 03 | 0/66 | 0 | 1/204 | 0·5 | 1/270 | 0·4 |
| 0 | 0 | 0/1 | 0 | |||
| 05 | 6/66 | 9·1 | 4/204 | 1·96 | 10/270 | 3·7 |
| 4/2 | 2 | 0/4 | 0 | |||
| 20 | 0/72 | 0 | 2/204 | 0·98 | 2/276 | 0·7 |
| 0 | 0 | 1/1 | 1 | |||
| 29 | 1/66 | 1·5 | 0/204 | 0 | 1/270 | 0·4 |
| 0/1 | 0 | 0 | 0 | |||
| 53 | 9/66 | 13·6 | 4/204 | 1·96 | 13/270 | 4·8 |
| 8/1 | 8 | 3/1 | 3 | |||
| 66 | 0/69 | 0 | 1/204 | 0·5 | 1/273 | 0·4 |
| 0 | 0 | 1/0 | ∞ | |||
| 74 | 0/66 | 0 | 0/204 | 0 | 0/270 | 0 |
| 0 | 0 | 0 | 0 | |||
| 117 | 0/66 | 0 | 1/204 | 0·4 | 1/270 | 0·4 |
| 0 | 0 | 0/1 | 0 | |||
| Heavy chains of MG | ||||||
| 13b | 9/66 | 13·6 | 8/204 | 3·9 | 17/270 | 6·3 |
| 7/2 | 3·5 | 4/4 | 1 | |||
| 14b | 0/66 | 0 | 2/204 | 0·98 | 2/270 | 0·7 |
| 0 | 0 | 1/1 | 1 | |||
| 16b | 3/66 | 4·5 | 5/204 | 2·5 | 8/270 | 3 |
| 3/0 | ∞ | 2/3 | 0·67 | |||
| 18a | 5/66 | 7·6 | 6/204 | 2·9 | 11/270 | 4·1 |
| 4/1 | 4 | 3/3 | 1 | |||
| 20a | 8/69 | 11·6 | 7/204 | 3·4 | 15/273 | 5·5 |
| 7/1 | 7 | 5/2 | 2·5 | |||
| 24a | 7/66 | 10·6 | 6/204 | 2·9 | 13/270 | 4·8 |
| 5/2 | 2·5 | 1/5 | 0·2 | |||
| 26a | 7/69 | 10·1 | 14/204 | 6·9 | 21/273 | 7·7 |
| 5/2 | 2·5 | 9/5 | 1·8 | |||
| 29a | 5/66 | 7·6 | 5/204 | 2·5 | 10/270 | 3·7 |
| 4/1 | 4 | 2/3 | 0·6 | |||
| 30b | 0/66 | 0 | 1/204 | 0·5 | 1/270 | 0·4 |
| 0 | 0 | 0/1 | 0 | |||
| Light chains of MG | ||||||
| 14b | 0/78 | 0 | 0/171 | 0 | 0/249 | 0 |
| 0 | 0 | 0 | 0 | |||
| 20a | 12/78 | 15·4 | 9/171 | 5·2 | 21/249 | 8·4 |
| 10/2 | 5 | 6/3 | 2 | |||
| 24a | 12/78 | 15·4 | 5/177 | 2·8 | 17/255 | 6·7 |
| 8/4 | 2 | 3/2 | 1·5 | |||
| 29a | 2/87 | 2·3 | 3/177 | 1·7 | 5/264 | 1·9 |
| 1/1 | 1 | 3/0 | ∞ | |||
| 30b | 13/87 | 14·9 | 14/177 | 7·9 | 27/264 | 10·2 |
| 9/4 | 2·25 | 9/5 | 1·8 | |||
No./total, Number of mutations/number of all bases in the region, R/S represents number of replacement/silent mutations, ratio is the R/S ratio. Although division by zero is not mathematically correct, ∞ and 0 are listed as results for clarity. Due to the complexity of the V(D)J joining, only the V genes are considered in these calculations.
