Abstract
Integrins play a major role in cell adhesion and migration. Previous work reported that a cleaved form of integrin α6 (α6p) was detected in invasive human prostate cancer tissue, absent in normal prostate tissue and was produced by urokinase-type Plasminogen Activator (uPA) in a plasmin-independent manner. Using site-directed mutagenesis we identified amino acid residues R594 and R595, located in the “stalk” region of integrin α6, as essential for cleavage. The cleavage site is located on the extracellular region of the protein between the β-barrel domain and the thigh domain. Prostate cancer cells (PC3N) were stably transfected to over express the cleavable, wild type (PC3N-α6-WT) or the non-cleavable form of integrin α6 (PC3N-α6-RR). The number of cells invading laminin 111 and laminin 332 coated filters by PC3N-α6-WT cells increased by three fold as compared to PC3N-α6-RR cells. Plasminogen Activator Inhibitor-1 (PAI-1) reduced the invasion of PC3N-α6-WT cells by approximately 42% through laminin 332 coated filters and plasmin inhibitor aprotinin had no significant effect. Linear cell migration increased production of integrin α6p in the PC3N-α6-WT cells and not in the PC3N-α6-RR cells and 32% of the PC3N-α6-WT cells migrated on laminin 111 in the linear migration assay as compared to the 5% PC3N-α6-RR cells. These data taken together suggest that the uPA mediated cell surface cleavage of the α6 integrin extracellular domain is involved in tumor cell invasion and migration on laminin.
Keywords: Integrin α6, cleavage, prostate cancer, ECM, migration, laminin, uPA, urokinase
INTRODUCTION
Integrins are heterodimeric cell surface proteins composed of an α and a β subunit. Each αβ combination has its own binding specificity and signaling properties [1]. Different integrin combinations act as receptors to different extracellular matrix (ECM) proteins [1, 2]. They are involved in many processes including cell migration, cell adhesion, differentiation, blood clotting, tissue organization and cell growth as well as cancer cell migration, invasion and metastasis. Integrins generally contain a large extracellular domain (α subunit ~1000 residues and β subunit ~750 residues), a transmembrane domain and a short cytoplasmic domain (~50 residues or less) with the exception of β4 whose cytoplasmic domain is large (more than 1000 residues) [3, 4].
The structure of the extracellular segment of the αv integrin subunit has been determined using protein crystals of soluble αvβ3 integrin fragment [5]. The amino terminus of the αv integrin subunit contains a seven-bladed β-propeller structure [6]. It is followed by a tail region composed of three β-sandwiched domains: an Ig-like “thigh” domain and two very similar domains that form the “calf” module [6]. It has been shown that the tail region can fold back at ~135 degree angle forming a V-shaped structure with a “genu” between the thigh domain and the calf module [6].
Our previous work has shown that a structural variant of the α6 integrin called the α6p exists that is missing over half of the extracellular segment, the presence of which has also been reported by others [7, 8]. This variant is present in prostate cancer tissue but is absent in normal prostate tissue [9]. The α6p is missing the extracellular β-propeller domain associated with ligand binding but remains bound to its β integrin partner [7]. We have also shown using four different experimental approaches that α6p is produced by proteolytic cleavage of the α6 integrin by Urokinase-type plasminogen Activator (uPA) in a plasmin-independent manner [9]. The α6p integrin is a 70 kDa protein, and mass spectrometry analysis has shown that the amino terminal end of the molecule contains amino acids starting at arginine 596 [7]. Using a multiple sequence alignment tool, this position in the α6 integrin has been shown to lie within an accessible loop upstream from the genu region described for the αV integrin subunit [5].
The protease that cleaves the α6 integrin, uPA, is a secreted 54-kDa serine protease which cleaves plasminogen as a primary substrate [10]. uPA has also been shown to catalyze the proteolytic cleavage of the extracellular matrix protein fibronectin [11], hepatocyte growth factor/scatter factor (HGF/SF) [12] and macrophage-stimulating protein (MSP) [13].
The aim of this study was twofold: first, to use site directed mutagenesis to determine the amino acid sequences necessary for the uPA dependent cleavage of the α6 integrin. Second, to determine the phenotypic properties of cells expressing the non-cleavable form of the integrin α6. Our results indicate that integrin α6 cleavage is a dynamic feature contributing to cell invasion and migration on laminin.
MATERIALS AND METHODS
Cells
All cells were incubated at 37°C in a humidified atmosphere of 95% air and 5% CO2. K562, PC3N and HaCaT cells were grown in Iscove’s Modified Dulbecco’s Medium (IMDM) (Gibco BRL, MD, USA) plus 10% fetal bovine serum (FBS). The wildtype (WT) and mutated (RR) integrin α6 transfected cell lines K562-α6-WT, K562-α6-RR, PC3N-α6-WT and PC3N-α6-RR were grown in IMDM plus 10% FBS in presence of 6μg/ml Blasticidin and 1 μg/ml Zeocin (Invitrogen Corp., CA, USA). These cells were induced using 0.02μg/ml Doxycycline (Sigma, St. Louis, MO, USA.). The PC3N cells are a variant of the PC3 prostate carcinoma cell line[14].
Antibodies and chemicals used
Anti-α6 integrin antibodies include: J1B5, rat monoclonal was a generous gift from Dr. Caroline Damsky (University of California, San Francisco, USA) [15] and AA6A rabbit polyclonal antibody was produced by Bethyl Laboratories Inc. (Montgomery, TX, USA). AA6A is specific for the last 16 amino acids (CIHAQPSDKERLTSDA) in the cytoplasmic region of the human α6A sequence [16]. The monoclonal β1 integrin antibody, TS2/16 [17] was produced by the hybridoma cell line obtained from the American Type Culture Collection. The rabbit anti-actin antibody, AAN01 was obtained from Cytoskeleton, Denver, CO, U.S.A. The monoclonal antibody against uPAR was purchased from American Diagnostica, Greenwich, CT, U.S.A. Rabbit anti-uPA antibody was purchased from Abcam (Cambridge, MA, U.S.A.). Urokinase was purchased from Chemicon, Temecula, CA, USA and consisted of a mixture of single chain and double chain forms of uPA. Human recombinant EGF was purchased from Invitrogen Corp., Carlsbad, CA, U.S.A. Plasminogen Activator Inhibitor-1 was from Calbiochem, San Diego, CA, U.S.A. Aprotinin was purchased from Sigma-Aldrich, St. Louis, MO, U.S.A.
