Abstract
We previously described autoantibodies against a UGA serine tRNA–protein complex (tRNP(Ser)Sec) in patients with type-1 autoimmune hepatitis [1] and now define the specificity and frequency of this autoantibody and the DNA sequence encoding the tRNA(Ser)Sec-associated antigenic protein. The presence of anti‐tRNP(Ser)Sec antibodies was highly specific for type-1 autoimmune hepatitis, as 47·5% of patients were positive compared with none of the control subjects. To characterize the antigenic protein(s), we immunoscreened a human cDNA library with anti-tRNP(Ser)Sec-positive sera. Two clones (19 and 13) were isolated. Clone 19 encodes a protein with a predicted molecular mass of 48·8 kD. Clone 13 is a shorter cDNA, almost identical to clone 19, which encodes a 35·9-kD protein. Expression of both cDNAs was accomplished in Escherichia coli as His-tagged recombinant proteins. Antibodies eluted from both purified recombinant proteins were able to immunoprecipitate the tRNA(Ser)Sec from a HeLa S3 cell extract, demonstrating their cross-reactivity with the mammalian antigenic complex. Recent cloning data relating to the target antigen(s) of autoantibodies in autoimmune hepatitis patients that react with a soluble liver antigen (SLA) and a liver-pancreas antigen (LP) have revealed that these two autoantibodies are identical and that the cloned antigen shows 99% amino acid sequence homology with tRNP(Ser)Sec.
Keywords: tRNP(Ser)Sec, UGA suppressor tRNA–protein complex, autoantibodies, autoantigen, autoimmune hepatitis
Introduction
Autoimmune hepatitis defines a subgroup of chronic liver diseases of unknown cause and encompasses a heterogeneous group of syndromes in which patients appear to lose immunological tolerance to the liver [2,3]. Many autoantibodies have been described in autoimmune hepatitis [1,4,5] and some define patients with distinctive clinical, laboratory and prognostic features [1,6–9]. Seropositivity for anti-nuclear antibodies (ANA) and/or smooth muscle antibodies (SMA) characterizes patients with type 1 autoimmune hepatitis (AIH), whereas seropositivity for antibodies to liver/kidney microsome type 1 (anti-LKM1) typifies patients with type 2 AIH [10]. As the presence of SMA and/or ANA has no prognostic value [11], new markers should be investigated to characterize further these subtypes of AIH. Patients in each of these subgroups have mutually exclusive autoantibodies with different clinical manifestations, genetic associations [12] and responses to therapy [13].
Antibodies anti-serine tRNA–protein complexes (tRNP(Ser)Sec) were described in an earlier paper [1] in a subgroup of patients with type-1 AIH, which is recalcitrant to corticosteroid therapy. These antibodies precipitate a 90-nucleotide RNA from human whole cell extracts and recognize a 48-kD polypeptide in immunoblot assays [1]. The RNA is a UGA suppressor serine tRNA (tRNA(Ser)Sec) (where Sec is selenocysteine) as shown by sequence analysis, and it functions in the pathway of selenoprotein synthesis in human cells. This tRNA is a requisite for the co-translational incorporation of selenocysteine into growing polypeptide chains [14]. The insertion of selenocysteine is directed by certain UGA triplets, which in other contexts act as termination codons. In brief, a specialized tRNA (tRNA(Ser)Sec) is initially charged with serine to form seryl-tRNASec, and is then converted to selenocysteyl-tRNASec by the action of a selenocysteine synthase, a selenophosphate synthetase, and factors not yet clearly defined. Moreover, a tRNASec-specific elongation factor, performing the function executed by the elongation factor Tu for all other aminoacyl-tRNAs, is required for the synthesis of selenoproteins [15]. As described in prokaryotes, strong evidence indicates the existence of a translational elongation factor in eukaryotes for insertion of selenocysteine into protein [14,16]. The antigenic 48-kD protein associated with the UGA suppressor tRNA may be a selenocysteine-specific elongation factor, or an enzyme involved in the conversion of seryl-tRNA(Ser)Sec to selenocysteyl-tRNA(Ser)Sec, some other unknown SECIS (selenocysteine-insertion sequence)-binding protein (SBP), or another unknown factor acting in the selenocysteine insertion pathway. In order to elucidate the precise nature of this antigenic protein and its relationship with some of these previously described factors, we cloned, sequenced and expressed the cDNA of the 48-kD antigenic protein recognized by autoantibodies from patients with type-1 AIH.
PATIENTS and METHODS
Sera
Fifty-nine patients who satisfied international criteria for the diagnosis of autoimmune hepatitis [17] were selected from 303 patients in the chronic hepatitis treatment programme of the Mayo Clinic because they fulfilled the following additional criteria: (i) all patients had been screened for the serologic markers of hepatitis B and C virus infection by second generation assays and had been found negative [7,8]; (ii) ANA (68%) and/or SMA (88%) had been demonstrated in each patient at admission, and the presence of one or both markers had justified their designation as type-1 AIH [11]; (iii) the observation period ranged from 7 to 348 months (mean 125·6 months). The mean age at diagnosis of AIH was 39 years (range 16–68 years); (iv) all patients had received immunosuppressive therapy consisting of prednisone monotherapy (n = 17) or combination therapy consisting of azathioprine and prednisone (n = 42) according to previously published protocols [18]. All patients were participants in a research programme that had been approved by the Institutional Review Board of the Mayo Clinic. Patients were evaluated at presentation and were followed in a uniform fashion in accordance with a pre-established protocol [18]. Complete examinations were performed every 6 months during and immediately after treatment and then at annual intervals if the clinical condition was stable. During a follow up of approximately 10 years, eight patients died of liver failure. Each treatment had been shown previously to be equally effective in the management of severe type-1 AIH and superior to placebo or non-steroidal regimens [19]. The average duration of treatment was 27 ± 3 months.