Four of eight of the different (4/12 in total) IgM VH were derived from the germ-line locus 3–23, while 4/9 different (10/15 in total) IgG VH originated from the 3–30 group. All but one of the light chains were from the loci 2a2, 3l, and 3r (Table 2).
As a control, eight randomly picked clones from the unpanned IgM library were sequenced. None was derived from locus 3–23. Three of eight clones had no functional VH and 4/8 were derived from germ-line genes different from the IVIG-selected clones. These loci were 3–7, 3–33, 4–30.1/4–31, and 4–59. Only 1/8 unselected clones was derived from a locus also observed among the IVIG-selected clones, namely 4–39. In contrast to IVIG-bound IgM, some of the unselected IgM contained apparently functional light chains, including those derived from germ-line loci 3l (1x) and 2a2 (2x).
No platelet binding of IVIG-selected Fab phages
The IVIG-selected IgG Fab-phage supernatants with both functional heavy and light chains were analysed for platelet binding in various ELISAs previously [15]. Applying the same criteria as for AITP-derived clones, all five clones from the library MG were negative. However, Fab phages from clones MG-24a and MG-29a showed a mean A450 of 0·108 and 0·133, respectively, on platelets from a donor with blood group A, rhesus negative. This was above the signal of the negative AITP clone LO19 (0·053) but well below the AITP clones NK22 (0·205) and LO46 (0·472) used as positive controls in this experiment [15]. Similar negative results were obtained with IVIG-selected Fab-phages from a patient with systemic lupus erythematosus (SLE) without symptoms of thrombocytopenia (unpublished results).
DISCUSSION
Previously we observed that IVIG-bound antibody Fab fragments cloned from patients with AITP originated from a limited set of germ-line genes. The aim of the current study was to analyse and compare at the molecular level IVIG-selected Fabs of both IgM and IgG class from a healthy individual. Therefore we selected clones from two phage-displayed antibody Fab libraries by biopanning with immobilized IVIG. The advantage of using Fab-phages instead of soluble Fab in the various assays allowed for specific detection of bound Fab-phages with labelled anti-M13 antibodies which did not cross-react with human IVIG.
Idiotypic interaction of IVIG with selected heavy and light chains
Our previous studies suggested that IVIG bound in an anti-idiotypic manner to the Fab molecules [14,15]. The method of biopanning with IVIG apparently did not select for original heavy and light chain pairing [14]. Instead, high affinity of IVIG molecules for either antibody chain seemed to result in enrichment of particular Fab-phages. Clones with certain combinations of two functional antibody chains often grew much more slowly in culture than other combinations or clones with only one insert [14], probably due to toxic effects of the bacteria [23]. Obviously, high affinity of IVIG for a particular heavy chain alone was often sufficient for selection, resulting in many clones without a functional light chain, especially in case of the IgM clones. The pComb3H system would not allow for surface display of light chains without a heavy chain that was in frame with the gene 3 fragment of the phage. The absence of IVIG-reactive clones among samples from the original, unpanned libraries on one hand and the enrichment of many identical clones, respectively, within the selected clones confirms a specific selection of phage-displayed heavy chains by IVIG, also in cases where a light chain is missing.