Site-directed mutagenesis
The cDNA for wild-type α6A was a generous gift from Dr. Isaac Rabinovitz. After cloning it into the pcDNA4/TO vector we used different sets of primers to induce the desired mutations in the α6 integrin. We used QuikChange Site-Directed Mutagenesis Kit (Stratagene, Cedar Creek Texas). The primers were designed using the recommendations of the manufacturer and were:
Primers for RR----> AA mutation 5′ggagatccaagagccaagctctGCTGCGcgagtgaattcacttccagaag 3′ 5′CttctggaagtgaattcactcgCGCaGCagagcttggctcttggatctcc 3′ Primers for RV-----> AA mutation 5′gccaagagccaagctctcgtagggcagcgaattcacttccagaagttcttcc3′ 5′GGAAGAACTTCTGGAAGTGAATTCGCTGCCCTACGAGAGCTTGGCTCTTGGC3′ Primers for R536-----> A mutation 5′AAAATCTGGGCTATCCTCAGCAGTTCAGTTTCGAAACCAAG3′ 5′CTTGGTTTCGAAACTGAACTGCTGAGGATAGCCCAGATTTT3′ Primers for R594-----> A mutation 5′GATCCAAGAGCCAAGCTCTGCAAGGCGAGTGAATTCACTTC3′ 5′GAAGTGAATTCACTCGCCTTGCAGAGCTTGGCTCTTGGATC3′ Primers for R595-----> A mutation 5′GAGCCAAGCTCTCGTGCGCGAGTGAATTCACTTCC3′ 5′GGAAGTGAATTCACTCGCGCACGAGAGCTTGGCTC3′
The reactions were carried out as recommended by the manufacturer. The K562 cells were transfected by electroporation as described previously [18]. Briefly, 30μg of plasmid and 10μg of repressor plasmid (pcDNA6/TR) were added to 10 million cells in a final volume of 800μl of serum free RPMI media in a 0.4cm electroporation cuvette. The cells were electroporated at 960 microfarads and 320 V. After 10 minutes on ice the cells were plated in IMDM media with 10% serum for 24 hours. The media was replaced with fresh media containing 0.2μg/ml doxycycline and the cells were incubated for another 24 hours. The cells were then analyzed for α6 integrin cleavage by immunoprecipitating the α6 integrin with J1B5 and adding 20μg of uPA in the immunocomplexes for 3 hours on ice. The samples were analyzed by immunoblotting using the anti-α6 integrin rabbit polyclonal antibody, AA6A. K562 stably transfected cells (K562-α6-WT expressing wildtype integrin α6 and K562-α6-RR expressing the non-cleavable integrin α6) were selected using the rat monoclonal anti-integrin α6 antibody J1B5 and magnetic microbeads (MACS, Miltenyi Biotech, Auburn, CA, USA) as per the manufacturer’s instructions. The PC3N cells were transfected using Effectene (Qiagen, Valencia, CA, U.S.A.) using the recommendations of the manufacturer. The stable clones (PC3N-α6-WT expressing wildtype integrin α6 and PC3N-α6-RR expressing the non-cleavable integrin α6) were isolated after treatments with 6 μg/ml Blasticidin and 1 μg/ml Zeocin (Invitrogen Corp., CA, USA).
Flow cytometry
Flow cytometric analysis was carried out as described earlier[19]. Briefly, K562, K562-α6-WT and K562-α6-RR cells were washed in PBS and incubated with rat anti-integrin α6 antibody J1B5 (1:10) for 30 min at 4 °C, washed two times with PBS + 1% BSA + 10mM Sodium Azide. The cells were then incubated with secondary anti-rat antibody Alexa 488 for 30 min at 4 deg C in the dark, washed two times with PBS + 1% BSA + 10mM Sodium Azide and pellet suspended in 1% Formaldehyde solution and analyzed on the BD FACScan (BD Biosciences, San Jose, CA USA).
Immunoprecipitations
Cells were washed three times with HEPES buffer and lysed in cold RIPA buffer (150 mM NaCl, 50 mM Tris (Biorad, Hercules, CA), 5 mM EDTA, 1% (v/v) Triton X-100, 1% (w/v) deoxycholate, 0.1% (w/v) SDS, pH 7.5) plus protease inhibitors inhibitors (PMSF, 2 mM; leupeptin and aprotinin, 1 μg/ml). The lysate was briefly sonicated on ice and precleared by rotating it with 50μl of protein G sepharose beads at 4 °C for 1 hour. The beads were removed by centrifugation, and the supernatant was collected for immunoprecipitations. Each reaction mixture contained 35 μl of protein G sepharose and 5μg of antibody. Anti-integrin α6 rat monoclonal antibody J1B5, mouse anti-integrin β1 antibody TS216 and mouse anti-uPAR antibody were used. The final volume of the lysate was adjusted to 500 μl with RIPA buffer. The mixture was rotated for 18 hours at 4 °C and complexes were then washed four times with cold RIPA buffer. The pellet was dissolved in 2X non-reducing sample buffer and boiled for 5 minutes. The samples were analyzed by immunoblotting procedures.
Immunoblotting
Cells were lysed in RIPA and the resulting protein solubilized in non-reducing sample buffer and analyzed by a 10 % SDS-PAGE gel. Proteins resolved in the gel were electrotransferred to Millipore Immobilon-P polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA), incubated with anti-integrin α6 antibody AA6A and detected using a secondary antibody conjugated to horseradish peroxidase. Rabbit anti-uPA antibody was used to probe for uPA. Membranes were probed with the anti-actin rabbit polyclonal antibody from Cytoskeleton (Denver, CO, USA) as loading control. Visualization was by chemiluminescence (ECL Western Blotting Detection System, Amersham, IL, USA).
Integrin α6 model
The homology model of the human integrin α6 was created using Modeller (version 7) maintained at the California Institute for Quantitative Biomedical Research, UCSF, San Francisco, CA, USA. The model is based on the crystal structure of integrin αvβ3 (pdb:1JV2). Energy minimization was done using Sybyl 7.0 (Tripos Inc. St. Louis, MO, USA.).
Transwell cell migration assay
Cell invasion studies were done in 24-well transwell cell culture chambers with the upper chamber containing filters of 8.0-μm pore size (BD Falcon Cell Culture Inserts, 8.0 μm 24 well and BD Falcon Cell Culture Companion Plates for Inserts, 24 well, from BD, NJ, U.S.A.). The upper surface of the filters was coated with a different matrices including 20 μg/ml laminin 111, 40 μg/ml fibronectin (BD Biosciences, NJ, U.S.A.), 40 μg/ml collagen IV (Gibco, BRL, MD, USA) or laminin 332 (HaCaT cell serum free conditioned media) and dried overnight at RT. Approximately 50,000 cells were seeded on the upper chamber wells together with or without 3 μg/ml PAI-1 or 10μg/ml aprotinin in serum free conditions. 1ng/ml EGF was placed in the lower chamber as a chemo-attractant. After 24h of incubation at 37 °C, cells that invaded into the lower compartment were stained with 0.5% Crystal Violet/20% methanol and counted. All experiments were done twice in triplicate.