Liver tissue was obtained by needle biopsy in all patients at the time of presentation. Additional assessments were made as indicated to document histological remission or to clarify clinical status. Specimens were interpreted under code and the diagnosis of cirrhosis required fibrosis and the presence of a complete regenerative nodule. The histological designations of interface hepatitis, bridging necrosis and multilobular necrosis required satisfaction of previously published criteria [20]. Moderate to severe interface hepatitis was the most advanced histological pattern at presentation in 25 (42%) patients; bridging necrosis was present in six (10%) patients; multilobular necrosis in 10 (17%) patients; and cirrhosis in 18 (30·5%) patients.
As control subjects, we studied 15 patients with type-2 AIH, 10 patients with chronic hepatitis B, 44 patients with chronic hepatitis C; 20 patients with anti-M2 antibody-positive primary biliary cirrhosis (PBC), three patients with primary sclerosing cholangitis who were positive for neutrophil-specific autoantibodies, five patients with alcoholic cirrhosis; 85 patients with organ-specific autoimmune diseases (60 thyroiditis and 25 diabetes mellitus type 1); 307 patients with non-organ-specific autoimmune diseases (32 patients with myopathy and/or pulmonary fibrosis, 25 patients with inflammatory bowel diseases, 80 patients with systemic lupus erythematosus (SLE), 75 patients with Sjögren's syndrome (SS), 70 patients with scleroderma, 25 patients with rheumatoid arthritis (RA)) and 20 healthy blood donors.
Laboratory assessments
Sera were screened for ANA, anti-mitochondrial (AMA) and anti-smooth muscle antibodies (ASMA) using the indirect immunofluorescence (IIF) technique as described previously [21]. Cryostat sections of rat liver, kidney and stomach were used as substrates. The fluorescein-labelled anti-human conjugate was purchased from Dako Labs (Santa Barbara, CA) and used at a dilution of 1:20.
Anti-LKM and AMA antibodies were also studied by immunoblot and ELISA tests. Anti-dsDNA antibodies were studied by Farr technique (Amersham Pharmacia Biotech, Uppsala, Sweden), and anti-thyroglobulin antibodies were studied by ELISA (Radim, Angleur, Belgium).
Analysis of immunoprecipitated ribonucleoproteins (RNPs)
To identify autoantibodies capable of binding specific small nuclear/cytoplasmic ribonucleoproteins (sn/scRNPs), sera or affinity-purified antibodies were tested for their ability to immunoprecipitate subsets of small RNAs from extracts of human HeLa S3 cells. The standard assay method was used [22]. Briefly, HeLa S3 cells growing in log phase at 4 × 105 cells/ml were labelled in vivo with 32P-orthophosphate (Amersham) as described [23]. Whole cell extracts were prepared as described [24] and precleared with 1/20th volume of 20% (w/v) suspension of Sepharose-protein A (Pharmacia) in NET-2 buffer (50 mm Tris pH 7·4, 0·15 m NaCl, 0·05% Nonidet P-40) plus 1 mg/ml bovine serum albumin (BSA); immunoprecipitations were performed as described by Lerner & Steitz [22] for 32P-labelled extracts. Deproteinized extracts were prepared by PCA extraction and by incubating 32P-labelled extract in 0·1 mg/ml proteinase K and 0·2% SDS for 2 h at 37°C, followed by extraction with phenol/chloroform/isoamyl alcohol (PCA) (50/49/1) with 0·1% 8-hydroxiquinoleine and ethanol precipitation.
Immunoprecipitated 32P-labelled RNAs were PCA extracted, ethanol precipitated, electrophoresed on 10% polyacrylamide denaturing gels in 0·1 m boric acid/0·1 m Tris base/2 mm EDTA (1× TBE) pH 8·3, and subjected to autoradiography.
Purification of tRNP(Ser)Sec antigen from HeLa S3 human cells
Affinity-purified protein–tRNA(Ser)Sec antigen complexes were prepared as described [1] with modifications, using the anti-tRNP(Ser)Sec pattern serum immobilized on Sepharose-protein A. Briefly, Sepharose-protein A was incubated with pattern serum for 2 h at room temperature. After washing three times with buffer IPP (10 mm Tris pH 8, 0·5 m NaCl, 0·1% NP-40) and one with Tris buffer saline (TBS) (40 mm Tris pH 7·4, 0·13 m NaCl), IgG coupled to Sepharose was treated with glutaraldehyde at 10% for 30 min at room temperature. The beads were then blocked with a solution of 1 mg/ml BSA in TBS for 2 h at 4°C. After washing with NET-2 buffer, the beads were ready for incubation with a NET-2 extract from HeLa cells prepared in the usual manner [24]. The antigen was eluted with glycine–HCl 0·2 m pH 2·5. After being neutralized the eluted antigen was dialysed with TBS and used for separation of proteins in a 10% SDS–PAGE as described [25] and transferred to nitrocellulose sheets in 25 mm Tris, 192 mm glycine pH 8·3, without SDS [26].
Western blot analysis
Crude extracts from total HeLa S3, CEM, and HL-60 cells, and rat liver, as well as bacterial lysates and purified recombinant proteins, were separated on 10% (w/v) polyacrylamide gels and transferred to nitrocellulose as described by Towbin et al. [26] with modifications. Immunoblots using cell and tissue extracts were performed as described previously [1] using 3% casein as blocking solution and 125I-protein A (Amersham Pharmacia Biotech) for detection of bound immunoglobulins. When bacterial lysates were used for immunoblotting, nitrocellulose was incubated with anti-tRNP(Ser)Sec-positive sera, normal human sera and with anti-Xpress antibody (Invitrogen, San Diego, CA) diluted 1:1000. Membranes were washed and consecutively incubated with alkaline phosphatase-conjugated anti-human rabbit immunoglobulins (Dako, Glostrup, Denmark). Positive reactions were performed using 5-bromo-4-chloro-3-indolyl phosphate (BCIP) with nitro blue tetrazolium (NBT) (Promega, Madison, WI) as a substrate.
Affinity purification of antibodies
Antibodies were affinity-purified as described in [21] and were used without dilution in Western blot assays and immunoprecipitation experiments.