Genetic origin of IVIG-selected heavy and light chains
All but one of the antibody chains (MG-30b VH) isolated by selection with IVIG (Table 2) were derived from putative germ-line genes that have been mapped recently [20] and were previously shown to be functionally expressed [20]. The gene locations 3–30 and 3–30/3–30.5 of the closely related germ-line genes V3–30 and DP49/1.9III, respectively, that are identical in all but one base were the predominant source for selected VH genes in the IgG library. In contrast, the VH gene selected most frequently among different clones from the IgM library was DP47/V3–23, which is located at 3.23 and observed often in the peripheral repertoire [24] as well as in the bone marrow [25]. Both germ-line segments were also the most frequently observed VH origins of IVIG-selected IgG from three patients with AITP [14] and from a patient with SLE lacking symptoms of thrombocytopenia (unpublished results). In case of the light chains, all but one were derived from the germ-line genes 3r9C5/DPL23, DPL16/VL3.1, or 2a2.27A12/DPL11 corresponding to the loci 3r, 3l, and 2a2, respectively. Again, the same light chain germ-line genes have also been detected most frequently in AITP [14]. However, the light chain origin appears to be of only minor importance for the selection by IVIG, because many Fab-phages without functional light chains were strongly bound by IVIG, and light chain loci 3l and 2a2 were also observed among 3/8 unpanned clones.
A literature search of publications on autoantibodies derived by combinatorial libraries revealed that the same germ-line gene loci dominating in the IVIG selected clones were also frequently used for coding other autoantibodies [14]. Despite their origin from the same germ-line genes, many of these patient-derived autoantibodies were highly specific for different autoantigens. These included dsDNA, the acetylcholine receptor, thyroid peroxidase, the pyruvate dehydrogenase E2 subunit, and SS-A/Ro60 [14]. In addition, autoantibodies from healthy, non-immunized individuals and a synthetic library were also derived from those gene loci. More recent articles on sequenced autoantibodies [26–28] confirmed our initial observation, showing that a growing list of autoantibodies isolated with different phage display systems were derived from VH germ-line genes 3–30, including 3–30/3–30.5 and 3–23 or VL segments 3l, 3r, and 2a2. Interestingly, these germ-line genes were also observed in the immune response to viruses [29].
Autoantibodies derived from 3–30 and 3l were also observed by single-cell PCR of B cells from the salivary glands of patients with Sjögren's syndrome [30]. Both 3–30 and 3–23 homologous sequences have also been reported for various autoantibodies including rheumatoid factors derived from Epstein–Barr virus (EBV)-transformed B cells and hybridomas, and were frequently observed in the fetal repertoire [31,32]. It was suggested that this correlation between autoantibody VH and early repertoire VH may be due to positive selection by self-antigens [31].
In control ELISAs, clones from the unpanned libraries did not react with IVIG, and sequencing of randomly picked clones from several libraries in our laboratory, including the IgM library of this study, revealed a variety of different germ-line genes not observed among the IVIG-bound Fabs ([14] and unpublished results). Furthermore, when using other antigens such as melanoma cells [33], Fc fragments [13], or c-erbB-2 [34] for panning, antibodies were found to be derived from different germ-line genes, arguing against a common skewing of combinatorial libraries.
A recent study by De Wildt et al. [35] investigated the pairings of VH and VL from 365 human IgG+ B cells. Interestingly, the VH germ-line genes observed most often in this report were also 3–23 and 3–30, while the VL segments 3l, 3r and 2a2 were in the medium range [35]. The over-representation of certain VH genes, especially those derived from the large VH3 family, was also observed in previous analyses in which a few gene segments dominated the investigated B cell repertoire [24,25,31,36–40]. Among the VH3 family, the 3–23 gene dominated early pre-B, as well as immature and mature B cell repertoires investigated from VH gene libraries [24], and from bone marrow and peripheral blood of adults [25]. A study of Huang et al. analysing over 4500 independent VH3-containing rearrangements from 12 subjects also observed 3–23 and 3–30 as the most frequent rearrangements, with a mean percentage of 24·3 and 16·1, respectively [36].
The importance of the 3–30 germ-line gene group for the regulation of autoreactivity is further documented in reports by Chen's group. They observed that the developmentally regulated Humhv3005 gene, an allelic variant from locus 3–30 that is 99% homologous to the 1.9III gene, was absent in 24% of patients with SLE. The same gene was also homozygously deleted in 31·8% of patients with chronic AITP, but only in 8% of normals [41].