Migration Assay
Doxycycline induced cells were grown to confluency on 20 μg/ml laminin 111, 40 μg/ml fibronectin (BD Biosciences, NJ, U.S.A.), 40 μg/ml collagen IV (Gibco, BRL, MD, USA) or laminin 332 (HaCaT cell conditioned media) coated square coverslips in IMDM (Gibco BRL, MD, USA) plus 1% fetal bovine serum (FBS). The source of laminin 332 was serum free conditioned medium from HaCaT cells and contains the pro-migratory form of laminin 322 (data not shown). A scratch was made diagonally across the square coverslip with a plastic cell scraper (Fisherbrand Cat. # 08-773-2). The coverslips were then rinsed in sterile PBS and placed in fresh serum free IMDM medium containing the different ligands (laminin 111, laminin 322, fibronectin or collagen IV) and 1 ng/ml EGF to induce migration. PAI-1 (3μg/ml) was used to inhibit uPA activity in the migration assay. The cells were incubated for 3, 6, 9 or 12 hours. The cells were fixed with cold methanol for 10 min followed by cold acetone. On drying, the cells were then stained with 0.5 μg/ml DAPI for 10 min. The coverslips were washed in PBS, post-fixed in Ethanol for 4 min and mounted using Prolong Antifade (Molecular Probes, Or, USA). The cells were observed using a Zeiss Axiovert microscope (10X magnification) and images were collected using a CCD camera. Quantification was done using the particle counting feature of Scion Image Software.
RESULTS AND DISCUSSION
The uPA mediated α6 integrin cleavage required amino acid residues R594 and R595
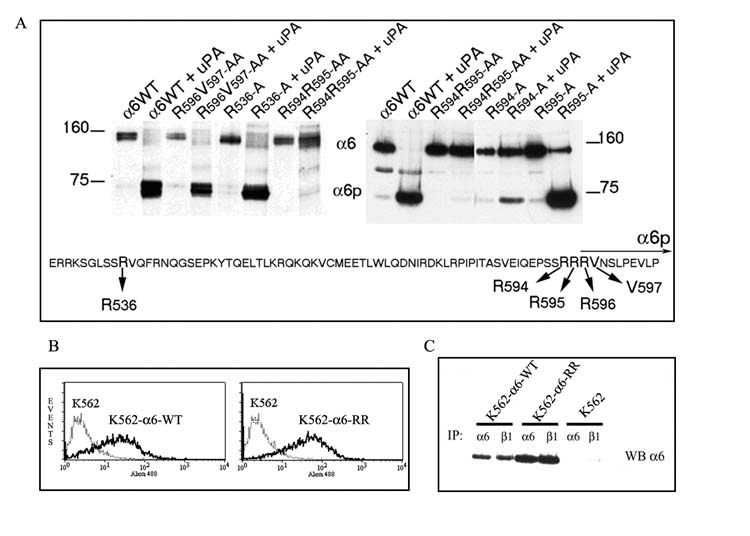
Site-directed mutagenesis was performed on selected amino acid residues that were identified as candidates for the cleavage site. These residues were selected on the basis of our previous work in which MALDI mass spectrometry and liquid chromatography-tandem mass spectrometry identified ten non-continuous amino acid fragments within the α6p variant corresponding exactly to the predicted trypsin fragments coded by exons 13–25 of the α6 integrin sequence [7]. Further, the sequencing data confirmed that both the heavy and light chains of the α6p variant contained identical portions of the full-length α6 integrin and that α6p contained at least amino acid R596. This information coupled with the knowledge that the protease that cleaves the α6 integrin is uPA, led us to mutate different arginine residues upstream from R596 (Fig. 1A). We excluded sequences that were conserved between the α6 integrin and the non-cleavable α3 integrin. We first mutated residues R536, R594, R595, followed by inducing double mutations R594R595 and R596V597. All residues were mutated to alanine. The constructs were transfected into an α6 null erythroleukemia cell line K562 resulting in K562-α6-WT cells expressing the wild type integrin α6 and the K562-α6-RR cells expressing the mutated, non-cleavable form of integrin α6. After immunoprecipitation of the mutated or wild type α6 integrin, uPA, which consisted of a mixture of single chain and double chain form [20], was added to the immunocomplexes and the samples were analyzed by SDS-PAGE analysis followed by western blotting for the α6 integrin (Fig. 1A). The data showed that mutation of arginine 594 to alanine significantly reduced the ability of uPA to cleave the α6 integrin as compared to the wild-type α6 or other mutants (R536-A and R596V597-AA). Interestingly mutation of both arginine residues 594 and 595 was required to completely abolish the cleavage of the α6 integrin by uPA.
Fig. 1. Site-directed mutagenesis analysis of the α6 integrin.
A. Five different residues (R536, R594, R595, R596 and V597) were replaced by alanine in the α6 integrin extracellular domain. K562 cells were transfected with different mutated α6 integrin constructs and the α6 variants induced by Doxycycline. After 24 hours the α6 integrin was immunoprecipitated from the cells and the immunocomplexes were treated with 20 μg/ml uPA. The resulting α6 and α6p integrin was detected by western blot analysis using the AA6A rabbit polyclonal antibody. A partial sequence of the integrin α6 integrin flanking the cleavage site is shown for illustrative purposes and the mutated residues are indicated with arrows. The horizontal arrow labeled α6p indicates the amino terminal end of the truncated integrin as analyzed by mass spectrometric analysis in our earlier work (7).
B. Surface expression of wild type and mutated integrin α6 in transfected K562 cells by flowcytometry. Untransfected cells (K562-light line) or transfected cells (K562-α6-WT or K562-α6-RR - heavy lines) were treated with primary Rat anti-integrin α6 antibody J1B5 followed by Alexa 488 anti-rat antibody and visualized using the BD FACScan.
C. The wild type and mutated integrin α6 binds to integrin β1. Immunoprecipitations (IP) of integrin α6 and integrin β1 in doxycycline induced K562-α6-WT and K562-α6-RR and untransfected K562 using rat anti-integrin α6 antibody J1B5 and mouse anti-integrin β1 TS2/16 were done. Rabbit anti-integrin α6 antibody AA6A was used for western blotting.