Statistical analysis
Quantitative variables were compared by the Fisher test or χ2 test. Since the variables for comparison had been formulated a priori and then assessed systematically in each group, an unadjusted P value of 0·05 was used to determine statistical significance.
Cloning
Screening of a human liver cDNA library in Uni-ZAP XR (Stratagene, La Jolla, CA) was performed [27] using anti-tRNP(Ser)Sec-positive sera. Detection of antigen–antibody complexes was performed using alkaline phosphatase-conjugated anti-human rabbit immunoglobulins (Dako). Positive clones were purified by repeated screening until all progeny plaques were positive. In a final screening, each filter was divided into eight pieces that were incubated with four different anti-tRNP(Ser)Sec-positive sera, and with anti-M2, anti-Ro, anti-NOR 90 and normal human sera, as unrelated and negative controls. Two positive clones (13 and 19) were isolated, and pBluescript plasmids containing cDNA inserts were obtained by in vivo Exassist/SOLR excision according to the protocol provided by Stratagene.
DNA sequencing and sequence analysis
Sequencing of double-stranded DNA templates was achieved by a modified dideoxy chain-termination method [28] using the ALFexpress Autoread Sequencing kit and the ALFexpress DNA Sequencer (Amersham Pharmacia Biotech). For priming, 5′-cyanine-labelled primers were used. Specific internal oligonucleotides were designed as the sequencing progressed. Sequences of the recombinant clones were compared with the non-redundant databases at the National Center for Biotechnology Information (NCBI) using the BLAST program [29].
Expression and purification of His-tagged recombinant proteins
The cDNA insert from clone 19 (3378 bp) was SacI/KpnI digested and gel-purified using GeneClean II kit (√Bio 101; Vista, CA). The DNA fragment was ligated in frame to SacI/KpnI digested and gel-purified (√Bio 101; Vista) expression vector pTrcHisA (Invitrogen, San Diego, CA), allowing the translation of a fusion protein bearing a 6-histidine tail at the NH2-terminus. The insert from clone 13 (2911 bp) was subcloned into pTrcHisC (Invitrogen) at BamHI and KpnI sites. In order to express His-tagged recombinant proteins, the constructs were transformed into the Escherichia coli strain TOP10 and selected on ampicillin-containing plates. Each recombinant clone was verified by sequence analysis. The pTrcHis vector with no insert was used as negative expression control, and the pTrcHisBCAT construct (Invitrogen) as positive expression control. Cells were grown in LB at 37°C to an OD600 = 0·6, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 1 mm and incubated for a further 3 h. Preparation of cell lysates and purification of recombinant proteins were performed under native conditions as specified in the manufacturer's instructions (Invitrogen). Briefly, TOP10 bacteria containing each recombinant protein were harvested and resuspended in buffer A (20 mm sodium phosphate, 500 mm NaCl, pH 7·8), treated with white lysozyme (100 μg/ml) and lysed by four rapid freeze–thaw and sonication cycles. Debris was spun down, and supernatants were loaded onto pre-equilibrated nickel-charged ProBond columns (Invitrogen). Washes were performed with buffer A at several pH: 6·3, 6·0 and 5·5. Elution was performed with a pH gradient from 4·5 to 2·5 in buffer A.
Results
AIH sera immunoprecipitate an opal suppressor tRNA(Ser)Sec
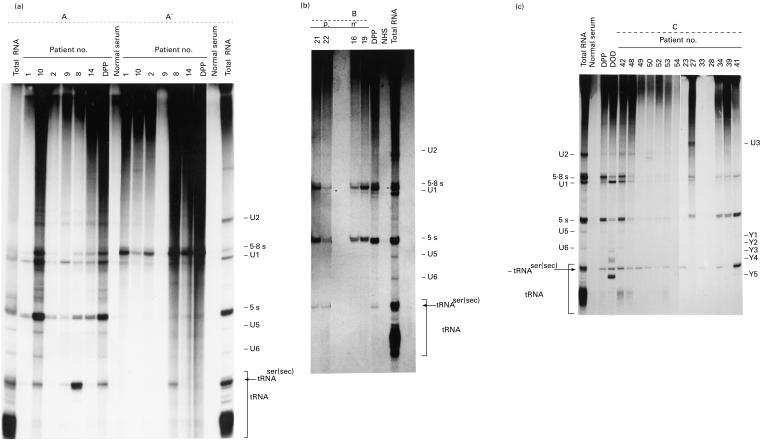
Sera randomly collected during follow up from 59 selected patients with type-1 AIH were studied by immunoprecipitation. Of the 59 analysed sera, 28 had antibodies which immunoprecipitated RNA species migrating at 4·5S region in a denaturing 10% urea–PAGE, between hY4 and hY5. This RNA had earlier been identified as the UGA suppressor serine tRNA (tRNA(Ser)Sec). Moreover, and as shown in Fig. 1, some sera immunoprecipitated other sn/scRNAs. Figure 1(a–c) shows three representative experiments of the analysis of the RNAs immunoprecipitated by patients with severe AIH.
Fig. 1.
(See next page) Immunoprecipitation of small RNAs by sera from patients with autoimmune hepatitis (AIH). 32P-labelled HeLa cell sonicates were immunoprecipitated and gel fractionated. Numbers at the top of each lane correspond to the number given to each patient. DPP and DOD are the identification names of two prototype anti-tRNP(Ser)Sec sera. Known RNAs are indicated on the side; tRNA(Ser)Sec (shown by an arrow) denotes the tRNA immunoprecipitated by AIH sera. (a,b,c) Three autoradiograms of the RNAs immunoprecipitated by sera from a representative number of AIH patients studied. Panel A′ shows the RNAs immunoprecipitated by the same sera described in panel A, from a deproteinized HeLa cell extract. Lanes Total RNA show the total RNAs extracted from the whole (a,b,c) and from the deproteinized (panel A′) HeLa cell sonicates prior to immunoprecipitation; lanes Normal serum and NHS show the immunoprecipitated RNAs by sera from healthy non-autoimmune donors; lanes DPP and DOD show the RNAs immunoprecipitated by two anti-tRNP(Ser)Sec reference sera.