In our current study, 3–23 was only observed among the IVIG-bound IgM clones, while 3–30 was only among the IgG. In contrast, in the previous study with AITP patients we observed both germ-line segments among IgG clones [14]. The reason for these differences is not clear at the moment and its significance will be the focus of future studies. In this regard, a single IgM IVIG binder selected among many IgGs from another library in our laboratory also originated from 3–23. This IgM Fab-phage resulted from a mismatch of the cG1z IgG constant region primer to IgM during PCR and therefore competed with the excess of IgG-presenting phages during the biopanning on IVIG (unpublished results).
Implications for the specific functions of IVIG
In conclusion, our current results in combination with the growing information from the literature on the expressed B cell repertoire extend our earlier conclusions [14] regarding normal B cell repertoires. The preferred binding of IVIG to antibodies derived predominantly from the two germ-line gene segments 3–30 and 3–23 that (i) code for many different autoantibodies and (ii) are the most frequently used in building normally expressed B cell repertoires suggests a basic principle. It supports the model of a V-region-connected, anti-idiotypic network interaction of antibodies and B cell receptors [6,7,42,43].
However, our current data show for the first time the genetic origin of these IgG and IgM antibodies targeted by IVIG in a healthy individual. They implicate a favoured interaction of IVIG, and normal IgG repertoires, with antibodies and B cell receptors derived from the VH 3–30 germ-line group and the 3–23 gene segment. Because these genes are the most frequently expressed germ-line genes and exist in duplications and many allelic variations [36], this interaction may be very important for the development and control of the normal B cell repertoire, in addition to biased rearrangements [44] during V–DJ recombination. IVIG and normal IgG may regulate 3–30 and 3–23 derived B cells either by activation or, when activating T cell signals are absent, by down-regulation of the autoreactive fraction [45]. The exact mechanism and potential clinical usefulness may now be further analysed at a molecular level by cloning and identification of the individual, regulatory anti-idiotypes with the help of the described IVIG targets.
Acknowledgments
We thank Dr C. Barbas from The Scripps Research Institute (La Jolla, CA) for providing the pComb3H vector and Dr R. Handgretinger (University Tübingen) for providing blood samples. This research was supported by grant GA 167/6-1 and 6-2 from the Deutsche Forschungsgemeinschaft. The paper contains data from the dissertation of M.H.
References
- 1.Imbach P, Wagner HP, Berchtold W, et al. Intravenous immunoglobulin versus oral corticosteroids in acute immune thrombocytopenic purpura in childhood. Lancet. 1985;2:464–8. doi: 10.1016/s0140-6736(85)90400-3. [DOI] [PubMed] [Google Scholar]
- 2.Fischer P, Uttenreuther-Fischer MM, Naoe S, Gaedicke G. Kawasaki disease: update on diagnosis, treatment and a still controversial etiology. Pediatr Hematol Oncol. 1996;13:487–501. doi: 10.3109/08880019609030864. [DOI] [PubMed] [Google Scholar]
- 3.Debre M, Bonnet MC, Fridman WH, et al. Infusion of Fc gamma fragments for treatment of children with acute immune thrombocytopenic purpura. Lancet. 1993;342:945–9. doi: 10.1016/0140-6736(93)92000-j. [DOI] [PubMed] [Google Scholar]