It was possible that ectopic expression of the mutant integrin could occur without correct localization or heterodimerization. The presence of the mutant and wild type integrin α6 on the cell surface of the K562-α6-RR and K562-α6-WT stably transfected cell lines and the parental K562 cells was analyzed using flow cytometry. As expected, integrin α6 was not detected in the parental K562 cells, however a significant amount of fluorescence was recorded in the K562-α6-WT and K562-α6-RR cells indicating the presence of integrin α6 on the cell surface (Fig. 1B). We next determined if the mutant and the wild-type integrin α6 expressed in the K562-α6-RR and K562-α6-WT cells were able to pair with the β1 subunit that is constitutively expressed in the K562 cells. Immunoprecipitations (IPs) were performed on doxycycline induced K562-α6-RR and K562-α6-WT cells using anti-integrin α6 and anti-integrin β1 antibodies and analyzed by western blotting to detect the α6 integrin. IP of the cell lysates with either α6 or β1 integrin antibodies will retrieve the α6 integrin. The results indicated that both the wild type and mutant α6 interacted with the β1 subunit (Fig. 1C). The data further indicates that the R594 R595 to A594 A595 substitution does not affect the ability of the integrin to bind to its β partner or become resident on the cell surface.
In order to spatially localize R594 and R595 on the α6 integrin, we created a homology model based upon the αvβ3 integrin (Fig. 2A). The crystal structure of αvβ3 is known [5]. A model of integrin α6 was created using integrin αv structure as a template (Fig. 2A). The model spans residues 24–1004, consisting of the calf-1, calf-2 and genu region (shown in black) and the thigh and the β-propeller domain (shown in white). Integrin α6 undergoes uPA-mediated cleavage at the RR site, which is located at the beginning of the thigh region (Fig. 2A). The location of this site in the model indicated that the majority of the thigh region and the entire β-propeller region were removed. The integrin α6p contains calf-1, calf-2, genu and a segment of the thigh region. The uPA mediated cleavage site within the two arginine residues is potentially accessible for cleavage, as it lies exposed in the outer region of integrin α6 as seen in Fig. 2A.
Fig. 2. Homology Model of the extracellular portion of the human integrin alpha 6 subunit (residues 24–1004) based on the crystal structure of the human alpha v beta 3 integrin (pdb: 1JV2) and sequence comparison of integrin α6.
A. The first 23 residues in the integrin α6 sequence undergo post-translational cleavage before the integrin reaches the surface and hence are not shown in the model. The model illustrates the location of the protease cleavage site within two consecutive arginine residues 594–595 (RR) shown along with their side chains. The beta propeller domain and a majority of the thigh domain (shown in white) are not contained in integrin α6p while the Calf-1 and Calf-2 domains of the stalk region remain (shown in black).
B. Comparison of the human integrin α6 to the integrin α6 sequence in mouse, rat, chicken and the integrin α chain sequence of C. elegans and the S40311 integrin sequence of D. melanogastor.
C. Comparison of the human integrin α6 to sequences of closely related integrins, integrin α7 and integrin a3. The boxes indicate the R594R595 residues that are required for cleavage of integrin α6 to integrin α6p. AliBee Multiple Alignment Release 2.0 was used to align the sequences.
The cleavage residues used by uPA are found within human, mouse and rat integrin α6
To determine if the cleavage residues used by uPA on the human integrin α6 were present in integrin α6 from other species, we aligned sequences from rat, mouse, chicken, worm and fly. The results indicate that the RR sequence was identical in human, rat and mouse integrin α6 (Fig. 2B). The RR sequence was not found in the integrin α chain sequence of C. elegans and the S40311 integrin sequence of D. melanogastor (Fig. 2B). The rat and mouse integrin α6 contains both the R594 and R595 residues present in human integrin α6, suggesting that rat and mouse integrin α6 would undergo uPA-mediated cleavage. Simplified model systems may be useful to study the physiological effects of uPA-mediated cleavage of integrin α6 to α6p in vivo.
Integrins α3, α6 and α7 are laminin receptors. Integrin α3 and α6 are found predominantly in epithelial cells and integrin α7 is found predominantly in skeletal and cardiac muscles and neural cells [21, 22]. Our earlier work has shown that uPA does not cleave integrin α3 [9]. Comparison of the cleavage site residues between integrin α6 and integrin α3 shows the absence of the RR sequence in integrin α3 (Fig. 2C). Integrin α7 does have the two arginine residues required for the uPA-mediated cleavage but is not expressed in epithelial cells [21]. It remains to be determined whether integrin α7 is processed on the cell surface by uPA.
Human prostate cancer cell lines expressed cleavable and non-cleavable integrin α6
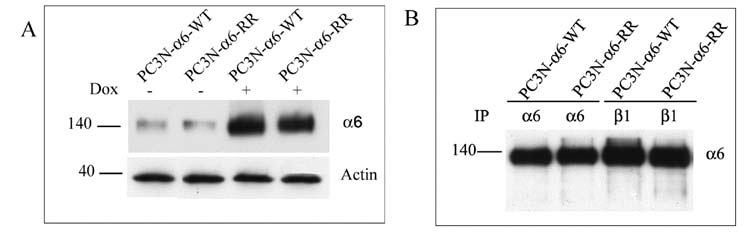
The cleaved form of integrin α6, α6p, has been observed in human prostate cancer specimens and not in the adjacent normal human prostate tissue [9]. In order to study the biological significance of this cleavage, we transfected prostate cancer cell line PC3N with wild type cleavable integrin α6 generating the PC3N-α6-WT. We also transfected PC3N cells with the mutated non-cleavable integrin α6 in which the residues R594 and R595 were substituted by residues A594 and A595 resulting in the generation of the PC3N-α6-RR cell line. These cells over express the wild type or the mutated integrin α6 respectively, in presence of doxycycline, to equivalent levels as estimated by western blot (Fig. 3A). It should be noted that in the absence of doxycycline, both the PC3N-α6-WT and the PC3N-α6-RR cells contain similar levels of endogenous integrin α6.
Fig. 3. Induction and expression of integrin α6 heterodimers in transfected cells.
A. Cell lysates were analyzed for expression levels of integrin α6 in PC3N-α6-WT and PC3N-α6-RR cells under unstimulated ((−) Dox) or stimulated ((+) Dox) conditions. The doxycycline concentration was 0.02μg/ml. The α6 integrin was detected using the AA6A rabbit polyclonal antibody on a western blot. Actin was used as a loading control.
B. The wild type and mutated integrin α6 binds to integrin β1. Immunoprecipitations were done for integrin α6 and integrin β1 in doxycycline (0.02μg/ml) induced PC3N-α6-WT and PC3N-α6-RR cells using rat anti-integrin α6 antibody J1B5 and mouse anti-integrin β1 TS2/16. Rabbit anti-integrin α6 antibody AA6A was used for western blotting.
In order to determine if the transfected wild type and mutated integrin α6 could pair with the endogenous integrin β1, immunoprecipitations were performed on doxycycline induced PC3N-α6-RR and PC3N-α6-WT cells using anti-integrin α6 or β1 antibodies and analyzed by western blotting to detect the α6 integrin. The results indicated that both the induced wild type and mutant α6 interacted with the β1 subunit (Fig. 3B).