Since most small RNAs precipitated by autoimmune sera are associated with antigenic proteins, we tested the ability of these sera to immunoprecipitate the tRNA(Ser)Sec from deproteinized extracts. Approximately 5% of all the patients studied had antibodies in their sera that recognized the structure of the RNA. Figure 1a shows a representative result of the immunoprecipitation assay performed with the same sera shown in Fig. 1a and deproteinized a HeLa cell extract. Serum 8 precipitated RNA about 50% as efficiently from deproteinized extracts. The other five sera as well as a prototype anti-tRNP(Ser)Sec control serum (DPP) showed no detectable immunoprecipitation of tRNA(Ser)Sec.
No patients from the control groups studied (patients with organ-specific autoimmune diseases, patients with non-organ-specific autoimmune diseases and normal blood donor volunteers) were found positive in the immunoprecipitation assay for anti-tRNP(Ser)Sec antibodies.
Immunoblotting assays; anti-tRNP(Ser)Sec autoantibodies recognize a 48/52-kD protein
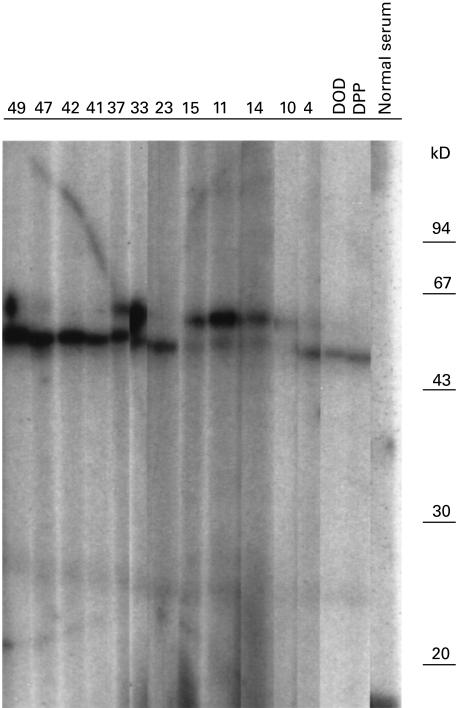
Autoantibodies against tRNA(Ser)Sec-associated protein(s) were studied by immunoblot using affinity-purified antigen from HeLa cell extracts prepared as described earlier. Fifteen of the 59 AIH sera were positive. Of these, 12 immunoprecipitated the tRNA(Ser)Sec. Figure 2 shows a representative number of sera with a positive immunoblot reaction. These antibodies recognized protein(s) of 48 and/or 52 kD. None of the sera from the control groups was positive (data not shown).
Fig. 2.
Immunoblotting of the affinity-purified tRNP(Ser)Sec antigen from HeLa cell sonicate, with autoimmune hepatitis (AIH) sera. The immunopurified HeLa cell extract was fractionated on 10% polyacrylamide/SDS gels, blotted onto nitrocellulose, and probed with antisera as indicated. The positions of molecular weight markers (in kD) are shown on the right. Numbers at the top of each lane correspond to the number given to each patient, lanes DOD and DPP show the immunoblot reaction of anti-tRNP(Ser)Sec reference sera, and lane Normal serum shows the immunoblot reaction of a serum from a healthy blood donor volunteer.
Sensitivity and specificity of anti-tRNA(Ser)Sec antibodies and AIH
Patients with and without anti-tRNP(Ser)Sec antibodies had similar clinical, laboratory, histological, and immunological features at presentation. There were no significant differences in the conventional laboratory indices of inflammatory activity or immunoreactivity in the patients with type-1 AIH who did and those who did not have anti-tRNP(Ser)Sec antibodies (data not shown). The frequencies of anti-tRNP(Ser)Sec were statistically different between AIH and control groups. Moreover, the patients who died of liver failure were more commonly seropositive for anti-tRNP(Ser)Sec than patients who did not have these antibodies (25% versus 3%, P = 0·04). Both groups of patients were followed for a similar period of time (121 versus 130 months) (Table 1) and received similar treatment.
Table 1.
Clinical and serological features in patients with type-1 autoimmune hepatitis (AIH)
| AIH patients* | |||
|---|---|---|---|
| Anti-tRNA(Ser)Sec-positive | Anti-tRNA(Ser)Sec-negative | P | |
| Number | 28 | 31 | |
| Age (years) | 36 ± 2 | 42 ± 3 | 0·107 |
| Female:male | 22:6 | 24:7 | 0·915 |
| Concurrent autoimmune diseases | 11 (39) | 15 (48) | 0·953 |
| SMA ≥ 1:40 | 24 (86) | 28 (90) | 0·630 |
| ANA ≥ 1:40 | 19 (68) | 21 (68) | 0·449 |
| Duration of follow up (months) | 121 ± 14·5 | 130 ± 17 | 0·709 |
| †Fatal outcome | 7 (25) | 1 (3) | 0·040 |
Two groups of type-1 AIH patients with or without antibodies to tRNA(Ser)Sec in their sera.
Liver-related death.
Numbers in parentheses are percentages. Significantly different from each other at levels of P. Differences between groups (< 0·05) are in bold.
Cloning and sequence analysis
Four anti-tRNPSer(Sec)-positive sera from type-1 AIH patients (all of them reacting with the 48/52-kD tRNA(Ser)Sec-associated protein from HeLa cells) were used to screen a human liver cDNA expression library to isolate and characterize the cDNA encoding the antigenic protein associated with the tRNA(Ser)Sec. A total of 5 × 105 plaques was screened and two positive clones (number 19 and number 13) recognized by all anti-tRNP(Ser)Sec (4/4) and by none of the control sera (one anti-Ro, one anti-M2, one anti-NOR 90 control sera and four normal human sera) were isolated. Both clones were converted into plasmids and used for further analysis. Nucleotide sequences and deduced amino acid sequences are shown in Fig. 3. Clone 19 had an insert of 3378 bp including 174 bp of 5′-untranslated region, a putative initiating ATG codon at nucleotide 175 followed by an open reading frame of 1326 bp, and 1878 bp of 3′-untranslated region. The open reading frame was predicted to encode a protein of 441 amino acids. The theoretical molecular mass was 48·8 kD, close to the estimated molecular mass from SDS–PAGE of the antigenic protein associated with tRNA(Ser)Sec (48 kD) [1]. The putative polypeptide showed a high content of basic amino acids, resulting in a theoretical pI of 9·40. This protein showed one zinc finger motif (amino acids 156–161) which has been associated with nucleic acid and protein interaction. Clone 13 had an insert of 2911 bp including 16 bp of 5′-untranslated region, a potential ATG start codon at nucleotide 17, followed by a 972-bp open reading frame coding for a putative protein with a predicted molecular mass of 35·9 kD, and a long 3′-untranslated region of 1923 bp.