- 4.Bjorkholm M. Intravenous immunoglobulin treatment in cytopenic haematological disorders. J Intern Med. 1993;234:119–26. doi: 10.1111/j.1365-2796.1993.tb00720.x. [DOI] [PubMed] [Google Scholar]
- 5.Levy Y, Sherer Y, George J, et al. Serologic and clinical response to treatment of systemic vasculitis and associated autoimmune disease with intravenous immunoglobulin. Int Arch Allergy Immunol. 1999;119:231–8. doi: 10.1159/000024199. [DOI] [PubMed] [Google Scholar]
- 6.Lacroix-Desmazes S, Mouthon L, Spalter SH, Kaveri S, Kazatchkine MD. Immunoglobulins and the regulation of autoimmunity through the immune network. Clin Exp Rheumatol. 1996;14:S9–S15. [PubMed] [Google Scholar]
- 7.Jerne NK. Towards a network theory of the immune system. Ann Immunol (Paris) 1974;125C:373–89. [PubMed] [Google Scholar]
- 8.Imholz B, Imbach P, Baumgartner C, et al. Intravenous immunoglobulin (i.v. IgG) for previously treated acute or for chronic idiopathic thrombocytopenic purpura (ITP) in childhood: a prospective multicenter study. Blut. 1988;56:63–68. doi: 10.1007/BF00633464. [DOI] [PubMed] [Google Scholar]
- 9.Abu-Shakra M, Buskila D, Shoenfeld Y. Idiotypes and anti-idiotypic antibodies. In: Peter JB, Shoenfeld Y, editors. Autoantibodies. Amsterdam: Elsevier; 1996. [Google Scholar]
- 10.Barbas CFI, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991;88:7978–82. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitling F, Dübel S, Seehaus T, Klewinghaus I, Little M. A surface expression vector for antibody screening. Gene. 1991;104:147–53. doi: 10.1016/0378-1119(91)90244-6. [DOI] [PubMed] [Google Scholar]
- 12.Duchosal MA, Eming SA, Fischer P, et al. Immunization of hu-PBL-SCID mice and the rescue of human monoclonal Fab fragments through combinatorial libraries. Nature. 1992;355:258–62. doi: 10.1038/355258a0. [DOI] [PubMed] [Google Scholar]
- 13.Yang YY, Fischer P, Leu SJ, Zhu M, Woods VL, Jr, Chen PP. Possible presence of enhancing antibodies in idiopathic thrombocytopenic purpura. Br J Haematol. 1999;104:69–80. doi: 10.1046/j.1365-2141.1999.01144.x. [DOI] [PubMed] [Google Scholar]
- 14.Jendreyko N, Uttenreuther-Fischer MM, Lerch H, Gaedicke G, Fischer P. Genetic origin of IgG antibodies cloned by phage display and antiidiotypic panning from three patients with autoimmune thrombocytopenia. Eur J Immunol. 1998;28:4236–47. doi: 10.1002/(SICI)1521-4141(199812)28:12<4236::AID-IMMU4236>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 15.Fischer P, Jendreyko N, Hoffmann M, Lerch H, Uttenreuther-Fischer MM, Chen PP, Gaedicke G. Platelet reactive IgG antibodies cloned by phage display and panning with IVIG from three patients with autoimmune thrombocytopenia. Br J Haematol. 1999;105:626–40. doi: 10.1046/j.1365-2141.1999.01407.x. [DOI] [PubMed] [Google Scholar]
- 16.Barbas CFI, Burton DR. 1993 Cold Spring Harbor Laboratory course on: monoclonal antibodies from combinatorial libraries. La Jolla: The Scripps Research Inst.; 1993. [Google Scholar]
- 17.Barbas CFI, Lerner RA. Combinatorial immunoglobulin libraries on the surface of phage (Phabs): rapid selection of antigen-specific Fabs. Methods Companion Methods Enzymol. 1991;2:119–24. [Google Scholar]
- 18.Fischer P, Leu SJC, Yang YY, Chen PP. Rapid simultaneous screening for DNA integrity and antigen specificity of clones selected by phage display. Biotechniques. 1994;16:828–30. [PubMed] [Google Scholar]