We next tested the ability of exogenously supplied uPA to cleave the induced integrin forms. In the presence of doxycycline, PC3N-α6-WT and PC3N-α6-RR cells were treated with 20μg/ml uPA for up to 120 min. Consistently increasing levels of integrin α6p, from 0 time to 120 min, in the PC3N-α6-WT cells were observed. The increase in integrin α6p levels was observed with a corresponding decrease in the full-length integrin α6 levels (Fig. 4A). In the PC3N-α6-RR cells, however, no significant formation of integrin α6p was seen (Fig. 4B). The faint bands that are visible in the blot are due to α6p generated from endogenous expression of the wild type integrin α6 (Fig. 4B). The induced wild type integrin α6 was cleavable by exogenously supplied uPA in a time dependent manner (Fig. 4B). A majority of the α6-WT integrin is cleavable by uPA. Theα6-WT that is not cleavable by uPA is either protected from cleavage while resident on the cell surface or based upon our previous findings, has not yet reached the cell surface (7). The mutant integrin was found to be refractory to uPA mediated cleavage. The data shows that the mutation due to the change from the arginine residues (R594, R595) to alanine residues prevents uPA-mediated cleavage of integrin α6 to α6p (Fig. 4B).
Fig. 4. PC3N-α6-RR cells express the uPA non-cleavable form of the integrin α6.
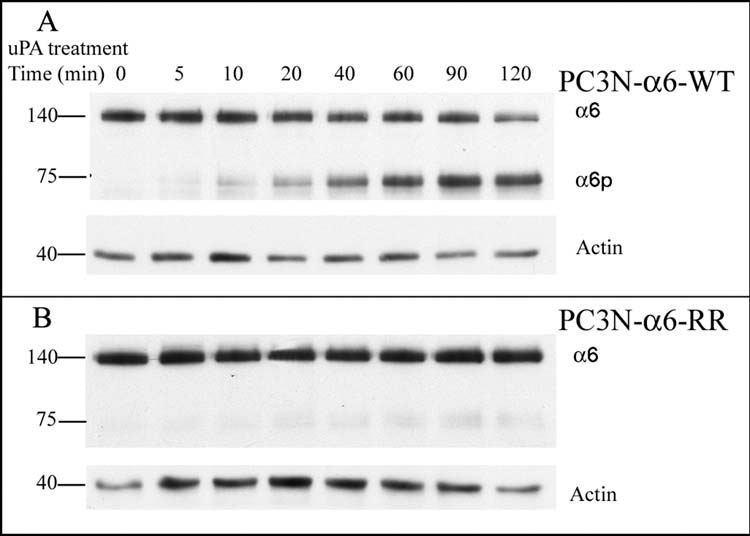
A. PC3N-α6-WT cells were treated with 20 μg/ml uPA over time and the lysates analyzed by PAGE. The integrins α6 (140 kD) and α6p (~75 kD) were detected using the AA6A rabbit polyclonal antibody recognizing the α6 integrin cytoplasmic domain by western blot analysis. Actin (~40kD) was used as a loading control.
B. PC3N-α6-RR cells were treated with 20 μg/ml uPA over time and the lysates analyzed by PAGE. The integrins α6 (140 kD) was detected using the AA6A rabbit polyclonal antibody recognizing the α6 integrin cytoplasmic domain by western blot analysis. Actin (~40kD) was used as a loading control.
Blocking integrin α6 cleavage reduces cell invasion and migration
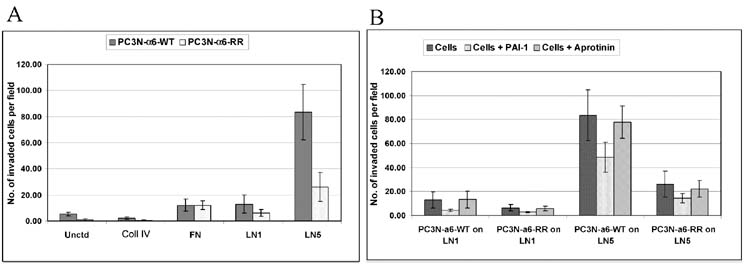
Several previous studies have implicated uPA in tumor cell invasion [23–26]. We next determined the influence of the cleavage of integrin α6 to α6p on cell invasion. PC3N-α6-WT and PC3N-α6-RR cells were placed on laminin 111, fibronectin, collagen IV or laminin 332 coated and uncoated 8.0 μm pore filters and 1 ng/ml EGF was placed in the lower wells to serve as a chemo-attractant in serum free media. The cells were incubated 24 hours and the number of cells that had invaded through the filter per microscopic field was quantified. The results are shown in Fig. 5A. Minimal, if any, invasion was observed on uncoated and collagen IV coated filters. Although some PC3N-α6-WT as well as PC3N-α6-RR cells invaded the fibronectin and laminin 111 coated filters, maximum invasion was observed on Laminin 332 coated filters. Approximately 84 cells per microscopic field of PC3N-α6-WT cells had invaded the filters as compared to 26 cells per microscopic field of PC3N-α6-RR (Fig. 5A).
Fig. 5. Comparison of invasive potential of PC3N-α6-WT and PC3N-α6-RR cells. Doxycycline (0.02 μg/ml) induced PC3N-α6-WT and PC3N-α6-RR cells were placed onto different ligands and incubated for 24 hrs at 37°C. 1 ng/ml EGF was placed in the bottom wells. The cells that had invaded the filters were stained with crystal violet and counted using 40X magnification. All experiments were done twice in triplicate.
A. Invasion of PC3N-α6-WT and PC3N-α6-RR cells through collagen IV (Coll IV), fibronectin (FN), laminin 111 (LN1) or laminin 332 (LN5) coated and uncoated transwell inserts.
B. Comparison of invasive potential of PC3N-α6-WT and PC3N-α6-RR cells through laminin 111 (LN1) or laminin 332 (LN5) coated transwell inserts in presence of uPA inhibitor PAI-1 (3 μg/ml) and plasmin inhibitor aprotinin (10 μg/ml).
In order to determine the effect of the uPA inhibitor PAI-1 on the invading cells, the cells were placed on laminin 111 or laminin 332 coated filters and incubated with 3 μg/ml PAI-1. Consistent with the results in Fig. 5A, laminin 332 coated filters provided the most effective support for invasion by either cell line. The uPA inhibitor PAI-1, inhibited invasion of laminin 332 coated filters by 42% and 45% in the PC3N-α6-WT and the PC3N-α6-RR cells respectively (Fig. 5B). We demonstrated in earlier work that uPA cleaves integrin α6 in a plasmin independent manner [9]. However, in order to determine whether membrane bound plasmin affects the integrin cleavage, the cells were incubated with a plasmin inhibitor, aprotinin [27, 28]. The addition of aprotinin to the cells did not significantly reduce PC3N-α6-WT or PC3N-α6-RR cell invasion on laminin 111 or laminin 332 coated filters (Fig. 5B), indicating that the observed invasion is independent of membrane bound plasmin.