Fig. 3.
(See next page) Nucleotide and deduced amino acid sequence of clone 19. The amino acid sequence was predicted from the nucleotide sequence of clone 19 cDNA. A 5′-untranslated region of 174 bp precedes the open reading frame. The initiator methionine codon is surrounded by the sequence GCAATCATGG at nucleotides 169–178, which closely approximates to the ideal ribosomal binding sequence GCCACCATGG [40] and one zinc finger motif (amino acids 156–161), which has been associated with nucleic acid and protein interactions. Similar sequence has been observed in RNA binding proteins [41, 42]. The TGA stop codon (*) is followed by a long 3′ non-coding region. This region includes putative polyadenylation signals AATAAA at nucleotides 3309–3314. First nucleotide of cDNA number 13 is underlined. Bold letters indicate the location of minor differences with clone 19. The symbol arrowhead indicates the site where the open reading frame of clone 13 starts.
Comparative analyses of nucleotide sequences of both clones demonstrated that the nucleotide sequence of clone 19 from nucleotide 513 was identical to the complete nucleotide sequence of clone 13 with minor exceptions. Minor differences were: five nucleotides of the 5′-untranslated region of clone 13, a single nucleotide in the 3′-untranslated region of both clones (indicated with bold letters in Fig. 3), and the presence of 72 nucleotides before the poly(A) tail of clone 13 which did not exist in clone 19. The alignment of the two nucleotide sequences showed that the ATG start codon of the cDNA number 13 perfectly matched a second in frame ATG codon at position 529 of the cDNA number 19. These data support the finding that clone 13 encoded for a shorter protein which lacked the first 118 amino acids of the protein encoded by clone 19.
As the sequences of the two clones were not previously recorded in the GenBank, EMBL, DDBJ and SwissProt databases, sequence of the cDNA number 19 was submitted to the EMBL Data library (accession number: AJ238617).
A BLASTN search in non-redundant databases revealed an identity of cDNA number 19 from nucleotide 629 to nucleotide 3331 (just before the poly(A) tail) with a region of human chromosome 4 (GenBank accession number AC007073). With regard to cDNA number 13, the identity region included nucleotide 180 to nucleotide 2891, just before the poly(A) signal of this cDNA.
The alignment of the predicted amino acid sequences with databases demonstrated 44% identity (clone 19) and 41% (clone 13), with a hypothetical 53·4-kD protein (481 amino acids) of Caenorhabditis elegans, whose function has not yet been described (SWALL accession number: Q18953).
Expression and purification of His-tagged recombinant proteins
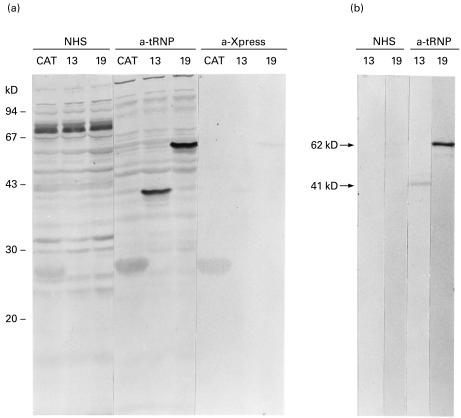
In order to demonstrate that we had isolated the cDNA encoding the antigenic protein, which associates with the tRNA(Ser)Sec, we proceeded with the expression, purification and analysis of both recombinant proteins. The expressions of a specific 62-kD protein (clone 19) and a specific 41-kD protein (clone 13) were demonstrated in Western blots using anti-tRNP(Ser)Sec-positive serum (Fig. 4a). Each protein correlated with a band obtained with anti-Xpress antibodies raised against an NH2-terminal X-press epitope of pTrcHis vectors (Fig. 4a). As His-tagged recombinant protein translation started at the ATG of pTrcHis vector, there was an increment in molecular weight of the recombinant proteins in comparison with the theoretical molecular mass estimated from the cDNA. This increment in the molecular weight was due to a fragment of pTrcHis vector, a fragment of pBluescript vector and the 5′-untranslated region of cDNA included in the expressed His-tagged recombinant protein.
Fig. 4.
Antigenicity of fusion proteins characterized by Western blot. (a) Escherichia coli TOP10 lysates with the cDNA sequences expressed from the bacterial vector pTrcHis were fractionated by SDS−10% PAGE, transferred to nitrocellulose membrane, and allowed to react with: normal human serum (NHS), type-1 autoimmune hepatitis (AIH) patient serum containing anti-48-kD associated tRNA(Ser)Sec antibodies (a-tRNP), and anti-Xpress antibodies that recognize the amino acid sequence -Asp-Leu-Tyr-Asp-Asp-Asp-Asp-Lys- found in the NH2-terminus of the vector pTrcHis (a-Xpress). CAT, E. coli TOP10-pTrcHisBCAT extract; 13, E. coli TOP10-pTrcHisC cDNA number 13 extract; 19, E. coli TOP10-pTrcHisA cDNA number 19 extract. Molecular weight markers are indicated on the left. (b) Purified His-tagged proteins from bacterial lysates were assayed by Western blot with a NHS and a type-1 AIH patient serum with anti-tRNA(Ser)Sec antibodies (a-tRNP). Arrows indicate the molecular weight of the recombinant proteins reactive with anti-tRNP(Ser)Sec antibodies.