- 19.Graus YF, Debaets MH, Parren PWHI, Berrihaknin S, Wokke J, Vriesman PJV, Burton DR. Human anti-nicotinic acetylcholine receptor recombinant Fab fragments isolated from thymus-derived phage display libraries from myasthenia gravis patients reflect predominant specificities in serum and block the action of pathogenic serum antibodies. J Immunol. 1997;158:1919–29. [PubMed] [Google Scholar]
- 20.Tomlinson IM, Williams SC, Ignatovich O, Corbett SJ, Winter G. Cambridge: MRC Centre for Protein Engineering; 1998. V BASE sequence directory. [Google Scholar]
- 21.Kabat EA, Wu TT, Perry HM, Gottesmann KS, Foeller C. 5. Bethesda: NIH; 1991. Sequences of proteins of immunological interest. [Google Scholar]
- 22.Lefranc MP, Giudicelli V, Ginestoux C, et al. IMGT, the international ImMunoGeneTics database. Nucl Acids Res. 1999;27:209–12. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knappik A, Plückthun A. Engineered turns of a recombinant antibody improve its in vivo folding. Protein Eng. 1995;8:81–89. doi: 10.1093/protein/8.1.81. [DOI] [PubMed] [Google Scholar]
- 24.Stewart AK, Huang C, Stollar BD, Schwartz RS. High-frequency representation of a single VH gene in the expressed human B cell repertoire. J Exp Med. 1993;177:409–18. doi: 10.1084/jem.177.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraj P, Rao SP, Glas AM, Hardy RR, Milner ECB, Silberstein LE. The human heavy chain Ig V region gene repertoire is biased at all stages of B cell ontogeny, including early pre-B cells. J Immunol. 1997;158:5824–32. [PubMed] [Google Scholar]
- 26.Hoet RMA, Raats JMH, de Wildt R, Dumortier H, Muller S, Van den Hoogen F, van Venrooij WJ. Human monoclonal autoantibody fragments from combinatorial antibody libraries directed to the U1snRNP associated U1C protein; epitope mapping, immunolocalization and V-gene usage. Mol Immunol. 1998;35:1045–55. doi: 10.1016/s0161-5890(98)00093-5. [DOI] [PubMed] [Google Scholar]
- 27.Rubinstein DB, Leblanc P, Wright DG, Guillaume T, Strotchevoi A, Boosalis M. Anti-CD34 (+) fabs generated against hematopoietic stem cells in HIV-derived combinatorial immunoglobulin library suggest antigen-selected autoantibodies. Mol Immunol. 1998;35:955–64. doi: 10.1016/s0161-5890(98)00075-3. [DOI] [PubMed] [Google Scholar]
- 28.Guo J, Mcintosh RS, Czarnocka B, Weetman AP, Rapoport B, McLachlan SM. Relationship between autoantibody epitopic recognition and immunoglobulin gene usage. Clin Exp Immunol. 1998;111:408–14. doi: 10.1046/j.1365-2249.1998.00492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikematsu W, Kobarg J, Ikematsu H, Ichiyoshi Y, Casali P. Clonal analysis of a human antibody response. III. Nucleotide sequences of monoclonal IgM, IgG, and IgA to rabies virus reveal restricted VK gene utilization, junctional V kappa J kappa and V lambda J lambda diversity, and somatic hypermutation. J Immunol. 1998;161:2895–905. [PubMed] [Google Scholar]
- 30.Stott DI, Hiepe F, Hummel M, Steinhauser G, Berek C. Antigen-driven clonal proliferation of a cells within the target tissue of an autoimmune disease—the salivary glands of patients with Sjögren's syndrome. J Clin Invest. 1998;102:938–46. doi: 10.1172/JCI3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda F, Shin EK, Nagaoka H, et al. Structure and physical map of 64 variable segments in the 3′0.8-megabase region of the human immunoglobulin heavy-chain locus. Nat Genet. 1993;3:88–94. doi: 10.1038/ng0193-88. [DOI] [PubMed] [Google Scholar]