Previous studies have shown that TPA (12-O-Tetradecanoyl-phorbol 13-Acetate) induces cell migration in migration assays [29]. Our earlier work has shown that the cleavage of integrin α6 to α6p can be induced by TPA in a uPA mediated manner [9]. Further, uPA has been shown to be present at the leading edge in wound healing experiments [30, 31]. These observations suggest that the cleavage of the α6 integrin may play a role in cell migration. In order to determine the significance of the cleavage of integrin α6 to α6p on linear cell migration, PC3N-α6-WT and PC3N-α6-RR cells were grown on a laminin 111, fibronectin, collagen IV and laminin 332 coated coverslips. Migration was induced by scratching [32] and EGF stimulation in the absence of serum in the media. After 12 hours of incubation, the images of the cells entering into the scratch on either collagen IV, fibronectin, laminin 111 or laminin 332 were collected (Fig. 6A). The quantification of the images are shown in Fig. 6B. In this assay, the greatest migration by the PC3N-α6-WT cells was observed on laminin 111 (32%) followed by fibronectin (12%) and laminin 332 (10%), with virtually no migration (approximately 4%) observed on collagen IV. In particular, the laminin 111 coated coverslips resulted in the most migration with approximately 32% of the PC3N-α6-WT cells entering the scratch whereas approximately 5% of the PC3N-α6-RR cells were observed (Fig. 6A, B). These data indicate that by comparing the best migration promoting ligand in this assay (i.e. laminin 111), there was approximately 6.4 fold difference between the number of PC3N-α6-WT and PC3N-α6-RR cells migrating into the scratch. It should be noted that the mutant cells as compared to the wild type cells also were hindered in migrating on fibronectin (approximately 2.4 fold difference) and lamnin 332 (1.4 fold difference) with no significant difference on collagen IV (Fig. 6B). We next tested if blocking uPA activity would inhibit migration on the best migrating promoting ligand, laminin 111. The cells were induced to migrate by generating the scratch and 3 μg/ml PAI-1 was added. Only 2% PC3N-α6-WT cells migrated into the scratch in the presence of PAI-1 as compared to 32% in absence of PAI-1 (Fig. 6C). These observations indicate that uPA mediated cleavage of integrin α6 to α6p facilitated migration on laminin 111.
Fig. 6. Comparison of linear migration of PC3N-α6-WT and PC3N-α6-RR cells on different ligands. PC3N-α6-WT and PC3N-α6-RR cells were grown to confluency on glass coverslips precoated with collagen IV (Coll IV), fibronectin (FN), laminin 111 (LN1) or laminin 332 (LN5). Migration was induced by scratching across the cell monolayer on the glass coverslip and addition of 1ng/ml EGF. The cells were fixed and stained with DAPI and incubated for 12 hours at 37°C. Images were collected using a Zeiss Axiovert microscope at 10 X magnification. The percentage of migrating cells was determined by using the particle counting feature of Scion Image software to detect the number of cells entering the scratch/total number of cells X 100. All experiments were done twice in triplicate.
A. Digital images of PC3N-α6-WT and PC3N-α6-RR cells on collagen IV, fibronectin (FN), laminin 111 (LN1) and laminin 332 (LN5) 12 hours after scratching cell monolayer.
B. Comparison of percentage migration of PC3N-α6-WT (solid bars) and PC3N-α6-RR (open bars) cells on collagen IV, fibronectin (FN), laminin 111 (LN1) and laminin 332 (LN5), 12 hours after scratching cell monolayer.
C. Inhibition of linear migration of PC3N-α6-WT by PAI-1 (3 μg/ml) on laminin 111 (LN1).
It is interesting to note that independent of the migration type assay, the PC3N-α6-WT are better equipped to migrate as compared to the PC3N-α6-RR cells. However, it appears that the assays are ligand dependent, i.e. laminin 332 coated filters facilitate invasion whereas laminin 111 coated coverslips support linear cell migration of prostate cancer cells. Taken together, the data suggests that integrin α6 cleavage assists cell invasion on laminin 332 and cell migration on laminin 111.
Integrin α6p is induced in migrating cells
Since the data suggested that the ability to cleave A6 integrin facilitated migration, we next determined if the A6 integrin was indeed cleaved under conditions of induced migration. The doxycycline induced PC3N-α6-WT and PC3N-α6-RR cells were analyzed for the presence of the integrin α6p during migration. An increase in integrin α6p levels was observed in the PC3N-α6-WT cells 18 hours after induced migration. Comparable levels of full-length integrin α6 were seen in the PC3N-α6-WT cells with or without migration. As expected, a comparable low level of integrin α6p was observed in the mutant containing PC3N-α6-RR cells with or without migration due to the cleavage of endogenous integrin α6 (Fig. 7A). Densitometric analysis was carried out on the blot to determine if the ratio of the α6p signal to the full-length form was altered under conditions of induced migration (Fig. 7B). An increased ratio of α6p to α6 integrin occurs in the PC3N-α6-WT cells that have been induced to migrate as compared to the PC3N-α6-WT that are not induced. The ratio of α6p to α6 in the uncleavable mutants (PC3N-α6-RR cells) was not influenced by the scratch procedure. We note with interest that integrin α6p appears as a doublet at approximately 75 kDa when migration is induced by scratching in the PC3N-α6-WT cells (Fig. 7A). These two forms are presumed to represent altered levels of post-translational modifications like glycosylation and palmitoylation resulting in altered molecular weight [34–36]. We have previously reported that five of the nine putative glycosylation sites on integrin α6 are retained on integrin α6p [7]. Hence the two forms of integrin α6p observed in the western blot may correspond to such modified integrins. It is important to note that the acute cellular response to uPA treatments (Fig. 4) over a period up to 120 minutes, results in an increase in α6p levels that corresponds to a drop in the full length form of α6 integrin. This is not observed if the analysis occurs after 18 hours, since enough time has elapsed for newly synthesized α6, not yet on the cell surface and not susceptible to cleavage, to be represented in the total full-length band.
Fig. 7. Induction of α6p in scratched PC3N-α6-WT cells.