Lysates from bacterial clones containing the recombinant proteins were loaded onto nickel-charged agarose resin columns. Western blots of the eluted fractions demonstrated that both recombinant proteins were highly purified at pH 2·5 eluted fraction (Fig. 4b).
Immunoprecipitation and Western blot analysis of antibodies eluted from His-tagged recombinant proteins
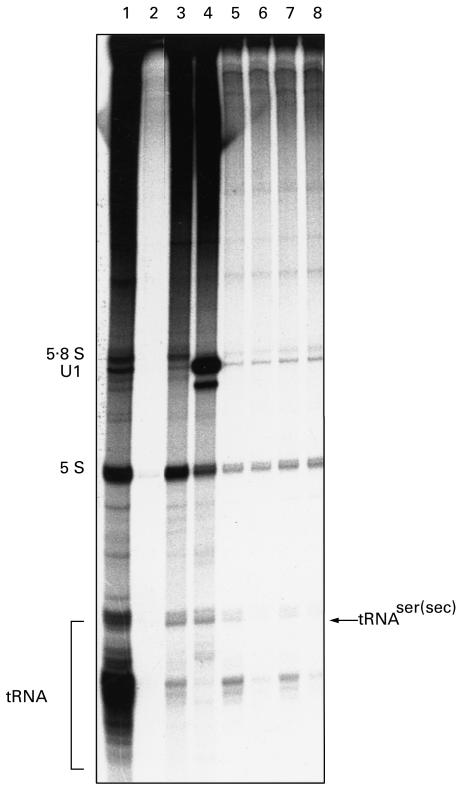
Using the purified fraction of the 62-kD recombinant protein (clone 19), a Western blot with an anti-tRNP(Ser)Sec-positive serum was performed, and antibodies eluted from the 62-kD band were assayed by immunoprecipitation of a 32P-labelled HeLa S3 cell extract. These affinity-purified antibodies clearly precipitated the tRNA(Ser)Sec (Fig. 5, lane 5). The same experiment was performed with antibodies eluted from the 41-kD protein encoded by clone 13, showing that these antibodies were also capable of immunoprecipitating tRNA(Ser)Sec (Fig. 5, lane 7). Control experiments using eluates from unrelated regions of the nitrocellulose did not precipitate the tRNA(Ser)Sec (Fig. 5, lanes 6 and 8). When these affinity-purified antibodies were used to re-test a Western blot, they detected the 62- and 41-kD proteins, respectively (data not shown).
Fig. 5.
Immunoprecipitation of tRNA(Ser)Sec by affinity-purified anti-62- and anti-41-kD recombinant protein antibodies. A non-immune serum from a healthy blood donor (lane 2), whole sera with anti-tRNP(Ser)Sec antibodies (lanes 3 and 4), anti-62-kD recombinant protein antibodies eluted from a Western blot of purified expressed protein from clone 19 (lane 5), anti-41-kD recombinant protein antibodies eluted from a Western blot of purified expressed protein from clone 13 (lane 7), a control eluate from an unrelated region of the Western blot from expressed clone 19 (lane 6), and a control eluate from an unrelated region of the Western blot from expressed clone 13 (lane 8) were used to immunoprecipitate a 32P-labelled HeLa S3 cell sonicate, and the RNAs were analysed. The mobility of known RNAs is given on the left, and tRNA(Ser)Sec precipitated by type-1 autoimmune hepatitis (AIH) sera are indicated on the right. Total RNA (lane 1) is RNA from the HeLa S3 cell sonicate prior to immunoprecipitation.
Moreover, the sera that immunoprecipitated the tRNA(Ser)Sec and strongly reacted in immunoblot with a 48/52-kD protein from partially purified HeLa cell extracts, recognized the 62-kD His-tagged recombinant protein.
Discussion
We previously identified anti-tRNP(Ser)Sec autoantibodies, specific for a subgroup of AIH [1]. These antibodies recognized tRNA(Ser)Sec-associated protein(s) and/or directly reacted with the tRNA(Ser)Sec itself. In this study we extend our previous report to 59 patients with AIH, include a more extensive group of controls, and demonstrate the high specificity and frequency (47·5%) of the anti-tRNP(Ser)Sec autoantibodies for severe forms of type-1 AIH. The complete nucleotide sequence and expression of an antigenic protein is reported.
The clinical relevance of these autoantibodies arises from the high specificity of the response of AIH patients, with a higher frequency in patients who died of liver disease. Autoantibodies against this antigen are not found in patients with other autoimmune diseases or in normal human blood donor volunteers.
The biological relevance of this antigen arises from its association with the human UGA suppressor tRNA(Ser)Sec.
In the present study we cloned and expressed a cDNA encoding a human protein, with a predicted molecular mass of 48·8 kD, which is specifically recognized by sera from type-1 AIH patients. Affinity-purified antibodies from this protein specifically immunoprecipitated the tRNA(Ser)Sec from HeLa S3 cells.
The deduced amino acid sequence, although novel, has a 44% homology with a hypothetical 53·4-kD protein of C. elegans, described as part of the C. elegans genome study, and whose function is not yet known.
Data in our previous study [1] and in the present paper strongly suggest that tRNP(Ser)Sec complex is involved in the selenocysteine insertion pathway in eukaryotes. However, the cloning and sequencing of the 48·8-kD protein did not completely elucidate its precise molecular nature. No relevant similarity was found with any synthetase [30], synthase [31], or elongation factors when compared with nucleotide and protein Data/Banks. However, recent data suggest that the 48·8-kD protein could be the Selenium-tRNA protecting factor (SePF) earlier described as a 50-kD tRNA(Ser)Sec-binding protein protecting mammalian 75Se-tRNA(Ser)Sec from alkaline hydrolysis [16,32]. Even though the sequence of SePF is not known, our data suggest that the 48·8-kD protein could be this selenocysteine-specific factor: it has a similar molecular weight, its sequence contains a zinc finger motif which has been associated with nucleic acid and protein binding region, and data not yet reported have demonstrated that the tRNP(Ser)Sec is more resistant to alkaline hydrolysis than the tRNA(Ser)Sec itself. Moreover, it has a putative ribosomal binding sequence.