- 32.Adderson EE, Shikhman AR, Ward KE, Cunningham MW. Molecular analysis of polyreactive monoclonal antibodies from rheumatic carditis: human anti-N-acetylglucosamine anti-myosin antibody V region genes. J Immunol. 1998;161:2020–31. [PubMed] [Google Scholar]
- 33.Noronha EJ, Wang XH, Desai SA, Kageshita T, Ferrone S. Limited diversity of human scFv fragments isolated by panning a synthetic phage-display scFv library with cultured human melanoma cells. J Immunol. 1998;161:2968–76. [PubMed] [Google Scholar]
- 34.Schier R, Bye J, Apell G, Mccall A, Adams GP, Malmqvist M, Weiner LM, Marks JD. Isolation of high-affinity monomeric human Anti-c-erbB-2 single chain Fv using affinity-driven selection. J Mol Biol. 1996;255:28–43. doi: 10.1006/jmbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 35.Dewildt RMT, Hoet RMA, Vanvenrooij WJ, Tomlinson IM, Winter G. Analysis of heavy and light chain pairings indicates that receptor editing shapes the human antibody repertoire. J Mol Biol. 1999;285:895–901. doi: 10.1006/jmbi.1998.2396. [DOI] [PubMed] [Google Scholar]
- 36.Huang SC, Jiang RH, Glas AM, Milner ECB. Non-stochastic utilization of Ig V region genes in unselected human peripheral B cells. Mol Immunol. 1996;33:553–60. doi: 10.1016/0161-5890(95)00162-x. [DOI] [PubMed] [Google Scholar]
- 37.Huang C, Stollar BD. A majority of Ig H chain cDNA of normal human adult blood lymphocytes resembles cDNA for fetal Ig and natural autoantibodies. J Immunol. 1993;151:5290–300. [PubMed] [Google Scholar]
- 38.Pascual V, Victor K, Randen I, et al. Nucleotide sequence analysis of rheumatoid factors and polyreactive antibodies derived from patients with rheumatoid arthritis reveals diverse use of VH and VL gene segments and extensive variability in CDR- 3. Scand J Immunol. 1992;36:349–62. doi: 10.1111/j.1365-3083.1992.tb03108.x. [DOI] [PubMed] [Google Scholar]
- 39.Sanz I, Casali P, Thomas JW, Notkins AL, Capra JD. Nucleotide sequences of eight human natural autoantibody VH regions reveals apparent restricted use of VH families. J Immunol. 1989;142:4054–61. [PubMed] [Google Scholar]
- 40.Schroeder HW, Jr, Hillson JL, Perlmutter RM. Early restriction of the human antibody repertoire. Science. 1987;238:791–3. doi: 10.1126/science.3118465. [DOI] [PubMed] [Google Scholar]
- 41.Mo L, Leu SJC, Berry C, et al. The frequency of homozygous deletion of a developmentally regulated Vh gene (Humhv3005) is increased in patients with chronic idiopathic thrombocytopenic purpura. Autoimmunity. 1996;24:257–63. doi: 10.3109/08916939608994718. [DOI] [PubMed] [Google Scholar]
- 42.Coutinho A. The network theory: 21 years later. Scand J Immunol. 1995;42:3–8. doi: 10.1111/j.1365-3083.1995.tb03619.x. [DOI] [PubMed] [Google Scholar]
- 43.Chen PP, Carson DA, Fischer P, et al. Molecular genetics of human autoantibodies. Pathol Biol. 1994;42:798–800. [Google Scholar]
- 44.Rao SP, Riggs JM, Friedman DF, Scully MS, Lebien TW, Silberstein LE. Biased V-H gene usage in early lineage human B cells: evidence for preferential Ig gene rearrangement in the absence of selection. J Immunol. 1999;163:2732–40. [PubMed] [Google Scholar]
- 45.Tighe H, Warnatz K, Brinson D, Corr M, Weigle WO, Baird SM, Carson DA. Peripheral deletion of rheumatoid factor B cells after abortive activation by IgG. Proc Natl Acad Sci USA. 1997;94:646–51. doi: 10.1073/pnas.94.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]