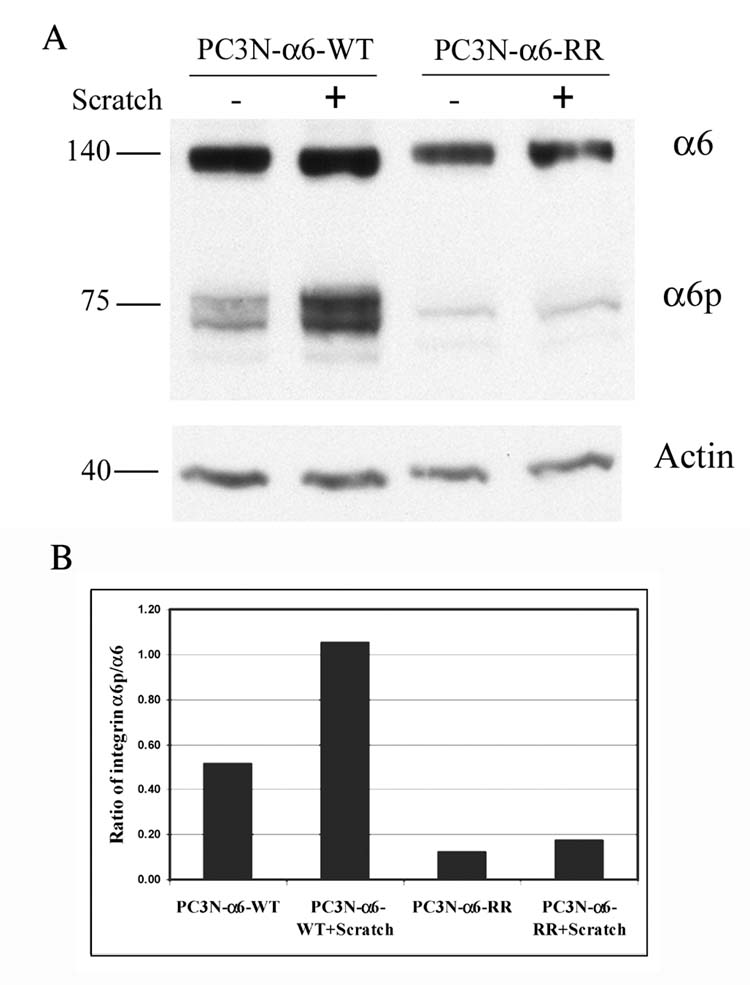
A. Doxycycline (0.02 μg/ml) induced PC3N-α6-WT and PC3N-α6-RR cells were grown to confluency in a tissue culture dish, scratched and incubated overnight in fresh IMDM medium. The cells were then lysed and the integrin α6 detected using SDS-PAGE. The integrins α6 (~140 kD) and α6p (~75 kD) were detected using the rabbit antibody AA6A specific for the integrin α6 cytoplasmic domain. Actin (~40kD) was used as a loading control.
B. Densitometric analysis was done on the blot in Fig. 7A and ratios of the integrin α6p to full length integrin α6 were calculated and graphed.
Taken together, our data suggests that the integrin α6 undergoes uPA-mediated cleavage at residues R594 R595 in a time dependent manner enhancing cell invasion and migration on laminin. Further understanding of factors regulating integrin cleavage holds promise for either modulating or detecting early tumor cell invasion or migration. Future work will be directed towards understanding the regulatory features dictating the cleavage of the integrin α6 on the tumor cell surface.
Acknowledgments
Special thanks are extended to Dr. Daruka Mahadevan and Mr. Kishore Shakalya for building the homology structure of integrin α6. We are thankful to Dr. Jonathan Jones, Northwestern University for suggesting the linear migration assay and insightful discussions. This work was supported in part by the following grants: CA 75152, CA 56666 and CA 23074.
The abbreviations used are
- uPA
urokinase-type plasminogen activator
- PAI-1
Plasminogen Activator Inhibitor –1
- EGF
Epidermal Growth Factor
- SDS
Sodium dodecyl sulphate
- PAGE
Polyacrylamide gel electrophoresis
- DOX
Doxycycline
Footnotes
First two authors contributed equally.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Sangita C. Pawar, Dept. of Cell Biology & Anatomy, University of Arizona, Tucson, AZ.
Manolis C. Demetriou, Dept. of Cell Biology & Anatomy, University of Arizona, Tucson, AZ.
Raymond B. Nagle, Dept. of Pathology, University of Arizona, Tucson, AZ
G. Tim Bowden, Dept. of Cell Biology & Anatomy, University of Arizona, Tucson, AZ.
Anne E. Cress, Dept. of Cell Biology & Anatomy, University of Arizona, Tucson, AZ. Arizona Cancer Center, University of Arizona, Tucson, Arizona 85724.
References
- 1.Giancotti FRE. Integrin signaling. Science. 1999;285(5430):1028–32. doi: 10.1126/science.285.5430.1028. Review. [DOI] [PubMed] [Google Scholar]
- 2.Miranti CK, Brugge JS. Sensing the environment: a historical perspective on integrin signal transduction. Nature Cell Biology. 2002;4(4):E83–90. doi: 10.1038/ncb0402-e83. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz M, Schaller M, Ginsberg M. Integrins: emerging paradigms of signal transduction. Annu Rev Cell Dev Biol. 1995;11:549–99. doi: 10.1146/annurev.cb.11.110195.003001. Review. [DOI] [PubMed] [Google Scholar]
- 4.Green L, Mould A, Humphries M. The integrin beta subunit. Int J Biochem Cell Biol. 1998;30(2):179–84. doi: 10.1016/s1357-2725(97)00107-6. Review. [DOI] [PubMed] [Google Scholar]
- 5.Xiong JP, et al. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296(5565):151–5. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 6.Arnaout MA, Goodman SL, Xiong JP. Coming to grips with integrin binding to ligands. Current Opinion in Cell Biology. 2002;14(5):641–51. doi: 10.1016/s0955-0674(02)00371-x. [DOI] [PubMed] [Google Scholar]
- 7.Davis TL, et al. Identification of a novel structural variant of the alpha 6 integrin. Journal of Biological Chemistry. 2001;276(28):26099–106. doi: 10.1074/jbc.M102811200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He B, et al. Tetraspanin CD82 attenuates cellular morphogenesis through down-regulating integrin alpha6-mediated cell adhesion. J Biol Chem. 2005;280(5):3346–54. doi: 10.1074/jbc.M406680200. [DOI] [PubMed] [Google Scholar]
- 9.Demetriou MC, et al. Extracellular alpha 6 integrin cleavage by urokinase-type plasminogen activator in human prostate cancer. Exp Cell Res. 2004;294(2):550–8. doi: 10.1016/j.yexcr.2003.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rickli EE. The activation mechanism of human plasminogen. Thrombosis et Diathesis Haemorrhagica. 1975;34(2):386–95. [PubMed] [Google Scholar]
- 11.Gold LI, Schwimmer R, Quigley JP. Human plasma fibronectin as a substrate for human urokinase. Biochemical Journal. 1989;262(2):529–34. doi: 10.1042/bj2620529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naldini L, et al. Extracellular proteolytic cleavage by urokinase is required for activation of hepatocyte growth factor/scatter factor. EMBO Journal. 1992;11(13):4825–33. doi: 10.1002/j.1460-2075.1992.tb05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyazawa K, et al. Structural organization and chromosomal localization of the human hepatocyte growth factor activator gene--phylogenetic and functional relationship with blood coagulation factor XII, urokinase, and tissue-type plasminogen activator. European Journal of Biochemistry. 1998;258(2):355–61. doi: 10.1046/j.1432-1327.1998.2580355.x. [DOI] [PubMed] [Google Scholar]