Knowledge of the biological nature of the antigen may help to elucidate the aetiopathogenesis of the anti-tRNP(Ser)Sec antibodies and their possible role in type-1 AIH.
In eukaryotes the tissue where the highest number of selenoproteins is found is the liver, and factors involved in selenoprotein synthesis may also possibly be predominant in this organ [33]. Anti-tRNP(Ser)Sec antibodies may arise following a breakdown in tolerance to ‘self-proteins’ in the liver, perhaps as a molecular mimicry mechanism, or to the ‘self modified’. Anti-tRNP(Ser)Sec antibodies could result from the direct interaction of the 48·8-kD protein with a virus RNA, thereby rendering it ‘foreign’ to the host immune system [34].
Other autoantibodies related to AIH have been described although their nature has not yet been elucidated. These include the soluble liver antigen (SLA) [35], the liver pancreas antigen (LP) [36], cytokeratins 8 and 18 [37] and the glutathione transferase (GTS) [38].
Almost simultaneously the 48-kD tRNP(Ser)Sec clone was registered (accession number AJ238617), the LP antigen also related to AIH was cloned (accession number AF146396). The reported sequence of the latter shows 99% homology with the 48-kD tRNP(Ser)Sec cloned protein. Moreover, it has recently been reported that the LP antigen is identical to the SLA antigen [39]. We studied eight sera from AIH patients with anti-SLA antibodies and found that all of them precipitated the tRNA(Ser)Sec and reacted with the recombinant 48·8-kD tRNP(Ser)Sec protein. We concluded that the tRNP(Ser)Sec antigen is the same as the LP antigen [36], is different from cytokeratins [37] and glutathione transpherase [38], and is included in the complex SLA [35].
This study demonstrated that the screening of anti-tRNP(Ser)Sec antibodies may be a very useful marker for type-1 AIH. Moreover, the improvement of an ELISA test with the recombinant protein expressed in eukaryote cells to screen the anti-tRNA-associated protein antibodies would increase both the sensitivity and utility of the assay. Antibodies that immunoprecipitated the tRNA(Ser)Sec included at least two subsets of autoantibodies, those that reacted with a 48·8-kD antigenic protein and those that recognized the tRNA itself or another tRNA-associated protein different from the 48·8-kD protein. At present, the screening of AIH patients by immunoprecipitation and analysis of RNAs is the most sensitive and specific test for anti-tRNP(Ser)Sec antibodies.
Acknowledgments
We thank Ms Carolina Newey for her assistance with the preparation of the manuscript. This work was supported by a grant from the ‘Fondo de Investigaciones Sanitarias de la Seguridad Social’ (94/0751), a grant from the ‘Ministerio de Educación y Ciencia’ (SAF 97/0123) and a grant from the ‘Generalitat de Catalunya’ (1997/SGR/00352) (Spain).
References
- 1.Gelpí C, Sontheimer EJ, Rodriguez-Sanchez JL. Autoantibodies against a serine tRNA–protein complex implicated in cotranslational selenocysteine insertion. Proc Natl Acad Sci USA. 1992;89:9737–43. doi: 10.1073/pnas.89.20.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maddrey WC. Subdivisions of idiopathic autoimmune chronic active hepatitis. Hepatology. 1987;7:1372–5. doi: 10.1002/hep.1840070631. [DOI] [PubMed] [Google Scholar]
- 3.Maddrey WC, Willis C. Chronic hepatitis. In: Bone RC, editor. Disease-a-month. XXXIX. 1993. pp. 53–126. No. 2. [PubMed] [Google Scholar]
- 4.Manns M. Autoantibodies and antigens in liver diseases-updated. J Hepatol. 1989;9:272–80. doi: 10.1016/0168-8278(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 5.Czaja AJ. Autoantibodies. Bailliere's Clin Gastroenterol. 1995;9:723–44. doi: 10.1016/0950-3528(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 6.Homberg J-C, Abuaf N, Bernard O, et al. Chronic active hepatitis associated with anti-liver/kidney microsome antibody type 1: a second type of ‘autoimmune’ hepatitis. Hepatology. 1987;7:1333–9. doi: 10.1002/hep.1840070626. [DOI] [PubMed] [Google Scholar]
- 7.Czaja AJ, Pfeifer K, Decker RH, Vallari A. Frequency and significance of antibodies to asialoglycoprotein receptor in type 1 autoimmune hepatitis. Dig Dis Sci. 1996;41:1733–40. doi: 10.1007/BF02088738. [DOI] [PubMed] [Google Scholar]
- 8.Czaja AJ, Cassani F, Cataleta M, Valentini P, Bianchi FB. Frequency and significance of antibodies to actin in type 1 autoimmune hepatitis. Hepatology. 1996;24:1068–73. doi: 10.1002/hep.510240515. [DOI] [PubMed] [Google Scholar]
- 9.Manns MP. Autoantibodies in chronic hepatitis: diagnostic reagents and scientific tools to study aetiology, pathogenesis, and cell biology. Prog Liver Dis. 1994;12:137–56. [PubMed] [Google Scholar]
- 10.Czaja AJ, Manns MP, Homburger HA. Frequency and significance of antibodies to liver/kidney microsome type 1 in adults with chronic active hepatitis. Gastroenterology. 1992;103:1290–5. doi: 10.1016/0016-5085(92)91518-9. [DOI] [PubMed] [Google Scholar]
- 11.Czaja AJ. Behaviour and significance of autoantibodies in type 1 autoimmune hepatitis. J Hepatol. 1999;30:394–401. doi: 10.1016/s0168-8278(99)80096-8. [DOI] [PubMed] [Google Scholar]