- 14.Tran NL, et al. N-Cadherin expression in human prostate carcinoma cell lines. An epithelial-mesenchymal transformation mediating adhesion withStromal cells. Am J Pathol. 1999;155(3):787–98. doi: 10.1016/S0002-9440(10)65177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damsky CH, et al. Integrin switching regulates normal trophoblast invasion. Development Supplement. 1994;120(12):3657–66. doi: 10.1242/dev.120.12.3657. [DOI] [PubMed] [Google Scholar]
- 16.Tamura RN, et al. Epithelial integrin alpha 6 beta 4: complete primary structure of alpha 6 and variant forms of beta 4. J Cell Biol. 1990;111(4):1593–604. doi: 10.1083/jcb.111.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemler ME, et al. Glycoproteins of 210,000 and 130,000 m.w. on activated T cells: cell distribution and antigenic relation to components on resting cells and T cell lines. J Immunol. 1984;132(6):3011–8. [PubMed] [Google Scholar]
- 18.Kazarov AR, et al. An extracellular site on tetraspanin CD151 determines alpha 3 and alpha 6 integrin-dependent cellular morphology. J Cell Biol. 2002;158(7):1299–309. doi: 10.1083/jcb.200204056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witkowski CM, et al. Altered surface expression and increased turnover of the alpha6beta4 integrin in an undifferentiated carcinoma. Carcinogenesis. 2000;21(2):325–30. doi: 10.1093/carcin/21.2.325. [DOI] [PubMed] [Google Scholar]
- 20.Soberano ME, et al. Purification and characterization of two forms of urokinase. Biochim Biophys Acta. 1976;445(3):763–73. doi: 10.1016/0005-2744(76)90126-1. [DOI] [PubMed] [Google Scholar]
- 21.Ziober BL, Kramer RH. Identification and characterization of the cell type-specific and developmentally regulated alpha7 integrin gene promoter. J Biol Chem. 1996;271(37):22915–22. doi: 10.1074/jbc.271.37.22915. [DOI] [PubMed] [Google Scholar]
- 22.Previtali SC, et al. Schwann cells synthesize alpha7beta1 integrin which is dispensable for peripheral nerve development and myelination. Mol Cell Neurosci. 2003;23(2):210–8. doi: 10.1016/s1044-7431(03)00014-9. [DOI] [PubMed] [Google Scholar]
- 23.Al-Atrash G, et al. uPA and uPAR contribute to NK cell invasion through the extracellular matrix. Anticancer Res. 2001;21(3B):1697–704. [PubMed] [Google Scholar]
- 24.Rabbani SA, Xing RH. Role of urokinase (uPA) and its receptor (uPAR) in invasion and metastasis of hormone-dependent malignancies. Int J Oncol. 1998;12(4):911–20. doi: 10.3892/ijo.12.4.911. [DOI] [PubMed] [Google Scholar]
- 25.Schmitt M, et al. Urokinase-type plasminogen activator (uPA) and its receptor (CD87): a new target in tumor invasion and metastasis. J Obstet Gynaecol. 1995;21(2):151–65. doi: 10.1111/j.1447-0756.1995.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 26.Brunner N, et al. Urokinase plasminogen activator (uPA) and its type 1 inhibitor (PAI-1): regulators of proteolysis during cancer invasion and prognostic parameters in breast cancer. Cancer Treat Res. 1994;71:299–309. doi: 10.1007/978-1-4615-2592-9_16. [DOI] [PubMed] [Google Scholar]
- 27.Welling TH, et al. Tissue plasminogen activator increases canine endothelial cell proliferation rate through a plasmin-independent, receptor-mediated mechanism. J Surg Res. 1996;66(1):36–42. doi: 10.1006/jsre.1996.0369. [DOI] [PubMed] [Google Scholar]
- 28.Weide I, Romisch J, Simmet T. Contact activation triggers stimulation of the monocyte 5-lipoxygenase pathway via plasmin. Blood. 1994;83(7):1941–51. [PubMed] [Google Scholar]
- 29.Inyang AL, Tobelem G. Tissue-plasminogen activator stimulates endothelial cell migration in wound assays. Biochem Biophys Res Commun. 1990;171(3):1326–32. doi: 10.1016/0006-291x(90)90831-7. [DOI] [PubMed] [Google Scholar]
- 30.Romer J, et al. The receptor for urokinase-type plasminogen activator is expressed by keratinocytes at the leading edge during re-epithelialization of mouse skin wounds. J Invest Dermatol. 1994;102(4):519–22. doi: 10.1111/1523-1747.ep12373187. [DOI] [PubMed] [Google Scholar]
- 31.Grondahl-Hansen J, et al. Urokinase- and tissue-type plasminogen activators in keratinocytes during wound reepithelialization in vivo. J Invest Dermatol. 1988;90(6):790–5. doi: 10.1111/1523-1747.ep12461511. [DOI] [PubMed] [Google Scholar]
- 32.Kidd KR, Williams SK. Laminin-5-enriched extracellular matrix accelerates angiogenesis and neovascularization in association with ePTFE. J Biomed Mater Res A. 2004;69(2):294–304. doi: 10.1002/jbm.a.20133. [DOI] [PubMed] [Google Scholar]
- 33.Wehrle-Haller B, Imhof BA. Integrin-dependent pathologies. J Pathol. 2003;200(4):481–7. doi: 10.1002/path.1399. [DOI] [PubMed] [Google Scholar]
- 34.Chammas R, et al. Functionally distinct roles for glycosylation of alpha and beta integrin chains in cell-matrix interactions. Proc Natl Acad Sci U S A. 1993;90(5):1795–9. doi: 10.1073/pnas.90.5.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Lampe B, Stallmach A, Riecken EO. Altered glycosylation of integrin adhesion molecules in colorectal cancer cells and decreased adhesion to the extracellular matrix. Gut. 1993;34(6):829–36. doi: 10.1136/gut.34.6.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, et al. Palmitoylation supports assembly and function of integrin-tetraspanin complexes. J Cell Biol. 2004;167(6):1231–40. doi: 10.1083/jcb.200404100. [DOI] [PMC free article] [PubMed] [Google Scholar]