- 12.Czaja AJ, Manns MP. The validity and importance of subtypes of: a point of view. Am J Gastroenterol. 1995;90:1206–11. [PubMed] [Google Scholar]
- 13.Czaja AJ. Autoimmune hepatitis: evolving concepts and treatment strategies. Dig Dis Sci. 1995;40:435–56. doi: 10.1007/BF02065434. [DOI] [PubMed] [Google Scholar]
- 14.Jung J-E, Karoor V, Sandbaken MG, et al. Utilisation of selenocysteyl-tRNA(Ser)Sec and seryl-tRNA(Ser)Sec in protein synthesis. J Biol Chem. 1994;269:29739–45. [PubMed] [Google Scholar]
- 15.Bock A, Forchhammer K, Heider J, Baron C. Selenoprotein synthesis: an expansion of the genetic code. Trends Biochem Sci. 1991;16:463–7. doi: 10.1016/0968-0004(91)90180-4. [DOI] [PubMed] [Google Scholar]
- 16.Yamada K. A new translational elongation factor for selenocysteyl-tRNA in eucaryotes. FEBS Letters. 1995;377:313–7. doi: 10.1016/0014-5793(95)01352-0. [DOI] [PubMed] [Google Scholar]
- 17.Johnson PJ, McFarlane IG, Alvarez F, et al. Meeting Report. International Autoimmune Hepatitis Group. Hepatology. 1993;18:998–1005. doi: 10.1002/hep.1840180435. [DOI] [PubMed] [Google Scholar]
- 18.Czaja AJ. Diagnosis, prognosis, and treatment of classical autoimmune chronic active hepatitis. In: Krawitt EL, Wiesner RH, editors. Autoimmune liver disease. New York: Raven Press; 1991. pp. 143–66. [Google Scholar]
- 19.Summerskill WHJ, Korman MG, Ammon HV, Baggentoss AH. Prednisone for chronic active liver disease: dose titration, standard dose, and combination with azathioprine compared. Gut. 1975;16:876–83. doi: 10.1136/gut.16.11.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czaja AJ, Carpenter HA. Sensitivity, specificity and predictability of biopsy interpretations in chronic hepatitis. Gastroenterology. 1993;105:1824–32. doi: 10.1016/0016-5085(93)91081-r. [DOI] [PubMed] [Google Scholar]
- 21.Gelpí C, Algueró A, Martinez MA, Vidal S, Juarez C, Rodriguez-Sanchez JL. Identification of protein components reactive with anti-PM/Scl autoantibodies. Clin Exp Immunol. 1990;81:59–64. doi: 10.1111/j.1365-2249.1990.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerner MR, Steitz JA. Antibodies to small nuclear RNAs complex with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1979;76:5495–9. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mimori T, Hinterberger M, Pettersson I, Steitz JA. Autoantibodies to the U2 small nuclear ribonucleoprotein in a patient with scleroderma–polymyositis overlap syndrome. J Biol Chem. 1984;259:560–5. [PubMed] [Google Scholar]
- 24.Forman MS, Nakamura M, Mimori T, Gelpí C, Hardin JA. Detection of antibodies to small nuclear ribonucleoproteins and small cytoplasmic ribonucleoproteins using unlabeled extracts. Arthritis Rheum. 1985;28:1356–61. doi: 10.1002/art.1780281207. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1979;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E, Maniatis T. Molecular cloning. 2. New York: Cold Spring Harbor Laboratory Press; 1989. pp. 12.16–12.20. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucl Acids Res. 1984;12:857–72. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanas JS, Hazuda DJ, Bogenhagen DF, Wu FY-H, Wu C-W. Xenopus transcription factor A requires zinc for binding to the 5S RNA gene. J Biol Chem. 1983;258:14120–5. [PubMed] [Google Scholar]
- 32.Draper DE. Protein-RNA recognition. Ann Rev Biochem. 1995;64:593–620. doi: 10.1146/annurev.bi.64.070195.003113. [DOI] [PubMed] [Google Scholar]
- 33.Vincent C, Leberman R, Hartlein M. EMBL/GENBANK/DDBJ Data Banks Accession: P49591.
- 34.Mizutani T, Kurata H, Yamada K, Totsuka T. Some properties of murine selenocysteine synthase. Biochem J. 1992;284:827–34. doi: 10.1042/bj2840827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada K, Mizutani T, Ejiri S, Totsuka T. A factor protecting mammalian [75Se]SeCys-tRNA is different from EF-1α. FEBS Letters. 1994;347:137–42. doi: 10.1016/0014-5793(94)00523-0. [DOI] [PubMed] [Google Scholar]
- 36.Burk RF, Hill KE. Regulation of selenoproteins. Ann Rev Nutr. 1993;13:65–81. doi: 10.1146/annurev.nu.13.070193.000433. [DOI] [PubMed] [Google Scholar]
- 37.Keene JD. RNA surfaces as functional mimetics of proteins. Chem Biol. 1996;3:505–13. doi: 10.1016/s1074-5521(96)90139-8. [DOI] [PubMed] [Google Scholar]
- 38.Manns M, Gerken G, Kyriatsoulis A, Staritz M, Meyer zum Büschenfelde KH. Characterization of a new subgroup of autoimmune chronic active hepatitis by autoantibodies against a soluble liver antigen. Lancet. 1987;7:292–4. doi: 10.1016/s0140-6736(87)92024-1. [DOI] [PubMed] [Google Scholar]
- 39.Stechemesser E, Klein R, Berg PA. Characterization and clinical relevance of liver-pancreas antibodies in autoimmune hepatitis. Hepatology. 1993;18:1–9. [PubMed] [Google Scholar]
- 40.Wächter B, Kyriatsoulis A, Lohse AW, Gerken G, Meyer zum Büschenfelde KH, Manns M. Characterization of liver cytokeratin as a major target antigen of anti-SLA antibodies. J Hepatol. 1990;11:232–9. doi: 10.1016/0168-8278(90)90119-c. [DOI] [PubMed] [Google Scholar]
- 41.Wesierska-Gadek J, Grimm R, Hitchman E, Penner E. Members of the glutathione-S-transferase gene family are antigens in autoimmune hepatitis. Gastroenterology. 1998;114:329–35. doi: 10.1016/s0016-5085(98)70485-8. [DOI] [PubMed] [Google Scholar]